Abstract
Triple-helical collagen IV protomers associate through their N- and C-termini forming a three-dimensional network, which provides basement membranes with an anchoring scaffold and mechanical strength. The noncollagenous (NC1) domain of the C-terminal junction between two adjacent collagen IV protomers from human placenta was crystallized and its 1.9-Å structure was solved by multiple anomalous diffraction (MAD) phasing. This hexameric NC1 particle is composed of two trimeric caps, which interact through a large planar interface. Each cap is formed by two α1 fragments and one α2 fragment with a similar previously uncharacterized fold, segmentally arranged around an axial tunnel. Each monomer chain folds into two structurally very similar subdomains, which each contain a finger-like hairpin loop that inserts into a six-stranded β-sheet of the neighboring subdomain of the same or the adjacent chain. Thus each trimer forms a quite regular, but nonclassical, sixfold propeller. The trimer–trimer interaction is further stabilized by a previously uncharacterized type of covalent cross-link between the side chains of a Met and a Lys residue of the α1 and α2 chains from opposite trimers, explaining previous findings of nonreducible cross-links in NC1. This structure provides insights into NC1-related diseases such as Goodpasture and Alport syndromes.
Basement membranes are organized as tough, sheet-like networks to control morphogenesis and maintenance of orderly tissue structures. Their main collagenous constituent are type-IV collagen protomers, which provide the membrane with mechanical stability and anchoring sites for a variety of cellular receptors (1, 2). The collagen IV protomers are trimeric chain assemblies, which consist of a 400-nm-long triple-helical domain and a C-terminal globular noncollagenous (NC1) domain (3). When secreted into the extracellular matrix, these protomers assemble through their N-terminal triple-helical segment and their NC1 domain, respectively, into tetramers and dimers, giving rise to a dense network, which eventually becomes stabilized by covalent cross-links (4–6).
The major collagen IV protomers detected in all basement membranes consist of two α1(IV) chains and one α2(IV) chain (1, 2, 5). Recently, four less abundant α1-like [α3(IV) and α5(IV)] and α2-like [α4(IV) and α6(IV)] isoforms were discovered that assemble into similar protomers and seem to play a major role in alveolar and renal basement membranes (1, 7, 8). Several inherited disorders associated with collagen IV defects are known in humans including Alport syndrome, a fatal progressive nephropathy caused by mutations of the α3, α4, or α5 encoding genes (1, 9).
Besides their crucial role in the collagen IV dimerization, the NC1 domains seem to determine chain selection, assembly, and folding during biosynthesis of the collagen IV protomers (10, 11). Recombinant NC1 monomers from α2(IV), α3(IV), and α6(IV) chains were shown to regulate angiogenesis and tumor growth, as well as integrin-mediated cell adhesion and migration (12). The NC1 domains of α3(IV) and α5(IV) have been identified as major targets of autoantibodies such as in Goodpasture syndrome, impairing the functions of kidney, lung, and skin (13, 14).
The amino acid sequences of NC1 domains from 28 collagen IV isoforms of mammalian and invertebrate species show a remarkably high similarity, suggesting strongly conserved functions (7, 9). Each chain exhibits an internal repeat, each internally cross-connected via six invariant cysteines of partially known connectivity (15). Hexameric NC1 domains of about 170 kDa can be obtained from bacterial collagenase digests of tissues (8, 16). Electron micrographs show a compact oval shape of about 60 × 110 Å (4, 10). Exposure to acidic pH or denaturing agents dissociates the hexamers into a mixture of monomers and covalent dimers (11, 16–18). These findings led to the proposal of a disulfide reshuffling mechanism of mammalian collagen IV during basement membrane network formation (15) and to a NC1 hexamer model, in which monomeric NC1 chains from both interacting collagen IV protomers interdigitate, with the tandem subdomains from opposite chains crossing over each other (9).
A deeper understanding of the complex functions of NC1 depends on the elucidation of its atomic structure. Multiple anomalous diffraction (MAD) methods have now allowed us to overcome former non-isomorphism problems (4, 19) and to determine the high-resolution crystal structure of the NC1 hexamer, also indicating a covalent Met-Lys cross-link between opposing NC1 chains, explaining the covalent dimer formation.
Materials and Methods
Crystallization and Data Collection.
NC1 hexamers were isolated from human placenta and purified as described (16, 18). Using SDS/PAGE, both initial and crystallized hexamers separated into two dimer and two monomer bands staining at a ratio of ≈2:1. Protein solution (1.5 μl; 10 mg/ml in 20 mM NH4HCO3) and precipitant buffer (1.5 μl; 0.1 M Mes/NaOH, 3.2–3.6 M sodium acetate, pH 7.1–7.5) were mixed and equilibrated against the precipitant by vapor diffusion at room temperature. The irregularly shaped crystals of up to 0.4 mm belonged to the space group R3 and had cell constants a = b = 234.59 Å, c = 99.49 Å, α = β = 90°, and γ = 120°. The crystals were mounted in cryo-loops directly from the mother liquor and were shock frozen in liquid nitrogen. X-ray data were collected at the wiggler beamline BW6 at DORIS/Hamburg (20), using the 165-mm MarCCD detector (Mar-USA, Evanston, IL) at 100 K, and were processed and scaled (Table 1) using the HKL package (21). Initial phases were obtained by a three-wavelengths MAD experiment using a crystal soaked in 8 mM K2[OsCl6]. The final experimental phases were determined from a crystal soaked for 30 s in mother liquor supplemented with 1 M NaBr.
Table 1.
Data collection statistics
| Data set | Wavelength, Å | Resolution, Å | Number of reflections,* total/unique | Completeness†
|
Rmerge, %† | |
|---|---|---|---|---|---|---|
| Overall | Anomalous | |||||
| Native1 | 1.05 | 2.01 | 249,220/132,884 | 97.3 (97.1) | — | 5.1 (38) |
| NaBr1 (peak) | 0.91780 | 2.30 | 340,768/178,767 | 99.0 (99.5) | 96.7 (97.3) | 6.5 (40) |
| NaBr2 (edge) | 0.91970 | 2.47 | 276,358/144,687 | 98.8 (99.3) | 96.7 (97.1) | 4.4 (17) |
| NaBr3 (remote) | 0.850 | 2.49 | 271,310/141,435 | 98.2 (99.7) | 96.1 (97.7) | 5.3 (26) |
| Native2 | 1.05 | 1.90 | 441,349/155,867 | 96.9 (97.6) | — | 5.8 (30) |
For the MAD wavelength and the native datasets friedel pairs were counted as independent and identical.
Values in parentheses correspond to the last resolution shell of 0.1-Å width.
Because a strong R32 pseudo symmetry complicated the analysis of the derivative Patterson maps, the Osmium derivative was interpreted with a Patterson superposition technique [SHELXS (22)], using “peak” anomalous differences. Initial MAD phases from this Osmium derivative were calculated with MLPHARE (23) and used to locate the 25 strongest Br−-sites in an anomalous difference Fourier map. MAD phase calculations (MLPHARE) using these sites and all three NaBr MAD datasets resulted in phases with a figure of merit (FOM) of 0.25 (0.42) at 2.5 (3.0) Å resolution.
Model Building and Refinement.
The FOM was increased by solvent flattening/histogram matching (one NC1 hexamer per asymmetric unit, 65% solvent content) with DM (23) to 0.66 (2.3 Å), and was further improved to 0.72 by twofold noncrystallographic symmetry (NCS) averaging and by phase extension to 2.0 Å, using the data set Native1 (Table 1). Several cycles of automatic model building with ARP-WARP (24) and NCS averaging (DM) resulted in the autotracing of 23 partial chains comprising the main chain atoms of 1,250 residues. After sequence assignment with ARP-WARP, these chain fragments were combined to four almost complete α1 and to two less complete α2 chains using local symmetry [LSQKAB (23)].
This model was completed by several cycles of manual inspection, model building and refinement using MAIN (25) and CNS V1.1 (26). The target parameters of Engh and Huber (27), bulk solvent correction, overall anisotropic B-factor scaling, and strong NCS restraints between the two trimers (except a few side chains) were applied. For the final refinement, a new 1.9-Å data set keeping the same Rfree set (Native2, Table 1) was used. The final structure (Table 2), with an R factor of 18.2%, was analyzed with PROCHECK and SFCHECK (23). For bromide binding studies, further data sets were measured at varying NaBr concentrations.
Table 2.
Refinement statistics
| Resolution | 19.95–1.90 |
| Number of reflections | |
| Total | 148,362 |
| Test set | 7,569 |
| Completeness, %* | |
| Total | 92.3 (95.3) |
| Test set | 5.1 (5.2) |
| R factor, %* | 18.2 (27.7) |
| Rfree, %* | 20.2 (29.9) |
| Protein atom (nonhydrogen) | 10,514 |
| Solvens atoms (nonhydrogen) | 1,114 |
| Acetate atoms (nonhydrogen) | 36 |
| Rms deviation from standard bond length | 0.009 |
| Average B factor, Å2† | 32.63 (34.9) |
| Rms deviation between bonded B factors, Å2 | 2.370 |
Values for the last resolution shell (1.90–2.02 Å) are given in parentheses.
Wilson-plot B factor in parentheses.
The NC1 coordinates were deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1LI1).
Analysis of Cross-Linked Peptides.
To evaluate the nature of the six Met-93–Lys-211 contacts, the corresponding molecular regions were remodeled against a composite simulated annealing omit map (CNS) and refined, assuming the simultaneous existence of free and covalently cross-connected side chains with occupancies of 0.5 and connections via covalent Met-Sδ–Lys-Cɛ thioether bonds.
Reduced NC1 monomers and dimers were separated in the presence of 4 M guanidine on a TSK SWXL3000 column (TosoHaas, Stuttgart), and were digested with trypsin. These fragments were separated by size on a Superdex HR10/30 Peptide column (Amersham) and subjected to sequence analysis.
Results
Structure Determination.
Using a previously uncharacterized crystal form of human NC1 collagen IV and employing a NaBr derivative (28), three-wavelength MAD (Table 1) led to experimental phases of exceptional quality such that 90% of the amino acid residues comprising the NC1 hexamer could be built by an automated procedure. Eighty-three bromide binding sites per NC1 hexamer became defined in the NaBr-soaked crystals, explaining why experimental phases from an Osmium(IV) derivatized crystal were required to determine the 25 strongest bromide sites subsequently used to calculate the experimental phases. The Iβ6-Iβ7 loops (Ala-74 to Asp-78), especially, bind a bromide ion very tightly and are almost fully occupied even at 1 mM NaBr.
Overall Structure.
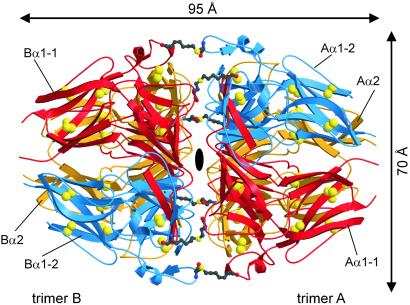
The asymmetric unit contains 1,359 aa in four α1(IV) and two α2(IV) NC1 chains, which are tightly packed into a distinct hexameric particle. This NC1 hexamer exhibits the shape of a prolate ellipsoid with a long 95-Å and a short 70-Å axis (Fig. 1). Two caps, made up of the trimeric NC1 ends of two opposing collagen IV protomers (in the following called A and B trimer, see Fig. 1), juxtapose each other via a central planar interface. In each cap, two α1 chains (A/Bα1-1 and A/Bα1-2) and one α2 chain (A/Bα2) are segmentally arranged, displaced by 120° around a central tunnel of varying width (Fig. 2a). The chains become defined with Asp-3 (A/Bα1-1, A/Bα1-2) and Ile-4 (A/Bα2) and can be fully traced up to the penultimate (A/Bα1-2) and the terminal residues (A/Bα1-1 and A/Bα2).
Figure 1.
Ribbon plot of the NC1 hexamer viewing along the pseudoexact twofold axis. The two α1(IV) chains and the α2(IV) chain of each trimer are shown in red (α1-1), blue (α1-2), and gold (α2). The intrachain disulfides are indicated by their bridging sulfur atoms (large yellow balls), and the covalent cross-links between Met-93 and Lys-211 are depicted as ball-and-stick representations. The figure was made with MOLSCRIPT (31) and RASTER3D (32).
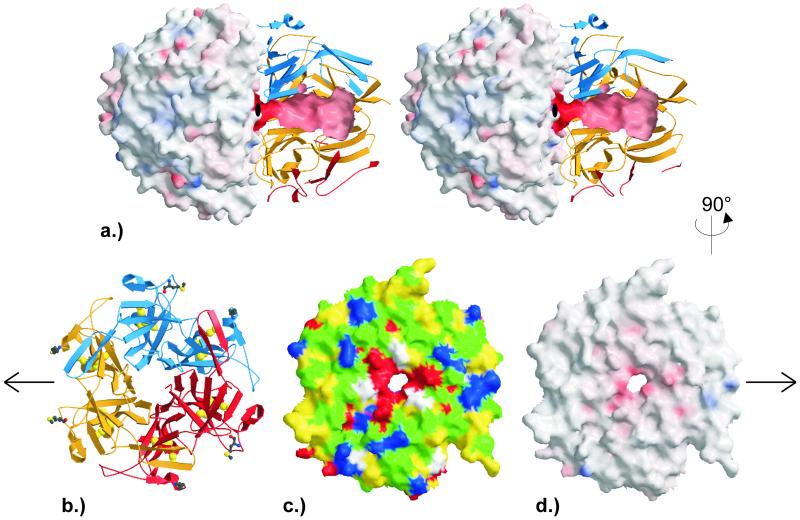
Figure 2.
Surface properties of the NC1 hexamers and trimers. (a) Stereo representation of the NC1 hexamer (orientation as in Fig. 1). The solid outer surface of trimer B and the internal tunnel surface of trimer A are colored according to the calculated electrostatic potential ranging from −50 e/kT and −150 e/kT (intense red) to 50 e/kT and 150 e/kT (intense blue), respectively. The ribbon plot for the back half of trimer A is as in Fig. 1. (b–d) Representations of trimer A along the pseudo sixfold as viewed from trimer B. (b) Ribbon representation as in Fig. 1. (c) Solid surface representation colored according to the physical properties of the surface forming amino acids (yellow, hydrophobic; green, hydrophilic; red, negatively charged; blue, positively charged; white, glycines). (d) Solid surface representation colored according to the calculated electrostatic potential ranging from −50 e/kT (intense red) to 50 e/kT (intense blue). The figure was made with GRASP [Nicholls, A., Bharadwaj, R. & Honig, B. (1993) Biophys. J. 64, A166 (abstr.)], MOLSCRIPT (31), and RASTER3D (32).
The NC1 hexamer exhibits a virtually exact twofold rotation axis (Fig. 1). The long NC1 particle axis exhibits a pseudo threefold rotation symmetry, transforming (on each trimer side) the α1-1 chain, the (chemically identical, but environmentally only similar) α1-2 chain and the (chemically different) α2 chain on each other (with rms deviations of 0.45 Å, 0.65 Å, and 0.66 Å for 223, 198, and 206 structurally equivalent amino acid residues, applying a threshold of 1.5 Å).
Single-Chain Geometry.
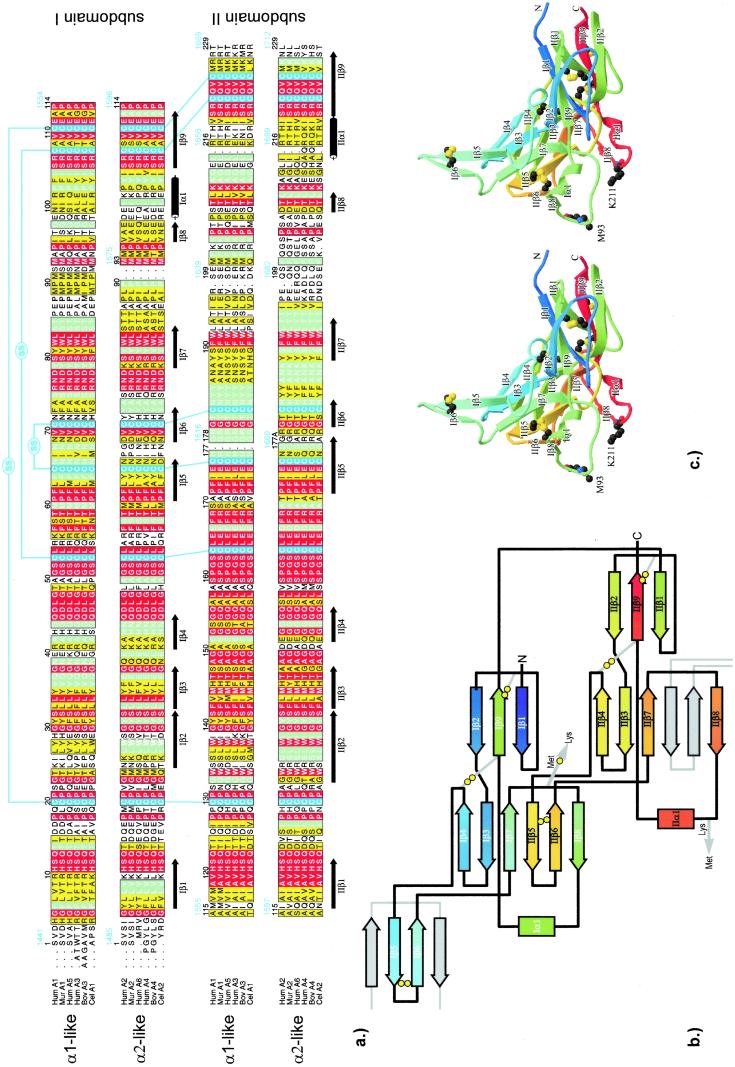
The crystallographically different α1-1 and α1-2 chains are folded virtually identically, and their topology closely resembles that of the α2 chain. A topologically optimal superposition suggests a two-residue deletion (residues 91 and 92) and a one-residue insertion (G178A) in the α2 chain (Fig. 3a), with significant differences occurring only between residues 197 and 202, where the alignment is somewhat arbitrary.
Figure 3.
Primary, secondary, and tertiary structure of the chain monomers. (a) Structure-based sequence alignment of the human NC1(IV) α1 and α2 chains with homologous sequences, grouped into α1-like (human α1, α3, α5, murine α1, bovine α3, and Caenorhabditis elegans α1) and α2-like sequences (human α2, α4, α6, murine α2, bovine α4, and C. elegans α2). Similar or identical amino acids within each group are colored in yellow and green, whereas residues completely conserved between both groups are shown in red. The sequence numbering (black) following the conventional NC1 numbering (15) is correlated with the total sequence numbering for human α1(IV) and α2(IV) chains (GenBank accession nos. P02462 and P08572; blue). The figure was made with ALSCRIPT (33). (b) Topological diagram of the α1-1 subunit of collagen type IV, orientated similar to Fig. 1. The spectral color coding is according to sequential order, from violet (N terminus) to red (C terminus), with disulfides (double circles) being explicitly shown. The β-strands provided from adjacent chain monomers to complete the six-stranded β-sheets are shown in gray. (c) Stereo ribbon plot of chain Aα1-1. Color coding is as in b. The Met-93 and Lys-211 residues involved in covalent cross-links are explicitly shown. The figure was made with MOLSCRIPT (31) and RASTER3D (32).
Each chain has the shape of a curved triangular plate spanning up to 180° around the small ellipsoid circumference but narrowing toward the poles, with a finger-like extension and a small incision (Fig. 3c). The NC1 chains start and end at the pole of the cap. As had been predicted from 35% sequence identity (9), they are folded in two successive subdomains I and II, which exhibit a similar topology (rms deviation of 0.64 Å for 76 Cα atoms, 1.5-Å threshold). The sequence lag between both chain halves varies from 110 (N terminus) to 114 amino acid residues (C terminus). Each subdomain is organized into nine β-strands (Iβ1 to Iβ9 and IIβ1 to IIβ9; Fig. 3b) comprising 54 amino acid residues, one helical six-residue turn and connecting segments. In each subdomain, three strands including the N- and the C-terminal segment are integrated in a small antiparallel 3-stranded β-sheet. Four strands of subdomain I (Iβ4, Ιβ3, Ιβ7, and Iβ8) form an incomplete β-sheet that is complemented to a six-stranded antiparallel sheet by the IIβ5–ΙΙβ6 finger-like hairpin loop from subdomain II, which inserts between Iβ7 and Iβ8. Equivalently, strands IIβ4, IIβ3, IIβ7, and IIβ8 of subdomain II form a fragmentary β-sheet that is completed by insertion of the Iβ5–Iβ6 hairpin loop from the “previous” chain monomer. The “own” Iβ5–Iβ6 finger, accordingly, is an integral part of the six-stranded IIβ-sheet of the “next” chain monomer (Fig. 3b). Both subdomains exhibit very similar disulfide connectivities, with all disulfide pairs exclusively made within a given subdomain (Fig. 3).
Trimer Structure and Tunnel.
In each trimer, the two α1 and one α2 triangular chain monomers are radially packed together forming a cap-like structure that exhibits a closed outside surface but leaves an internal, axial tunnel (Fig. 2a). Both subdomains of a given chain are displaced by 60°, giving rise to a pseudo sixfold rotation symmetry of the NC1 particle (Fig. 2b). The inter-subdomain contacts within a given monomer are similar to the 2,200-Å2 interface between adjacent monomers. About 40% of this inter-subdomain interface is provided by the β-hairpin finger, which grips into the premade six-stranded β-sheet (Fig. 3b).
The internal tunnels of opposing trimers have a funnel-like shape, with the diameters shrinking from a 15-Å cavity close to the particle poles down to a 4-Å narrow bottleneck before merging in a central cavity (Fig. 2a). These tunnels are mainly negatively charged and are lined by the three Iβ4 (Arg/Lys-41–Gln-45) and three IIβ4 edge strands (Glu-152–Gln-156) per trimer. These edge strands, primarily comprising small residues, are alternatively hydrogen-bonded to internal solvent molecules and to the adjacent partner strand of their respective β-sheet, forming a closed six-stranded “barrel”-like structure lacking interconnecting hydrogen bonds, however. Thus, the six subdomains of each cap form a propeller-like structure with pseudo sixfold rotation symmetry, in which the twisted antiparallel β-sheets of the NC1 trimer are radially arranged and pack back-to-face, with primarily hydrophobic interactions between adjacent blades (Fig. 2b).
Trimer–Trimer Interface.
Across the quite planar trimer–trimer interface of approximately 4,000 Å (2) (Fig. 2 c and d) the two α2 chains mainly juxtapose each other, whereas the α1-1 chains mainly oppose the α1-2 chains and vice versa. In the center of this interface is the above-mentioned almost spherical cavity with a diameter of about 11 Å, lined by eight acidic residues (Glu-40, Glu-152) provided by the four α1 chains, and by eight residues (Glu-37, Glu-40, Asp-151, Glu-152) from the two α2 chains. All six Glu-40 side chains are oriented axially toward the opposite trimer and interdigitate with each other, in addition getting in close contact with the opposing two Glu-37 carboxylate (α2) and four Gln-37 carboxamide (α1) moieties. This central cavity contains about 20 localized and a few more disordered solvent molecules. Further toward the periphery, the charges are more balanced, with continuous hydrogen-bond networks being made between residues such as Asn-39⋅⋅⋅Asn-187⋅⋅⋅Arg-76⋅⋅⋅Glu-175, which are alternatively provided by chains from opposing trimers and are embedded in aromatic clusters (Fig. 2 c and d).
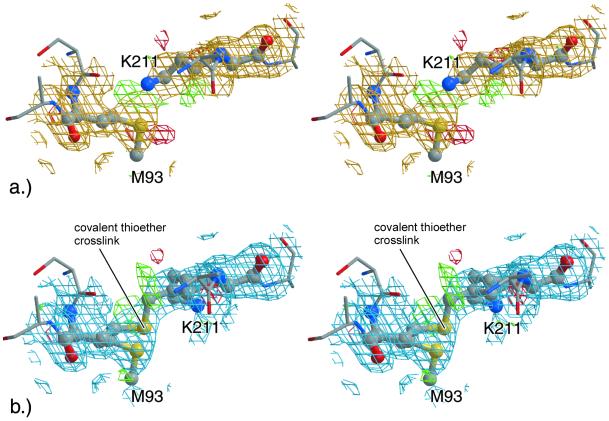
Interestingly, each α1-1, α1-2, and α2 chain makes very close contacts to the α1-2, α1-1, and α2 chain from the opposite trimer, respectively, via the IIβ8/IIα1 and Iβ7/Iβ8 loops. In the course of crystallographic refinement, the side chains of the surface located Lys-211 and Met-93 residues from opposing chains were pushed together beyond normal van der Waals contacts, still leaving a strong positive difference electron density between Met-93 Sδ and Lys-211 Cɛ/Nζ, which could not be interpreted by noncovalent contacts between these atoms alone. A simulated annealing omit electron density map (29) showed a continuous electron density level across these junctions (Fig. 4a). Thus, this region was also modeled as consisting of an ≈1:1 mixture of a noncovalent and a (Sδ–Cɛ) covalent form between these two side chains, resulting in a smoother difference density (Fig. 4b) and an unchanged R factor, in agreement with multiple conformations. However, the current resolution of 1.9 Å does not allow a definitive refinement of occupancy, B factors and coordinates.
Figure 4.
Electron densities around Met-93 and Lys-211. (a) Stereo representation of the section about Lys-211 (Bα1-1) and Met-93 (Aα2) in its open form (fully refined ball-and-stick model) with simulated annealing omit (gold, contoured at 1 σ) and fobs − fcalc (green, contoured at 2.5 σ; red, −2.5 σ) difference electron density maps. (b) Equivalent section for a modeled 1:1 mixture between the open and a potential thioether cross-linked form superimposed with the corresponding 2fobs − fcalc (blue, contoured at 1.0 σ) and the fobs − fcalc (green, contoured at 2.5 σ; red, −2.5 σ) difference electron density maps. The figure was made with BOBSCRIPT (34) and RASTER3D (32).
The suggested cross-links were investigated by chemical methods. As determined by amino acid analysis, 70/60% of the α1 and α2 chains of dissociated NC1 exist as covalent dimers before/after strong reduction. Sequence analysis of the size-separated tryptic peptides showed that only dimers, but not monomers, yielded the expected cross-linked peptides (α1: Asn-77–Arg-102/Lys–204–Arg-216; α2: Ser-78–Arg-107/Tyr-189–Arg-216) in the first peak, whereas later eluting peaks containing almost all other predicted peptides were identical for monomers and dimers.
Discussion
By applying MAD techniques to crystals soaked in 1 M NaBr, the non-isomorphism problem encountered 15 years ago with other collagen NC1(IV) crystals (4, 19) was overcome. The NC1 hexamer turns out to be an ellipsoidal particle of approximate 622 symmetry, corresponding in size and shape to the particles seen by electron microcopy (4, 16). This hexamer is made up of two trimeric caps packed against each other via their planar bases. Each trimeric cap consists of two α1 chains and one α2 chain, arranged around a tunnel of conical shape, which widens toward the poles ending in a closed cavity. This chain fold is rich in β-structures and is unlike any other protein of known structure. In particular, the suggested structural homology to the tissue inhibitors of matrix metalloproteinases (9) was not found.
The structurally very similar α1 and α2 chain monomers are composed of two similar subdomains, displaced by exactly 60° around the central NC1 particle axis. Each subdomain represents a compact disulfide-stabilized triangular structure, from which a finger-like hairpin loop projects into an incompletely formed six-stranded β-sheet of an adjacent subdomain of the same or of an adjacent chain clamping the subdomains tightly together.
In each cap, the completed six-stranded β-sheets from the six subdomains are radially arranged around the central tunnel like propeller blades. However, in contrast to typical β-propellers, where a single chain builds up in consecutive order four-stranded blades with strand order 1-2-3-4 (30), the three NC1 chains make up six blades consisting of six β-strands aligned in the strand order 4-3-7-5′-6′-8 (numbering according to Fig. 3, with the primed strands provided by the neighboring subdomain). In some of these typical β-propeller domains the first and the last blades are connected via N-terminal strand overlap, also called Molecular Velcro. In the NC1 propeller, in contrast, all adjacent blades are much more tightly clamped together by the extensive interactions between hairpin finger and the premade β-sheet, probably conferring enormous mechanical strength on each trimer cap.
In contrast to this tight chain interdigitation within each NC1 cap and contradictory to previous NC1 models that suggested domain swapping (9), both caps are connected via relatively planar opposing surfaces, coupling two collagen IV protomers together. The cap–cap interface is large, with mostly polar and favorable electrostatic interactions. Of particular note, however, is the strong accumulation of negatively charged residues in the central cavity, which must be compensated by a number of soluble cations so as not to disrupt the trimer–trimer interaction.
At the periphery, this cap–cap junction is further stabilized by a previously uncharacterized covalent cross-link, where the three Met-93 side chains from each cap are cross-connected with the three Lys-211 side chains from the opposing cap. The crystallographic evidence suggests the existence of multiple conformations, with separate and covalently linked side chains occurring simultaneously. This finding is in agreement with preliminary protein chemical data, showing that the predicted cross-linked peptides are found in dimers, but not in monomers, and that the implicated lysines were not recognized by trypsin. Our data also agree with the existence of nonreducible α1-α1 and α2-α2 dimers observed in various NC1 preparations (11, 15, 16). Because of a distance of about 20 Å between the respective cysteine residues, these dimers cannot result from disulfide exchanges between the two opposite caps as was previously suggested (15). The degree of cross-linking might change with tissue type and aging but should lead to an increased mechanical strength along the collagen IV fibers. The exact nature of this covalent bond remains to be established, however.
According to the structure-based sequence alignment (Fig. 3), the two Goodpasture epitope regions identified in α3(IV) chains map in a ring like fashion to the β1-β2 loop and strand β2 of subdomains I and II close to the cap pole, in agreement with their predicted close spatial proximity (14). They are, however, as opposed to previous suggestions (14), outside of the trimer–trimer interface. The nine listed mutations in α5(NC1) leading to Alport syndrome (9) affect strongly to strictly conserved residues, which are distributed over the whole monomer. Most of them seem to be important for structural integrity, whereas α5Arg1563 (corresponding to Arg-107) seems to modify surface properties of the NC1 globules—i.e., it may affect interactions with other extracellular matrix components.
Acknowledgments
We thank John Richardson for careful reading. The financial support by the SFB596 is kindly acknowledged.
Abbreviations
- NC1
noncollagenous
- MAD
multiple anomalous diffraction
Footnotes
Data deposition: The atomic coordinates for NC1 have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1LI1).
References
- 1.Hudson B G, Reeders S T, Tryggvason K. J Biol Chem. 1993;268:26033–26036. [PubMed] [Google Scholar]
- 2.Kühn K. Matrix Biol. 1994;14:439–445. doi: 10.1016/0945-053x(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 3.Oberbäumer I, Wiedemann H, Timpl R, Kühn K. EMBO J. 1982;1:805–810. doi: 10.1002/j.1460-2075.1982.tb01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timpl R, Oberbäumer I, von der Mark H, Bock W, Wick G, Weber S, Engel J. Ann NY Acad Sci. 1985;460:58–72. doi: 10.1111/j.1749-6632.1985.tb51157.x. [DOI] [PubMed] [Google Scholar]
- 5.Timpl R. Eur J Biochem. 1989;180:487–502. doi: 10.1111/j.1432-1033.1989.tb14673.x. [DOI] [PubMed] [Google Scholar]
- 6.Yurchenco P D. In: Extracellular Matrix Assembly and Structure. Yurchenco P D, Birk D E, Mecham R P, editors. San Diego: Academic; 1994. pp. 351–388. [Google Scholar]
- 7.Zhou J, Ding M, Zhao Z, Reeders S T. J Biol Chem. 1994;269:13193–13199. [PubMed] [Google Scholar]
- 8.Gunwar S, Ballester F, Noelken M E, Sado Y, Ninomiya Y, Hudson B G. J Biol Chem. 1998;273:8767–8775. doi: 10.1074/jbc.273.15.8767. [DOI] [PubMed] [Google Scholar]
- 9.Netzer K-O, Suzuki K, Itoh Y, Hudson B G, Khalifah R G. Protein Sci. 1998;7:1340–1351. doi: 10.1002/pro.5560070610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dölz R, Engel J, Kühn K. Eur J Biochem. 1988;178:357–366. doi: 10.1111/j.1432-1033.1988.tb14458.x. [DOI] [PubMed] [Google Scholar]
- 11.Boutaud A, Borza D-B, Bondar O, Gunwar S, Netzer K-O, Singh N, Ninomiya Y, Sado Y, Noelken M E, Hudson B G. J Biol Chem. 2000;275:30716–30724. doi: 10.1074/jbc.M004569200. [DOI] [PubMed] [Google Scholar]
- 12.Peticlerc E, Boutaud A, Prestayko A, Xu J, Sado Y, Ninomiya Y, Sarras M P, Hudson B G, Brooks P C. J Biol Chem. 2000;275:8051–8061. doi: 10.1074/jbc.275.11.8051. [DOI] [PubMed] [Google Scholar]
- 13.Netzer K-O, Leinonen A, Boutaud A, Borza D-B, Todd P, Gunwar S, Langeveld J P M, Hudson B G. J Biol Chem. 1999;274:11267–11274. doi: 10.1074/jbc.274.16.11267. [DOI] [PubMed] [Google Scholar]
- 14.Borza D-B, Netzer K-O, Leinonen A, Todd P, Cervera J, Saus J, Hudson B G. J Biol Chem. 2000;275:6030–6037. doi: 10.1074/jbc.275.8.6030. [DOI] [PubMed] [Google Scholar]
- 15.Siebold B, Deutzmann R, Kühn K. Eur J Biochem. 1988;176:617–624. doi: 10.1111/j.1432-1033.1988.tb14321.x. [DOI] [PubMed] [Google Scholar]
- 16.Weber S, Engel J, Wiedemann H, Glanville R W, Timpl R. Eur J Biochem. 1984;139:401–410. doi: 10.1111/j.1432-1033.1984.tb08019.x. [DOI] [PubMed] [Google Scholar]
- 17.Weber S, Dölz R, Timpl R, Fessler J H, Engel J. Eur J Biochem. 1988;175:229–236. doi: 10.1111/j.1432-1033.1988.tb14188.x. [DOI] [PubMed] [Google Scholar]
- 18.Ries A, Engel J, Lustig A, Kühn K. J Biol Chem. 1995;270:23790–23794. doi: 10.1074/jbc.270.40.23790. [DOI] [PubMed] [Google Scholar]
- 19.Stubbs M, Summers L, Mayr I, Schneider M, Bode W, Huber R, Ries A, Kühn K. J Mol Biol. 1990;211:683–684. doi: 10.1016/0022-2836(90)90066-U. [DOI] [PubMed] [Google Scholar]
- 20.Blume H, Boesecke P, Bourenkov G P, Kosciesza D, Bartunik H D. Nuclear Instr Meth A. 2001;467–468:1358–1362. [Google Scholar]
- 21.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Sheldrik G M. Methods Enzymol. 1997;276:628–640. [Google Scholar]
- 23.Collaborative Computational Project. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 24.Perrakis A, Morris R, Lamzin V S. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 25.Turk D. Dissertation. Munich: Technische Universität; 1992. [Google Scholar]
- 26.Brünger A T, Adams P D, Clore G M, Delano W L, Gros P, Grossekunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 28.Dauter Z, Dauter M, Rajashankar K R. Acta Crystallogr D. 2000;56:232–237. doi: 10.1107/s0907444999016352. [DOI] [PubMed] [Google Scholar]
- 29.Brünger A T, Adams P D, Rice L M. Structure(London) 1997;5:325–336. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 30.Fülöp V, Jones D T. Curr Opin Struct Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 32.Merrit E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 33.Barton G J. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 34.Esnouf R M. Acta Crystallogr D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]