Abstract
Many anticancer drugs interact directly with DNA to exert their biological functions. To date, all noncovalent, intercalating drugs interact with DNA exclusively by inserting their chromophores into base steps to form elongated and unwound duplex structures without disrupting the flanking base pairs. By using actinomycin D (ActD)-5′-GXC/CYG-5′ complexes as examples, we have found a rather unusual interaction mode for the intercalated drug; the central Watson–Crick X/Y base pairs are looped out and displaced by the ActD chromophore. The looped-out bases are not disordered but interact perpendicularly with the base/chromophore and form specific H bonds with DNA. Such a complex structure provides intriguing insights into how ligand interacts with DNA and enlarges the repertoires for sequence-specific DNA recognition.
Chemotherapy is widely used to combat and treat various types of cancers. Although numerous compounds have been developed as potential candidates for such a purpose, only few of them have become effective for clinical practice. Currently three major classes of DNA-acting antitumor drugs are available (1). Some form noncovalent complexes with DNA through intercalation (such as actinomycin D, Fig. 1). Others form covalent linkages with DNA (such as cisplatin), whereas some antibiotics bind to DNA and cleave its backbone in subsequent steps (such as bleomycin and enediyne).
Figure 1.
The structure of actinomycin D.
Because of its interesting sequence specificity and strong potency against tumor, the ActD/DNA complex has been well studied by thermodynamics (2), foot-printing (3), x-ray crystallography (4, 5), NMR (6, 7), and spectroscopic measurements (8, 9) to understand how this drug binds to DNA and, hence, interferes with the replication and transcription processes. These studies have revealed that 5′-GpC-3′ sequence is the major binding site, with the phenoxazone ring of ActD intercalating into the 5′-GC/CG-5′ step and the two cyclic pentapeptide lactone anchoring on both sides of the minor groove. The formation of four threonine-guanine H bonds possibly accounts for the preference of this drug for the 5′-GpC-3′ site. Furthermore, the flanking sequences of the GpC site also have been found to play important roles in determining the binding affinity of ActD, likely because of their interactions with the cyclic pentapeptide lactone rings of ActD (8–10).
Intriguingly, there have been reports from the thermodynamic as well as spectroscopic studies that ActD also interacts strongly with DNA sequences devoid of the classic 5′-GC/CG-5′ sequence (2, 11–13). We now report the complex structures of ActD binding with previously uncharacterized DNA recognition sites 5′-GXC/CYG-5′ (where X/Y is G⋅C or T⋅A Watson–Crick base pair) in DNA hairpins closed by a mini-ACC loop (14). Our results indicate that ActD interacts with the 5′-GXC/CYG-5′ sequences in an unusual way in which the central Watson–Crick X⋅Y base pair is completely disrupted. The looped-out bases are not disordered but interact perpendicularly with the stacked bases/chromophore and form specific hydrogen bonds with DNA. Such a dramatic manipulation of Watson–Crick base pair conformation by a potent anticancer drug also provides interesting potential for discovering new types of DNA–protein interaction.
Methods
Sample Preparation.
All DNA samples were synthesized in 3 μmol scale on an Applied Biosystems 380B DNA synthesizer with the final 5′-DMT groups attached. The samples were purified and prepared for NMR studies as described (15). The DNA was dissolved in 500 μl of buffer solution to make 3 mM concentration. ActD was first dissolved in methanol, and aliquots of it were added to the DNA samples to reach the desired DNA/drug ratio. The complexes were reannealed by heating to 95°C for several minutes before being chilled in ice water.
UV Melting Studies.
Absorbance (OD) vs. temperature profile was obtained at 260 nm with a Varian Cary 100 UV spectrometer equipped with a temperature controller. The temperature in each run was increased from 20°C to 95°C at a rate of 0.5°C per min. Melting temperatures were extracted from the maxima of the first derivatives of the melting curves.
Binding Constant Measurements.
Absorption spectral titrations were carried out by starting with a 2 ml solution of about 5 μM ActD, followed by progressive additions of stock oligomer solution at equal time intervals. Absorbance differences between 427 and 480 nm were used to obtain the binding isotherms. The association binding constants were deduced by means of nonlinear least-squares fits on these isotherms using the 1:1 drug:strand binding model (13).
NMR Experiments.
All NMR experiments were obtained on a Varian Unity Inova 600 MHz spectrometer. One-dimensional imino proton spectra at 0°C were acquired by using jump-return pulse sequence (16). The carrier frequency was set at the resonance of water, and the maximum excitation was set at 12.5 ppm. Two-dimensional (2D) nuclear Overhauser effect spectroscopy (NOESY) in 90% H2O/10% D2O was performed at 0°C in a pH 6.8 low salt (10 mM sodium phosphate/20 mM NaCl) buffer under a 200 ms mixing time. NOESY 2D experiments in D2O were carried out at 20°C in hypercomplex mode and collected by using two mixing times of 100 and 250 ms. The wet-TOCSY experiment was performed at 0°C under a mixing time of 75 ms. A double quantum filtered-correlated spectroscopy (DQF-COSY) spectrum was collected in the TPPI mode. A proton-detected 31P-1H heterocor spectrum (17) was collected in the TPPI mode. The acquired data were transferred to an IRIS 4D workstation and processed by felix software (MSI Inc. Accelerys) as described (18).
Structure Determination.
The final ActD/5′-GTCACCGAC-5′ complex structures were accomplished by first applying distance geometry calculations (DGII from MSI Inc. Accelerys) using constraints derived from NMR experiments to determine the initial DNA structures. Most distance constraints from NOESY/D2O were classified as very strong, strong, medium, or weak, based on their relative intensities at 100 ms mixing time and given generous distance bounds of 2.0–3.0 Å, 2.0–4.0 Å, 3.0–5.0 Å, or 4.0–6.0 Å, respectively. Canonical hydrogen bond distances with bounds of 1.8–2.1 Å were assigned to Watson–Crick base pairs. The distance constraints involving exchangeable protons also were derived from NOESY/H2O and were given only two wide distance bounds of either 2.0–5.0 Å or 3.0–6.0 Å. The β and γ torsional angle constraints were determined semiquantitatively from the 31P-1H heteronuclear correlation data (19) by using the in-plane “W rule” (20). Based on the absence of long-range 4JH2′-P coupling, all ɛ torsion angles were constrained to the trans domain (180° ± 30°) (21). The ζ and α dihedral angles were all left unconstrained. The DNA structures were generated by embedding the DNA bound matrix. About half of the initial DNA structures (15 of 30) have conformation suitable for subsequent docking and molecular dynamics studies. Because the majority of conformational parameters of the drug in the ActD/TA and ActD/GC (see next section on notations) complex structure were similar to those of the free drug in the crystal (4, 5, 22), ActD coordinates determined from the x-ray diffraction method (4) were used to dock against the initial DNA structures (6, 8). Energy minimization and constrained molecular dynamics then were applied with 56 intermolecular distance constraints (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org) as well as 167 intramolecular constraints (including 129 intra-DNA and 38 intra-ActD) to determine the final complex structures that best fit the experimental NMR data. Dynamics were initiated at 500 K with a 1-fs time step. After a total of 10 ps of molecular dynamics at 500 K, the system was slowly cooled to 300 K in 10 ps. The system then was equilibrated at 300 K for 5 ps. Well converged final structures with pair-wise rmsd values of ≈0.75 Å were obtained after the final molecular dynamics calculations. Similar strategy was applied to obtain the final ActD/5′-GGC ACCGCC-5′ complex structures.
Results
Complex Formation.
At low temperature (0°C) and low salt (20 mM NaCl and 10 mM, pH 6.8 sodium phosphate buffer) condition, single-stranded d(GXCACCGYC) sequences form stable hairpin structures containing three canonical Watson–Crick base pairs in the stem region closed by a miniACC loop (Fig. 2; ref. 14). The ActD/hairpin complexes then were generated by adding one equivalent of ActD to one equivalent of d(GXCACCGYC) sequence (the free DNA hairpins will be designated as TA, AT, GC, and CG for X⋅Y = T⋅A, A⋅T, G⋅C, and C⋅G, whereas the corresponding ActD/DNA complexes will be designated as ActD/TA, ActD/AT, ActD/GC, and ActD/CG, respectively). Only one major set of imino proton resonances (assigned by 2D NOESY/H2O experiments, to be described later) with significant changes in chemical shift was observed for the ActD/TA (Fig. 2A), ActD/GC (Fig. 2B), and ActD/CG (data not shown), whereas two major sets of signals were observed for the ActD/AT (data not shown). The reason for such a difference is not yet clear. These DNA nonamers were chosen for binding studies because they are the simplest GXC-containing oligomers that exhibit strong binding affinities with ActD to permit a detailed structural characterization and determination of a previously uncharacterized binding mode for this classic drug.
Figure 2.
The ActD titration imino proton spectra of the 5′-GTCACCGAC-3′ (A) and 5′-GGCACCGCC-3′ (B) hairpins at 0°C before (Top) and after (Bottom) addition of ActD at 1/1 ratio. Significant changes of imino proton chemical shifts were observed in both cases.
Thermodynamics and Binding Constant Studies.
The ActD-induced thermal stabilization of d(GXCACCGYC) hairpins containing the GXC/GYC binding sites and the control GC/GC site were carried out by UV melting studies. The results indicate that ActD binding at the conventional 5′-GC/CG-5′ site does cause a considerable stabilization of approximately 12°C for the DNA hairpin (51°C vs. 39°C). However, an even greater stabilization of more than 20°C was observed for the ActD/DNA complexes containing the 5′-GXC/CYG-5′ sites. We have obtained melting data for the ActD/hairpin complexes vs. free hairpins as following: ActD/TA, 68°C vs. TA, 45°C; ActD/GC, 74°C vs. GC, 50°C; ActD/CG, 73°C vs. CG, 50°C; ActD/AT, 65°C vs. AT, 45°C). These data correlate well with the previous report of a nonclassic high-affinity binding site in the 5′-GTC/CAG-5′ containing DNA sequence (2). These results also indicate that the extra X/Y pair inserted into the classic ActD recognition site in these hairpins does not interfere with ActD binding but even stabilize ActD binding by an additional 8–12°C.
Binding constants of ActD/XY complexes also were deduced by means of nonlinear least-squares fits on the differential absorbance isotherms at 427 nm/480 nm. The following binding constant data further confirm the stronger ActD binding at the 5′-GXC/CYG-5′ sites than at the classic 5′-GC/CG-5′ site. Although ActD binding at the classic 5′-GC/CG-5′ site of the 5′-GCACCGC-3′ hairpin has a binding constant of 0.36 ± 0.01 × 106 (M−1), those of ActD binding at the 5′-GXC/CYG-5′ sites of the corresponding hairpins have binding constants of 5.1 ± 0.2 × 106, 3.4 ± 0.3 × 106, 16.4 ± 2.9 × 106, and 3.7 ± 0.4 × 106 for the ActD/TA, ActD/AT, ActD/GC, and ActD/CG complexes, respectively. In other words, the ActD/XY complexes of these hairpins exhibit ActD affinities nearly an order of magnitude higher than their corresponding classic ActD/5′-(GC)/(CG)-5′ complex.
NMR Studies in D2O.
Further insights into the unusual structures of the ActD/GC and ActD/TA complexes come from 2D NMR studies. An expanded region of the NOESY (250 ms mixing time) contour plot of the ActD/TA complex correlating the base proton with the sugar H1′/H5/H3′/H4′ protons at 10°C is shown in Fig. 6, which is published as supporting information on the PNAS web site. Although interruptions occur during the sequential nuclear Overhauser effect (NOE) assignments, it was still possible to come to a complete and self-consistent set of assignments through the combined tracing of the base-base, base-H1′, base-H3′, base-H4′, and base-H2′/H2" connections. Interestingly, unlike those in B-DNA, the inter base-base, base-H3′, base-H1′, and base-H4′ NOE intensities were found to be dramatically different yet highly correlated with each other, possibly because of the different positions of the unpaired T2/A8 bases in the ActD/TA complex, as compared with those in the paired B-DNA duplex, as shown in Fig. 6b. In addition, we also observed consistent chemical shift changes for the T2/A8 and G2/C8 protons upon ActD binding (listed in Table 1, which is published as supporting information on the PNAS web site). The aromatic and H1′ protons of T2/A8 and G2/C8 bases all move downfield, because of their looped-out state and the swinging away from the shielding zone of the stacked bases/chromophore core. The fact that most protons in the 5′-end G1 and G7 residues experience upfield shifting, whereas those in the 3′-end C3 and C9 residues undergo downfield shifting, is also consistent with the idea of a stacked G1-phenoxazone-G7 core with unstacked cytosines (see Fig. 4c).
Figure 4.
(a) Superimposed wide-eye stereo views of the final ActD/TA complex structure. The guanosine residues are drawn in yellow, cytosines in green, ACC loop in white, ActD in gray, the looped-out residues A8 in purple, and the looped-residues T2 in blue. (b) The complex structure was shown in downthe-helical view with the top ACC loop removed and all hydrogen atoms deleted for reasons of clarity. The planar chromophore of ActD passes through to the major groove and exhibits excellent hydrophobic stacking with the G7 and G1 bases, whereas the C3 and C9 bases are almost unstacked from the hydrophobic core. The phenoxazone-2NH2 form a hydrogen bond with the T2O4′ atom (indicated by a red dotted line). Two perpendicular G7H8-A8(π-ring) and phenoxazone-3O-T2(π-ring) interactions were detected (indicated by dotted blue lines). (c) The complex structure was drawn from the major groove view. The C3 and C9 bases are tilted away from the hydrophobic stacking. The 6NH2 atom of the vertical A8 base forms a hydrogen bond with the G7 phosphate oxygen. (d) The space-filling wide-eye stereo view of the ActD/TA complex from the major groove (the ActD 3O and 5O atoms are drawn in orange). The two methyl groups of the chromophore 4-CH3 and 6-CH3 interact well with the surrounding A8-G7/C3-T2-G1/C9 bases. The ActD/GC complex structure drawn from the down-the-helical view and the major groove are shown in e and f, respectively. The looped-out C8 residue is drawn in red, and the looped-out G2 residue is drawn in blue. One hydrogen bond between the phenoxazone-2NH2 and G2O4′ atoms (indicated by a red dotted line in e), an H bond between the C8NH2 and G7 phosphate oxygen atom (by a red dotted line in f), as well as one perpendicular interaction between the phenoxazone-CH3 proton and the G2 base (indicated by a blue dotted line in e) were observed.
NMR Studies in H2O.
The imino protons of the ActD/GC and ActD/TA complexes shown in Fig. 2 were assigned through the NOESY/H2O experiments. Fig. 7 (which is published as supporting information on the PNAS web site) shows the expanded NOESY contour plots establishing distance connectivities from the imino proton to imino proton (Bottom) and imino proton to amino proton and H5 proton (Top). In both spectra, the G1 imino proton exhibits NOEs to its own amino protons as well as to the amino protons of C9. Similarly, the G7 imino proton exhibits NOEs to its own amino protons and to the amino protons of C3. On the contrary, the G2 imino proton in the ActD/GC complex spectrum exhibits no NOE to its own amino protons or the C8 amino protons. A similar situation also occurs for the T2 imino proton in the ActD/TA complex spectrum, which exhibits no NOE to the A8 amino protons. Furthermore, no imino proton—imino proton NOE was detected for the G1/G2/G7 context in the ActD/GC complex or the G1/T2/G7 context in the ActD/TA complex spectrum (marked by dotted empty circles in the spectra), although such NOEs were clearly detected in the free DNA hairpins at 200 ms mixing time (data not shown). Such results indicate that although G1/C9 and C3/G7 bases in the ActD/GC and ActD/TA complexes remain paired, the T2/A8 base pair in the ActD/TA complex or the G2/C8 base pair in the ActD/GC complex has been disrupted upon ActD binding. This conclusion also is supported by the extraordinary upfield shifting of the T2 and G2 imino protons from the paired region at 13.7 and 12.9 ppm to the unpaired region at 11.4 and 11.3 ppm, respectively (Fig. 2).
All protons within each of the cyclic pentapeptide rings in the ActD/GC and ActD/TA complexes also were completely assigned by using the well established procedures (23). The expanded NOESY contour plots in water at a mixing time of 200 ms featuring the sequential assignments of the cyclic pentapeptide rings for both complexes are shown in Fig. 8, which is published as supporting information on the PNAS web site. The connectivity starts from the recognition of the HN protons of L-Thr on the quinoic (Q) and benzenoic (B) cyclic pentapeptide rings respectively at 7.43 and 7.38 ppm for the ActD/GC complex (Top) and 7.83 and 7.67 ppm for the ActD/TA complex (Bottom), based on their characteristic NOEs to the corresponding Hα proton, Hβ proton, and CH3γ protons (the complete assignments of the ActD protons are listed in Table 2, which is published as supporting information on the PNAS web site). We can connect inter-residue L-ThrHα→D-ValHN, D-ValHα→L-ProHα, L-ProHα→SarHα/Hα′, and SarHα/Hα′→ LMeValNCH3 NOE cross peaks each for the Q (in solid lines) and B rings (in dotted lines), respectively. Similar tracing also could be followed for the ActD/TA complex shown in the bottom figure. The peptide linkages in the cyclic pentapeptide rings also were determined to be trans between the L-Thr and D-Val residues and the Sar and L-MeVal residues based on the strong dHα-HN NOE cross peaks, whereas the linkages between the D-Val and L-Pro residues and the L-Pro and L-MeVal residues were determined to be cis based on the strong dHα-Hα NOE cross peaks (6, 7, 23).
The intermolecular NOEs in the NOESY spectra of the ActD/GC and ActD/TA complexes also have been identified after complete characterization of the DNA and ActD protons, which are listed in Table 3. Several such typical NOESY spectra are shown in Fig. 9, which is published as supporting information on the PNAS web site, and Fig. 3. In Fig. 9, the 4CH3 protons of the phenoxazone quinoid ring exhibit NOE cross peaks to the G1H8 and C3NH2 protons, whereas the 6CH3 protons of the benzenoid ring exhibit NOE cross peaks to the G7H8 and C9NH2 protons in both complexes. Such intermolecular NOEs unambiguously define the intercalation of the phenoxazone chromophores within the 5′-(G1C3)/(G7C9)-3′ step with looped-out T2/A8 or G2/C8 residues. Hence the phenoxazone chromophores are situated in such a way as to align their long axes with that of the flanking base pairs, with the benzenoid ring being sandwiched between the G7/C9 bases on one strand and the quinoid ring between the G1/C3 bases on the partner strand. This conclusion is also corroborated by the abundant intermolecular NOEs between the ActD amino acid protons and the DNA protons, as shown in Fig. 3, with the red lines connecting the NOE cross peaks between the quinoid pentapeptide lactone ring and the G1⋅C9 base pair protons and the blue lines connecting the NOE cross peaks between the benzenoid pentapeptide lactone ring and the C3⋅G7 base pair protons. Similar characteristic NOEs were observed for both spectra of the ActD/GC (Fig. 3 Top) and ActD/TA (Fig. 3 Middle) complexes, which are cartooned in Fig. 3 Bottom.
Figure 3.
Some of the important intermolecular NOE cross peaks illustrating the interactions of ActD protons with the H1′ protons of the nonadjacent G1⋅C9 and C3⋅G7 base pairs in the minor groove edge of the ActD/GC (Top) and ActD/TA (Middle) complexes. Red lines connecting the NOE cross peaks between the quinoid pentapeptide lactone ring with the G1⋅C9 base pair, whereas blue lines connecting the NOE cross peaks between the benzenoid pentapeptide lactone ring with the C3⋅G7 base pair. Thus, G1H1′ proton exhibits NOEs to the L-ThrQHN (a), L-ThrQCH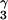 (b), L-MeValQHα (c), and L-MeValQNCH3 (d), whereas C9H1′ proton exhibits NOEs to the L-ProQHβ,γ,β′ protons (i–k, respectively). Similarly, G7H1′ proton exhibits NOEs to the L-ThrBHN (e), L-ThrBCH
(b), L-MeValQHα (c), and L-MeValQNCH3 (d), whereas C9H1′ proton exhibits NOEs to the L-ProQHβ,γ,β′ protons (i–k, respectively). Similarly, G7H1′ proton exhibits NOEs to the L-ThrBHN (e), L-ThrBCH (f), L-MeValBHα (g), and L-MeValBNCH3 (h), whereas C3H1′ proton exhibits NOEs to the L-ProBHβ,γ,β′ protons (l–n, respectively). The green lines connect the NOEs exhibited by the C9H5 proton to the H1′ proton of the looped-out residue C8 or A8 (o) and the C3H5 proton to the H1′ proton of the looped-out residue G2 or T2 (p). The orange lines also connect NOEs exhibited by the ActD chromophore phenoxazone 4CH3 protons to the G1H8 and C3NH2 protons and the 6CH3 protons to the G7H8 and C9NH2 protons shown in Fig. 9. Such NOEs are cartooned in the bottom figure.
(f), L-MeValBHα (g), and L-MeValBNCH3 (h), whereas C3H1′ proton exhibits NOEs to the L-ProBHβ,γ,β′ protons (l–n, respectively). The green lines connect the NOEs exhibited by the C9H5 proton to the H1′ proton of the looped-out residue C8 or A8 (o) and the C3H5 proton to the H1′ proton of the looped-out residue G2 or T2 (p). The orange lines also connect NOEs exhibited by the ActD chromophore phenoxazone 4CH3 protons to the G1H8 and C3NH2 protons and the 6CH3 protons to the G7H8 and C9NH2 protons shown in Fig. 9. Such NOEs are cartooned in the bottom figure.
Structural Features of the ActD/TA and ActD/GC Complexes.
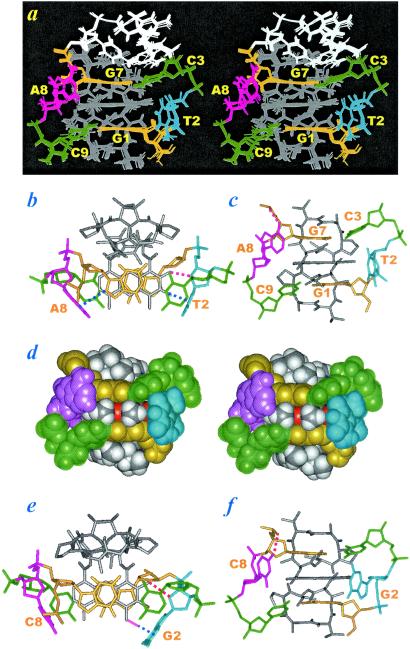
As shown in Fig. 4a of the determined ActD/TA structure, many unexpected features were detected. Firstly, instead of being hydrogen-bonded, stacked and parallel, the central T2⋅A8 base pairing is now disrupted, the bases are looped out and are perpendicular to the long axis of the flanking Watson–Crick base pairs. Intriguingly, the ActD chromophore still prefers stacking with the G1/G7 bases by pushing out the T2⋅A8 base pair in the 5′-G1T2C3/C9A8G7-5′ sequence because of the strong intermolecular ActD-DNA hydrogen bonds between the G7NH2–ThrBC⩵O and the G1NH2–ThrQC⩵O atoms, and the excellent stacking within the G1/phenoxazone/G7 bases (Fig. 4b), which also were observed in the x-ray (4, 5) and NMR structures (6, 8) of the classic ActD/DNA complexes. Unlike the almost planar C⋅G base pairs present in the classic ActD/5′-(GC)/(CG)-5′ complexes, the C3⋅G7 and C9⋅G1 base pairs surrounding the phenoxazone chromophore are, however, highly buckled, because of the extrusion of the unpaired T2/A8 residues in the present case. Interestingly, the buckle is not evenly distributed in the C3⋅G7 base pair or the C9⋅G1 base pair, with the C3 or C9 base in each pair playing a major role of buckling and being almost completely unstacked from the phenoxazone chromophore and tilted away from their complementary G7 or G1 base (Fig. 4 a and c). This phenomenon indicates the strong stacking preference for the G7/phenoxazone/G1 bases. Such a result in turn produces two cavities in the C9-A8 and C3-T2 backbone (Fig. 4 a and c). However, these two cavities are not formed in an equal manner; in the C9-A8 backbone, the cavity is caused by the A8 ζ torsional angle change from the gauche− to the close trans (−150°) domain, whereas in the C3-T2 backbone, it is caused by the C3 β and γ torsional angle changes from the trans/gauche+ domains to the gauche+/trans domains, respectively. Such nonsymmetric backbone torsional angle alterations are supported by the 1H-31P heterocor data (20, 21, 24), in which the (n)P-(n)H4′ cross peak was clearly detected for the C9 residue but was completely absent for the C3 residue (see Fig. 10, which is published as supporting information on the PNAS web site). Some sugar puckers also were found to differ from the regular C2′-endo domain to form the backbone cavities; although A8 sugar pucker is in the C2′-endo domain, that of T2 is switched into the O4′-endo domain because of the formation of quinoid-2NH2–T2-O4′ hydrogen bond. Such sugar-pucker change is fully supported by the DQF-COSY data.
Secondly, the looped-out A8 and T2 residues are not disordered but are rigid, with some tertiary interactions involved. Other than the hydrogen bond formation mentioned above for the quinoid-2NH2 and the T2-O4′ (marked by the red dotted line in Fig. 4b), one hydrogen bond also was detected for the A8NH2-G7O2P (1.8 Å, marked by red dotted line in Fig. 4c). Besides, two perpendicular base–base interactions also were observed. One interaction is between the G7H8 and A8(π-ring) and another is between the quinoid-3O and T2(π-ring) (marked by blue dotted lines in Fig. 4b). Although it is not clear whether the perpendicular C⩵O/π-ring interaction is significant, the C-H/π-ring interaction has been calculated to be of equal importance to a regular base–base stacking (25) and has been found to exist in several DNA (26–28) and protein structures (29). Such unusual tertiary interactions can account for the ordered structure of the looped-out A8 and T2 residues and the extra stability of the ActD/TA complexes compared with the corresponding classic ActD/DNA complexes.
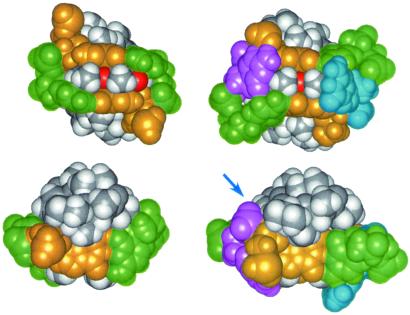
Thirdly, the two methyl groups attached to the phenoxazone ring (4-CH3 and 6-CH3) exhibit good contact with the surrounding A8-G7/C3-T2-G1/C9 bases (Fig. 4d).
Also, we have used similar strategies to determine the ActD/GC complex structure (Fig. 4 e and f). Basically, the ActD/GC complex structure is very similar to the ActD/TA complex structure; the amino group of the looped-out C8 residue forms a hydrogen bond with the G7 backbone phosphate oxygen, and the looped G2 base forms a perpendicular interaction with the ActD chromophore 4CH3 group.
The intermolecular distance constraints applied and the distances measured from the determined ActD/TA and ActD/GC complex structures are listed in Tables 4 and 5 (which are published as supporting information on the PNAS web site), respectively.
Structural Comparison Between the ActD/5′-(GGC)/(CCG)-5′ Complex and the Classic ActD/5′-(GC)/(CG)-5′ Complex.
The almost exclusive formation of the ActD/5′-(G1G2C3)/(C9C8G7)-5′ complex with disrupted G2⋅C8 base pair and excellent G1/phenoxazone/G7 stacking is intriguing to note, because it is also possible for the phenoxazone chromophore of ActD to insert between the G2⋅C8 and C3⋅G7 base pairs in the 5′-(G1G2C3)/(C9C8G7)-5′ sequence to form a classic ActD/5′-(G2C3)/(C8G7)-5′ complex with intact G2⋅C8 base pair. However, from the detailed NMR studies described in this manuscript, it is unambiguously determined that the ActD chromophore does push away the central G2⋅C8 base pair to stack with the nonadjacent G1⋅C9 and C3⋅G7 base pairs. Because such a structural formation with disrupted G2⋅C8 base pair is even more stable than the classic structure with intact G2⋅C8 base pair (as revealed by the larger melting temperature increase and the stronger binding constant of the ActD/GC complex), we have, therefore, solved the ActD/5′-(G1G2C3)/(C9C8G7)-5′ complex structure and compared it with the classic ActD/5′-(GC)/(CG)-5′ complex structure published (ref. 6; Fig. 5). Interestingly, the looped-out G2/C8 residues in the ActD/GC complex do not change the G1/phenoxazone/G7 stacking core and the H bonding between the DNA guanosines and the ActD-L-Thr residues. But they do cause the C3 and C9 bases (shown in green in Fig. 5 Upper Right) to tilt away from the ActD phenoxazone plane, opposite to the direction in the ActD/5′-(GC)/(CG)-5′ complex structure (Fig. 5 Left). The two looped-out perpendicular G2/C8 bases do interact well with the ActD phenoxazone 6CH3 and 4CH3 groups (Fig. 5 Upper Right) and also participate in perpendicular interaction with the G1/phenoxazone/G7 stacking core (Fig. 5 Upper Right) in the major groove. In the minor groove, some extra hydrophobic interaction between the C8H5′/H5" protons with the L-ThrBCH in the benzenoid side and the L-ProQHγ and Hδ protons in the quinoid side are also observed (marked by a blue arrow in Fig. 5 Lower Right). Furthermore, two H bonds between the C8NH2-G7PO5′ atoms and the phenoxazone-2NH2-G2O4′ atoms are also observed (Fig. 4 e and f). Altogether, these factors may explain why the melting temperature of the ActD/GC complex is ≈10°C higher than that of the classic ActD/5′-(GC)/(CG)-5′ complex, and the binding constant is stronger by roughly an order of magnitude.
in the benzenoid side and the L-ProQHγ and Hδ protons in the quinoid side are also observed (marked by a blue arrow in Fig. 5 Lower Right). Furthermore, two H bonds between the C8NH2-G7PO5′ atoms and the phenoxazone-2NH2-G2O4′ atoms are also observed (Fig. 4 e and f). Altogether, these factors may explain why the melting temperature of the ActD/GC complex is ≈10°C higher than that of the classic ActD/5′-(GC)/(CG)-5′ complex, and the binding constant is stronger by roughly an order of magnitude.
Figure 5.
Space-filling structures of the classic ActD/5′-(GC)/(CG)-5′ complex (Left) and the ActD/5′-(GGC)/(CCG)-5′ (Right) complex. The color scheme used is the same as that in Fig. 4. The coordinates for the ActD/5′-(GC)/(CG)-5′ complex are taken from Liu et al. (6). The views into the minor groove are shown in Top, whereas the down-to-the-helical views are shown in Bottom. The extra hydrophobic contact region between the ActD and DNA in the minor groove is indicated by a blue arrow.
Discussion
The Watson–Crick base pair in a double helical DNA can undergo distortion when bound by a protein (30–32), by a drug (33, 34), or when one of the paired bases is modified by a carcinogen (35) to result in a completely looped-out base (30–32), with the flanking base pairing modified to a Hoogsteen style (33, 34) or the carcinogen replacing the disrupted base pair for stacking (35). The present report describes another distorted DNA motif that contains a disrupted looped-out Watson–Crick base pair displaced by the chromophore of a potent chemotherapeutic drug containing cyclic pentapeptide rings. The looped-out bases are situated perpendicularly to the DNA bases/chromophore, akin to what also have been observed in the structures of DNA oligomers modified by carcinogens (35). An x-ray crystal structure of ActD bound to a slipped DNA duplex was recently reported to contain two looped-out nucleotides in one strand, with one of the looped-out base perpendicular to the ActD chromophore (22).
Although ActD has been found to bind at DNA sequences other than the classic 5′-GC/CG-5′ site (2, 11–13), the character of such a binding is still not well understood. In contrast, we have unambiguously characterized the strong binding of ActD with the 5′-GXC/CYG-5′ triplets in the present article. The exclusive looping out of the central G⋅C base pair rather than the intercalation into the 5′-GC/CG-5′ steps in the 5′-GGCACCGCC-3′ sequence by the ActD chromophore is also intriguing to note. These results indicate that ActD binds with stronger affinity toward the 5′-GGC/CCG-5′ recognition site than toward the classic 5′-GC/CG-5′ site. Similar specific binding to the 5′-GTC/CAG-5′, 5′-GAC/CTG-5′, and 5′-GCC/CGG-5′ triplets also were recognized. These results suggest that 5′-GXC/CYG-5′ motifs can serve as the preferred DNA-binding site at certain sequence contexts for this extensively studied anticancer drug.
Although only one major set of proton signals was detected for the ActD/TA (Fig. 2A), ActD/GC (Fig. 2B), and ActD/CG (data not shown) complexes, two major sets of signals were detected for the ActD/AT (data not shown) complex. These two forms most likely stem from the different insertion orientation of the unsymmetrical ActD phenoxazone chromophore. It is not clear why only one major form is present in the ActD/GC and ActD/TA complexes, but their simpler NMR spectra allow us to determine their structures with confidence.
Because the ActD binding to the GXC/CYG site requires the disruption of the central X⋅Y base pair, a worthwhile future inquiry may be to investigate whether this drug also can bind to a site that is imbedded in the interior of a stable duplex DNA.
Supplementary Material
Acknowledgments
We thank the National Science Council and the Chung-Zhen Agricultural Foundation Society of Taiwan, Republic of China for the instrumentation grants. This work was supported by National Science Council Grants 90–2113-M-005–027 (to S.-H.C.) and a subproject of National Institutes of Health-Minority Biomedical Research Support Program S06GM0892 (to F.-M.C.). S.-H.C. is a recipient of the Outstanding Research Award from the National Science Council.
Abbreviations
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- DQF-COSY
double-quantum filtered correlated spectroscopy
- TOCSY
total correlation spectroscopy
- ActD
actinomycin D
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1L1V).
References
- 1.Yang X-L, Wang A H-J. Pharmacol Ther. 1999;83:181–215. doi: 10.1016/s0163-7258(99)00020-0. [DOI] [PubMed] [Google Scholar]
- 2.Snyder J G, Hartman N G, D'Estantott B L, Kennard O, Remeta D P, Breslauer K J. Proc Natl Acad Sci USA. 1989;86:3968–3972. doi: 10.1073/pnas.86.11.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F-M. Biochemistry. 1988;27:6393–6397. doi: 10.1021/bi00417a030. [DOI] [PubMed] [Google Scholar]
- 4.Kamitori S, Takusagawa F. J Mol Biol. 1992;225:445–456. doi: 10.1016/0022-2836(92)90931-9. [DOI] [PubMed] [Google Scholar]
- 5.Kamitori S, Takusagawa F. J Am Chem Soc. 1994;116:4154–4165. [Google Scholar]
- 6.Liu X, Chen H, Patel D J. J Biomol NMR. 1991;1:323–347. doi: 10.1007/BF02192858. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Liu X, Patel D J. J Mol Biol. 1996;258:457–479. doi: 10.1006/jmbi.1996.0262. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Chen F-M. Biochemistry. 1996;35:16346–16353. doi: 10.1021/bi961060d. [DOI] [PubMed] [Google Scholar]
- 9.Chen F-M. Biochemistry. 1998;37:3955–3964. doi: 10.1021/bi972110x. [DOI] [PubMed] [Google Scholar]
- 10.Lian C, Robinson H, Wang A H-J. J Am Chem Soc. 1996;118:8791–8801. [Google Scholar]
- 11.Bailey S A, Graves D E, Rill R. Biochemistry. 1994;33:11493–11500. doi: 10.1021/bi00204a011. [DOI] [PubMed] [Google Scholar]
- 12.Wadkins R M, Vladu B, Tung C-S. Biochemistry. 1998;37:11915–11923. doi: 10.1021/bi9809730. [DOI] [PubMed] [Google Scholar]
- 13.Chen F-M, Sha F. Biochemistry. 2001;40:5218–5225. doi: 10.1021/bi010047c. [DOI] [PubMed] [Google Scholar]
- 14.Chou S-H, Tseng Y-Y, Wang S-W. J Mol Biol. 1999;287:301–313. doi: 10.1006/jmbi.1999.2564. [DOI] [PubMed] [Google Scholar]
- 15.Chou S-H, Tseng Y-Y. J Mol Biol. 1999;285:41–48. doi: 10.1006/jmbi.1998.2318. [DOI] [PubMed] [Google Scholar]
- 16.Plateau P, Gueron M. J Amer Chem Soc. 1982;104:7310–7311. [Google Scholar]
- 17.Sklenar V, Miyashiro H, Zon G, Miles H T, Bax A. FEBS Lett. 1986;208:94–98. doi: 10.1016/0014-5793(86)81539-3. [DOI] [PubMed] [Google Scholar]
- 18.Chou S-H, Cheng J-W, Fedoroff O, Reid B R. J Mol Biol. 1994;241:467–479. doi: 10.1006/jmbi.1994.1521. [DOI] [PubMed] [Google Scholar]
- 19.Chou S-H, Zhu L, Gao Z, Cheng J-W, Reid B R. J Mol Biol. 1996;264:981–1001. doi: 10.1006/jmbi.1996.0691. [DOI] [PubMed] [Google Scholar]
- 20.Sarma R H, Mynott R J, Wood D J, Hruska F E. J Am Chem Soc. 1973;95:6457–6459. doi: 10.1021/ja00800a055. [DOI] [PubMed] [Google Scholar]
- 21.Altona C. Recl Trav Chim Pays-Bas. 1982;101:413–433. [Google Scholar]
- 22.Robinson H, Gao Y-G, Yang X-L, Sanishvili R, Joachimiak A, Wang A H-J. Biochemistry. 2001;40:5587–5592. doi: 10.1021/bi002859z. [DOI] [PubMed] [Google Scholar]
- 23.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 24.Chou S-H, Zhu L, Reid B R. J Mol Biol. 1997;267:1055–1067. doi: 10.1006/jmbi.1997.0914. [DOI] [PubMed] [Google Scholar]
- 25.Sponer J, Gabb H A, Leszczynski J, Hobza P. Biophys J. 1997;73:76–87. doi: 10.1016/S0006-3495(97)78049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou S-H, Zhu L, Reid B R. J Mol Biol. 1994;244:259–268. doi: 10.1006/jmbi.1994.1727. [DOI] [PubMed] [Google Scholar]
- 27.Chou S-H, Chin K-H. J Mol Biol. 2001;312:753–768. doi: 10.1006/jmbi.2001.4962. [DOI] [PubMed] [Google Scholar]
- 28.Chou S-H, Chin K-H. J Mol Biol. 2001;314:139–152. doi: 10.1006/jmbi.2001.5131. [DOI] [PubMed] [Google Scholar]
- 29.Umezawa Y, Nishio M. Bioorg Med Chem. 2000;8:2643–2650. doi: 10.1016/s0968-0896(00)00197-8. [DOI] [PubMed] [Google Scholar]
- 30.Klimasauskas S, Kumar S, Roberts R J, Cheng X. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 31.Reinisch K M, Chen L, Verdine G L, Lipscomb W N. Cell. 1995;82:143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 32.Goedecke K, Pignot M, Goody R S, Scheidig A J, Weinhold E. Nat Struct Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 33.Quigley G J, Ughetto G, van der Marel G A, van Boom J H, Wang A H-J, Rich A. Science. 1986;279:1255–1258. doi: 10.1126/science.3704650. [DOI] [PubMed] [Google Scholar]
- 34.Addess K J, Sinsheimer J S, Feigon J. Biochemistry. 1993;32:2498–2508. doi: 10.1021/bi00061a006. [DOI] [PubMed] [Google Scholar]
- 35.Geacintov N E, Cosman M, Hingerty B E, Amin S, Boroyde S, Patel D J. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.