Abstract
The yeast chorismate mutase is regulated by tyrosine as feedback inhibitor and tryptophan as crosspathway activator. The monomer consists of a catalytic and a regulatory domain covalently linked by the loop L220s (212–226), which functions as a molecular hinge. Two monomers form the active dimeric enzyme stabilized by hydrophobic interactions in the vicinity of loop L220s. The role of loop L220s and its environment for enzyme regulation, dimerization, and stability was analyzed. Substitution of yeast loop L220s in place of the homologous loop from the corresponding and similarly regulated Aspergillus enzyme (and the reverse substitution) changed tyrosine inhibition to activation. Yeast loop L220s substituted into the Aspergillus enzyme resulted in a tryptophan-inhibitable enzyme. Monomeric yeast chorismate mutases could be generated by substituting two hydrophobic residues in and near the hinge region. The resulting Thr-212→Asp–Phe-28→Asp enzyme was as stable as wild type, but lost allosteric regulation and showed reduced catalytic activity. These results underline the crucial role of this molecular hinge for inhibition, activation, quaternary structure, and stability of yeast chorismate mutase.
Chorismate mutases (CMs) catalyze the Claisen rearrangement from chorismate to prephenate in the biosynthesis of tyrosine and phenylalanine (1). The eukaryotic CMs from Saccharomyces cerevisiae, Aspergillus nidulans, and Hansenula polymorpha exhibit a very similar pattern of regulation of enzyme activity (2). They are feedback inhibited by tyrosine, one of the end products of the biosynthetic branch, and activated by tryptophan, the end product of the parallel branch (3).
X-ray structure analysis revealed that the yeast homodimeric CM contains no β-sheet structures, but 71% of the protein is formed by α-helices (4). Two allosteric and two active sites occur per CM dimer. The allosteric ligands bind to a cleft in the dimer interface formed by H4 and H5 from one monomer and H8 and loop L130s from the other monomer. A twisted four-helix bundle structure is formed by helices H2, H8, H11, and H12 of each subunit at the end of which the active site is located.
Helices H2, H4, and H11 and loops L50s and L80s contribute to hydrophobic clusters that account for dimerization (4). In one cluster residues Phe-28, Ile-31, Val-211, and Tyr-212 from helices H2 and H11, respectively, from both subunits are packed against each other. Residue Leu-67 from helix H4 also forms a hydrophobic patch with its counterpart from the other monomer. In addition, residues from loop L50s and helix H4 from one subunit interact with loop L80s from the other subunit, leading to formation of another two hydrophobic cores. These interactions were so strong that dissociation of the dimers occurred only after addition of 4 M guanidine hydrochloride upon unfolding of the monomers (5).
Strong similarities on the primary structural level indicate conserved characteristics of the yeast enzyme with CMs from other fungi as A. nidulans (AnCM) and H. polymorpha (HpCM). Except for Asn-194 of the yeast's active site, which is equivalent to Asp-200 in AnCM, and Thr-145 of the yeast's allosteric site, which can be aligned to Met-143 in HpCM, respectively, all residues important for activity or binding of allosteric effectors are conserved among these three enzymes.
Loop L220s serves as a hinge that connects the allosteric domain with the catalytic domain within one polypeptide in the dimer interface. A constitutively activated yeast CM is known in which the last residue of this particular loop (Thr-226) is substituted by an isoleucine residue. This residue locks the enzyme structure in the allosteric R state, which completely prevents inhibition or further activation (6). The end of this loop, however, is not as strongly conserved among these homologous CMs. Sequence alignments show that the Thr residue is replaced by an aspartic acid residue in AnCM and a Lys residue in HpCM, which suggests somewhat different signaling pathways at least in this part of the homologous CMs from A. nidulans and H. polymorpha, respectively. Here, we report the construction and kinetic characterization of chimeric enzymes in which the whole loops homologous to L220s were exchanged to gain more insight in structure and function of this molecular hinge of CM. In addition, we present the generation of a stable yeast CM monomer by substituting amino acid residues in and around L220s to elucidate the process of dimerization and the characteristics of a single CM polypeptide.
Materials and Methods
Yeast Strains, Plasmids, Media, and Growth Conditions.
For overexpression of chimeric CMs, a derivative of plasmid p426MET25 (7) was used in S. cerevisiae strain RH2192 (MATα, pra1–1, prb1–1, prc1–1, cps1–3, ura3Δ5, leu2–3, 112, his−, aro7Δ∷LEU2), which is a derivative of the protease-deficient strain c13-ABYS-86 (8). For overexpression of wild-type (wt), constitutively activated, and monomeric CMs, derivatives of plasmid pME781 (9) were used in strain RH1242 (MATa, aro7, leu2–2). A PCR-based method with Pfu DNA polymerase (Promega) was applied for site-directed mutagenesis of ARO7 (10). The 5′- or 3′-terminal portions of the ARO7 gene in plasmid pME1459 were replaced by a PCR-generated NdeI/XbaI or XbaI/BamHI fragment, respectively, which encoded for one substitution each. Both mutated fragments were then combined in one plasmid to generate a double-mutant ARO7 gene. Chimeric CM genes were constructed by overlap extension by using PCR (11). The PCR-generated fragments were sequenced to confirm the presence of the substitutions and mutations and rule out second-site mutations (12). Strain RH1671 (MATa, ura3–251, ura3–328, ura3–375, aro7Δ∷URA3) was used for integration of linear wt and monomeric ARO7 fragments. Yeast transformation and cultivation were described (13, 14). Growth rates of strains RH1671, RH2698 (MATa, ura3–251, ura3–328, ura3–375), and RH2699 (MATa, ura3–251, ura3–328, ura3–375 aro7m) were determined as described (3).
Purification of CMs.
Plasmid-carrying yeast cells were cultivated and harvested as described (15). Enzymes derived from yeast CM were purified as described (16). CMs derived from the A. nidulans enzyme were purified according to Krappmann et al. (15).
Enyzme Assays and Data Evaluation.
For substrate saturation curves, CM activity was measured as described (15). Effector concentrations were 100 μM tyrosine and 10 μM tryptophan for enzymes derived from ScCM and 50 μM tyrosine and 5 μM tryptophan for enzymes generated from AnCM, respectively. kcat values were calculated by using the respective molecular weight of the enzymes as calculated by the LASERGENE Biocomputing software from DNAStar (Madison, WI). pH optima were determined as described (17) with 4 μg of purified enzyme and 2 mM chorismate concentration. Rate constants for thermal inactivation were determined with 1 μg of purified enzyme as described (18).
Determination of the Native Molecular Weight.
The native molecular weight of the double-mutant CM was determined by gel filtration on a Superdex 200-pg column with 50 mM potassium phosphate/150 mM NaCl, pH 7.6, as elution buffer (15). The void volume of the column was determined with blue dextran, and a calibration plot was defined by using thyroglobulin, bovine γ-globulin, chicken ovalbumin, equine myoglobin, and vitamin B12. In addition, the molecular weight was estimated by native PAGE by using a gradient from 10–20% polyacrylamide and chicken egg albumin, α-lactalbumin, carbonic anhydrase, BSA, and urease as standard (19).
Circular Dichroism Spectroscopy.
The CD spectra of the wt and mutant CMs were measured in the range of 200–250 nm with a Jobin Yvon CD6 spectrometer at 0.5-nm resolution. The path length of the cell was 0.1 cm. The spectra were recorded as an average of three scans at 20°C with 1.33 μM monomeric CM in 10 mM Tris⋅HCl, pH 7.6. Buffer baseline spectra were subtracted from the protein spectra.
Sequence Alignment and Homology Modeling Studies.
All sequence analyses were performed by using the LASERGENE Biocomputing software. Alignments were created based on the CLUSTAL W method (20) by using the Network Protein Sequence analysis service (21). For homology modeling, the deduced primary structure of the A. nidulans and H. polymorpha CMs were aligned to the crystallographic data of yeast CMs as described in the Brookhaven protein database (22) and refined by the SWISS-MODEL service (23, 24). By using the WEBLAB VIEWER software (Molecular Simulations, San Diego, CA), three-dimensional structure models could be generated by calculation of secondary structures.
Results
Fungal CMs with an Altered Hinge Connecting the Catalytic and the Regulatory Domain.
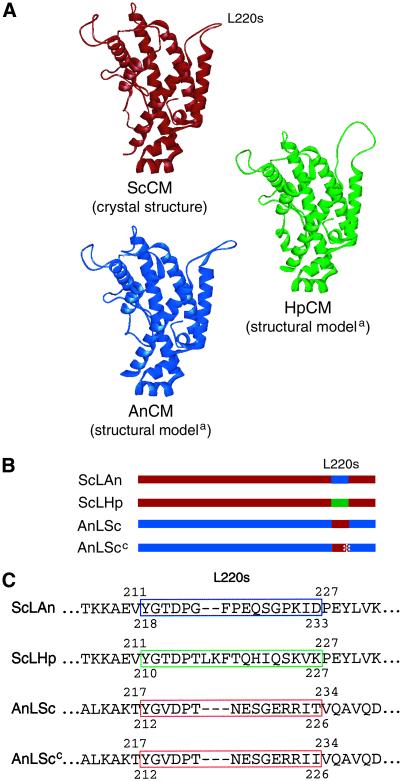
The crystal structure of yeast CM determined in complex with different ligands is an all-helical dimer composed of 256 amino acids forming 12 helices per monomer (22; Fig. 1A). The high similarities between the deduced primary structures of CMs from S. cerevisiae, A. nidulans, and H. polymorpha suggest homology of the corresponding genes. Alignments of the protein sequences show more than 50% identity between all three enzymes with long conserved stretches especially in helices 2, 8, 11, and 12. Information on the three-dimensional structures of AnCM and HpCM was gained by homology modeling based on crystal structures of the yeast enzyme by using SWISS-MODEL (refs. 23–25; Fig. 1A). The residues that in the primary structures are homologous to residues of loop L220s of ScCM also formed corresponding loops in these structures. Therefore, the corresponding regions of the encoding genes were exchanged (Fig. 1 B and C). The flexible L220s loop from the wt and the Thr-226→Ile loop of the mutant enzyme frozen in the activated state were introduced into the Aspergillus enzyme leading to chimeras AnLSc and AnLScc, and the corresponding loops from HpCM and AnCM were incorporated into the yeast enzyme (ScLHp and ScLAn). The ScLHp chimeric enzyme was highly unstable during purification, which suggests that this hinge region might be critical for stability and has to be well aligned to the rest of the molecule. This instability was surprising because the loop is positioned on the surface of the protein and the Hansenula loop has a similar size (18 aa) in comparison with the S. cerevisiae loop (15 aa). Thus, further analysis was only performed for the ScCM–AnCM chimeric enzymes that were stable and active.
Figure 1.
Structural models of wt and chimeric CM monomers. (A) The detailed structure of yeast CM (ScCM) was determined by x-ray crystallography; the model of the three-dimensional structures of the Hansenula (HpCM) and Aspergillus (AnCM) enzymes were created by using the SWISS-MODEL service(a). (B) Schematic drawings of the chimeras. Parts of the yeast enzyme are shown in red, parts of the Hansenula enzyme are shown in green, and fragments derived from the Aspergillus enzyme are shown in blue. The asterisk marks the Thr-226→Ile mutation in the loop from the constitutively activated yeast enzyme. (C) Sequences of the loop L220s regions of the chimeras. The amino acid sequences are presented as they were aligned with the newly introduced loop sequences shown in place of the wt sequences. The loop from the yeast enzyme is marked with a red box, that from the Hansenula enzyme with a green box and from the Aspergillus enzyme with a blue one. The numbers indicate the amino acid position in the respective wt protein.
Tyrosine Becomes a Yeast Enzyme Activator Instead of an Inhibitor When the L220s Hinge (212–226) Is Replaced by the Loop from Aspergillus CM.
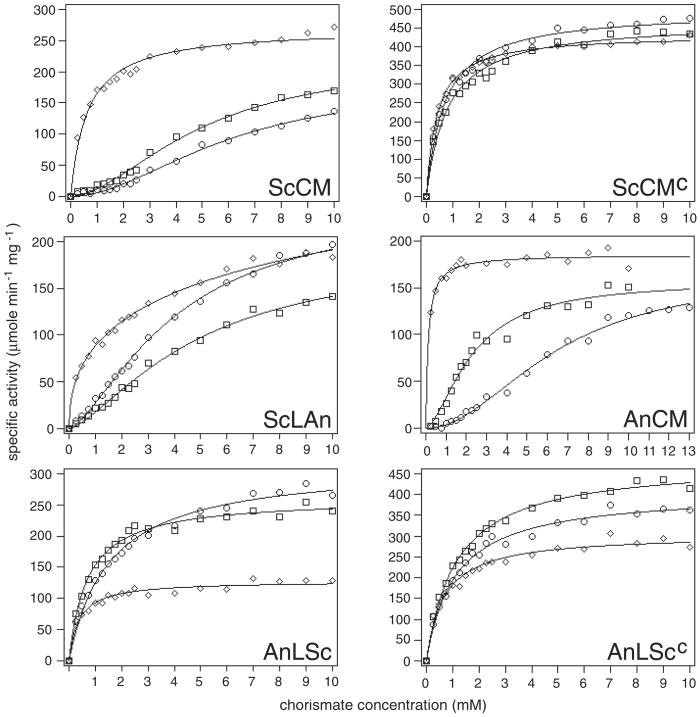
Substrate saturation curves were measured to determine the catalytic parameters (Fig. 2; Table 1). The sigmoid saturation curves of the wt yeast and Aspergillus CM were used as references. These curves are depressed under inhibition conditions and show Michaelis–Menten-like kinetics under activation conditions. Substrate affinity is reduced by tyrosine and increased by tryptophan. The Thr-226→Ile mutant yeast enzyme did not respond to the allosteric effectors. This variant showed all hyperbolic curves with Hill coefficients and Km values smaller than 1 with overall high activity.
Figure 2.
Substrate-saturation plots of wt and chimeric CMs. The enzymes were assayed in the presence of 100 μM tyrosine for the yeast enzyme and 50 μM tyrosine for the Aspergillus enzyme, respectively (○), or 10 μM tryptophan for the yeast enzyme and 5 μM tryptophan for the Aspergillus enzyme, respectively (⋄), or were assayed unliganded (□). The concentrations used for the chimeric enzymes depended on the origin of the protein's major part. Each point was measured at least five times and the collected data were fitted to functions describing either cooperative or Michaelis–Menten-type saturation.
Table 1.
Kinetic parameters of wt and chimeric CMs
| Protein | Tyrosine-liganded*
|
Unliganded
|
Tryptophan-liganded†
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat, s−1 | Km, S0.5, mM | nH‡ | kcat/Km, s−1⋅mM−1 | kcat, s−1 | Km, S0.5, mM | nH‡ | kcat/Km, s−1⋅mM−1 | kcat, s−1 | Km, S0.5, mM | nH‡ | kcat/Km, s−1⋅mM−1 | |
| ScCM | 91 | 6 | 2.0 | 15 | 110 | 5 | 1.9 | 22 | 134 | 0.6 | 0.8 | 210 |
| ScCMc | 246 | 0.7 | 0.8 | 357 | 232 | 0.8 | 0.8 | 289 | 214 | 0.4 | 0.9 | 530 |
| ScLAn | 139 | 5.5 | 1.3 | 27 | 96 | 4.9 | 1.4 | 19 | 217 | 16.5 | 0.5 | 13 |
| AnCM | 84 | 6.4 | 2.0 | 13 | 80 | 2.3 | 2.0 | 35 | 95 | 0.1 | 1.0 | 758 |
| AnLSc | 161 | 1.5 | 1.0 | 105 | 133 | 0.7 | 1.0 | 190 | 65 | 0.4 | 1.0 | 176 |
| AnLScc | 207 | 1.1 | 1.1 | 189 | 244 | 1.2 | 1.0 | 213 | 155 | 0.7 | 0.8 | 233 |
Tyrosine (100 μM) for the yeast-derived enzymes and 50 μM tyrosine for the Aspergillus-derived enzymes, respectively.
Tryptophan (10 μM) for the yeast-derived enzymes and 5 μM tryptophan for the Aspergillus-derived enzymes, respectively.
Determined as the slope of the Hill plot in the region of v = 0.5Vmax.
For the chimeric enzymes, substrate saturation curves were different. Introduction of the Aspergillus loop into the yeast enzyme led to an enzyme that had dramatically changed characteristics under activation conditions. ScLAn displayed a highly elevated Km value of 16.5 mM for the activated enzyme in comparison with 0.6 mM of the yeast wt enzyme. Under this condition, the Hill coefficient was well below 1 indicating negative cooperativity of substrate binding, and the catalytic efficiency was reduced from 210 s−1 mM−1 in ScCM to 13 s−1 mM−1 in ScLAn. Tyrosine acted as activator instead of inhibitor under all chorismate concentrations in the substrate saturation assay. In the presence of this effector the catalytic efficiency was elevated because kcat was increased whereas Km was near the wt value.
Aspergillus CM with the Yeast Hinge Between Catalytic and Regulatory Region Is Inhibited by Tryptophan.
A different situation was found for the Aspergillus-derived chimeric enzymes (Fig. 2; Table 1). For the tyrosine-bound and unliganded enzymes a decrease in Km was accompanied by a loss of cooperativity; kcat was increased compared with wt AnCM so that catalytic efficiency was strongly improved. For AnLSc, tyrosine proved to activate this enzyme at chorismate concentrations greater than 3.5 mM and only slightly inhibited the enzyme below 3.5 mM chorismate. This effector still led to enzyme inhibition for AnLScc. In the presence of tryptophan, however, the opposite occurred. For both chimeras, tryptophan acted as inhibitor of enzyme activity. The curve of the tryptophan-liganded chimeras was always lower than the curves of the unliganded chimeras in the substrate saturation plot. In the presence of tryptophan, cooperativity was lost as in the wt, and Km was slightly increased. kcat was reduced to 49% for AnLSc and to 64% for AnLScc compared with the unliganded enzyme. The catalytic efficiency dropped to 23 and 31%, respectively, compared with the activated wt AnCM. Similar to the yeast enzyme, the loop L220s-Thr-226→Ile prevented a stronger regulation like it was found for the chimera with the wt L220s loop.
Two Amino Acid Substitutions in the Hydrophobic Cluster Including the L220s Hinge Are Sufficient for Monomerization of Yeast CM.
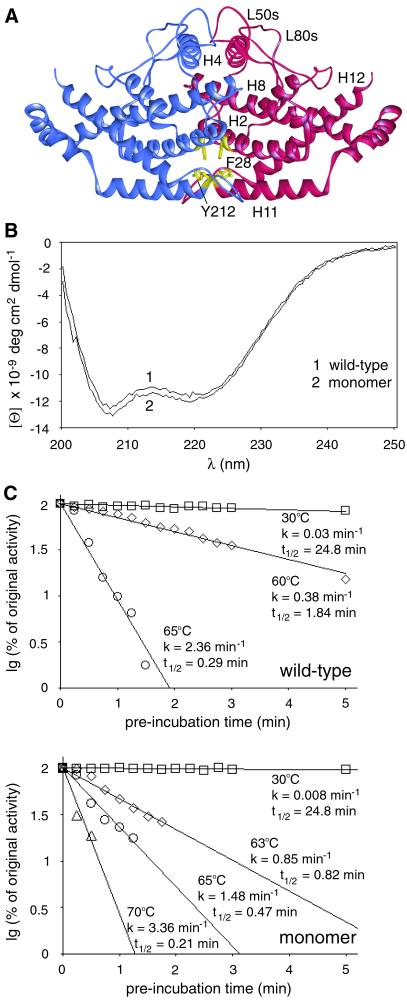
The instability of the ScLHp chimera proved the importance of an appropriate hinge region for enzyme stability. Furthermore, the C terminus of L220s is part of a dimerization domain in which hydrophobic amino acid residues establish contact between the two monomers, which prompted us to examine whether dimerization is essential for protein stability or whether an intact hinge mediates stable protein folding of a single polypeptide chain. Therefore, hydrophobic amino acid residues at positions 28 and 212 were substituted by neutral (alanine) and charged amino acid residues (aspartic acid), respectively (Fig. 3A). Single substitutions with either alanine or aspartic acid residues yielded enzymes that were impaired in inhibition and activation, but no clear results were obtained as far as their quaternary structures were concerned (data not shown). However, Tyr-212–Asp and Phe-28–Asp replacements in combination (aro7m) were sufficient to prevent any hydrophobic interactions between CM proteins. The purified double-mutant enzyme was electrophoresed in a native gradient polyacrylamide gel, which allowed the estimation of a molecular mass of approximately 30 kDa (data not shown). The determination of the native molecular weight by gel filtration analysis on a calibrated Superdex 200-pg column yielded a Kd of 0.75 as calculated from the protein's elution volume (data not shown). For global proteins this Kd value corresponds to a molecular mass of 25,449 Da. Compared with the molecular mass of one CM polypeptide of 29,746 Da, as calculated from the DNA sequence, this result confirms the monomeric state of the mutant enzyme variant.
Figure 3.
Structural characteristics and thermal stability of wt and monomeric CMs. (A) Wt ScCM with Phe-28 and Tyr-212 of the dimerization region labeled yellow. (B) CD spectra of monomeric mutant and dimeric wt CM. Each spectrum of the purified enzymes is an average of three independently obtained spectra, which were recorded in 10 mM Tris⋅HCl buffer, pH 7.6. (C) Determination of thermal inactivation. Rate constants for thermal inactivation and half-lives of the two CMs were determined in stop assays at 30°C with 1 μg of purified enzyme, 2 mM chorismate concentration, and 2 min of catalytic turnover. One hundred percent of original activity equaled 3.2 units per mg for monomeric and 28.2 units per mg for wt CM, respectively. Each value is an average of four independent measurements.
Monomeric CM Enables Growth of Yeast Cells in Media Lacking Aromatic Amino Acids.
The aro7m gene encoding the monomeric CM was integrated at the ARO7 locus of S. cerevisiae strain RH1671 by selection against a URA3 marker (RH2699). The wt ARO7 gene was reintegrated into the same strain to construct a control strain (RH2698). Growth rates were determined in minimal vitamins medium containing uracil or uracil plus the aromatic amino acids for comparison with the aro7Δ strain RH1671. The growth rates of strain RH2699 (aro7m) equaled 0.21 h−1 in both media. Reintegration of ARO7 did not improve the growth rate (0.24 h−1 and 0.25 h−1, respectively), whereas the aro7Δ strain did not grow on MV+ura, but best on the medium containing aromatic amino acids (0.28 h−1). Thus, compared with the strain containing the reintegrated ARO7 gene, no growth defect under nonstarvation conditions was observed for strain RH2699 (aro7m). A single-copy chromosomal aro7m gene was sufficient to restore prototrophy for tyrosine and phenylalanine.
Circular dichroism curves were determined for both the monomeric and dimeric CM (Fig. 3B). As expected, the spectrum of the wt enzyme showed a double minimum at 207 and 219 nm, which matches the minima of a typical all-helical protein. The spectrum recorded for the monomer also had two minima, and even the intensities of the minima were nearly identical. This result showed that a single CM polypeptide folds into an ordered tertiary conformation and is still all-helical. Furthermore, the monomer proved very stable in activity assays. The enzymes were preincubated at different temperatures for increasing periods of time and chilled on ice, and the residual activity was determined in stop assays at 30°C (Fig. 3C).
At temperatures above 60°C, activity decreased very rapidly. The rate constants of thermal inactivation at 65°C were 2.36 min−1 for the dimer and 1.48 min−1 for the monomer, respectively, with half-lives of 0.29 min and 0.47 min, respectively. These experiments show that the monomeric CM is nearly as stable at 70°C as the wt enzyme at 65°C. Thus, monomerization did not influence the protein's stability.
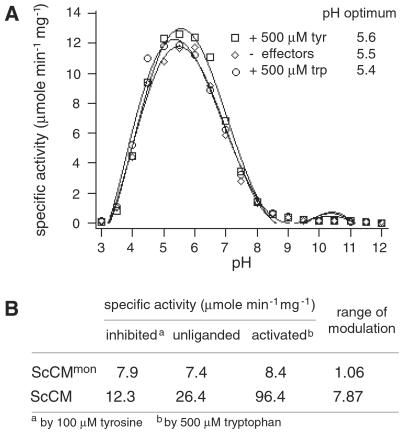
In a modulated stop assay the specific activity could be determined in dependence of the pH (Fig. 4A). The monomer has a clear optimum at pH 5.5 that is not altered by tyrosine or tryptophan. This value corresponds to that of the wt enzyme as determined (17), which shows that, although activity is strongly reduced, the obvious structural changes in the active site do not influence the pH dependence of the catalyzed reaction.
Figure 4.
Specific activity of monomeric CM. (A) pH optima for monomeric CM. Modified stop assays were performed with 4 μg of purified enzyme, 2 mM chorismate concentration, and 10 min of catalytic turnover. Each value is an average of four independent measurements. (B) Specific activities under different effector conditions. Enzyme activities were determined in a stop assay with 1 μg of purified enzymes, 2 mM chorismate concentration, and 3 min of catalytic turnover. Each value is an average of four independent measurements.
Regulation of Enzyme Activity Is Lost for Monomeric CM.
The monomeric CM has lost the oligomeric state of the protein and has therefore lost what is assumed to be the main prerequisite for allosteric regulation. Because the allosteric sites were formed by residues contributed from both polypeptides in the dimer interface, even the ability to bind the allosteric effectors is expected to be lost. A stop assay revealed that the range of modulation of enzyme activity under different effector conditions equaled a factor of nearly 1, whereas the wt enzyme's activity could be modulated by a factor of about 8 (Fig. 4B). Thus, essentially no regulation could be observed for the monomeric enzyme. In addition, the specific activity of the monomer was dramatically reduced. Activity dropped down to 30% compared with the unliganded wt enzyme. In fact, activity was so low that a substrate saturation plot could not be measured because standard deviations exceeded the differences of the measured activities. Thus, this monomeric yeast CM is as stable as the wt dimer, but has lost allosteric regulation. The catalytic efficiency was strongly reduced, but sufficient to complement an aro7 deletion under nonstarvation conditions and exhibited the same pH optimum. Indeed, a different unregulated CM was unsuitable for amino acid starvation conditions (3).
Discussion
The eukaryotic yeast (ScCM) and the bacterial Escherichia coli (EcCM) CMs seem to have one evolutionary origin (22). EcCM is a dimer composed of three helices per subunit that resembles a yeast CM monomer. It gains stability by the formation of four-helix bundles composed of helices H1, H2, and H3 of one subunit and H1′ of the other subunit, which also form the active-site cavities. Computer-modeling studies revealed that helices H8, H11, H12, and H2 of ScCM correspond to these helices of EcCM, whereas H4 and H7 of ScCM can be compared with H2′ and H3′ of the E. coli enzyme indicating an evolutionary gene duplication event (22). For stable folding of the two connected CMs additional helices and loops must have evolved, which contribute to the stability of the somewhat distorted four-helix bundle in the yeast monomer. The results presented here show the stability of this ancestral CM. The monomer generated by two amino acid substitutions can be regarded as an intermediate between a monomeric CM with two active sites and a dimeric CM with two active sites and two additional regulatory domains. The monomer is stable against heat and changes of pH and is still active.
The subsequent step in evolution might have been the generation of hydrophobic cores permitting dimerization. As described above, four regions evolved for dimerization including the formation of an additional four-helix bundle structure composed of H2, H2′, H8, and H8′. The stability of the artificial monomer suggests that dimerization occurs after folding of the single subunits and is not required for the correct and stable folding of the single polypeptides. Thus, for this enzyme system, formation of dimers evolved primarily for the ability to regulate enzyme activity.
After the evolutionary dimerization event, the protein's conformation must have changed to account for possible unfavorable rearrangements around the active sites. Comparing the EcCM and ScCM active sites, Xue et al. (4) found that although important residues were mainly conserved, the yeast enzyme has a more exposed active site than does the bacterial counterpart (4). This exposure is caused by the presence of additional helices (H9 and H10) near the active site, and it leads to a weaker binding affinity for the substrate. Nevertheless, the cooperativity of substrate binding which can be observed for the dimeric yeast enzyme seems to be sufficient for substantial activity. Perhaps, on the other hand, these rearrangements upon monomerization cause reduction of activity by reorientation of crucial residues of the active site. On the other hand, it cannot yet be excluded that the substitutions introduced at positions 28 and 212 account for the change in activity, because single substitutions in loop L220s can strongly affect enzyme activity in some cases (5, 26). However, because the pH optimum of the monomer equaled that of the unliganded wt enzyme, conformational rearrangements cannot be very pronounced. In addition to alterations in the active site, further rearrangements in the loops must have occurred during evolution to gain stability. These structural changes in the loops obviously contribute to stability to a different extent in a particular CM. Although all chimeric CMs investigated were dimeric enzymes, the introduction of the Hansenula loop into the yeast enzyme strongly reduced the protein's stability, which demonstrates not only the yeast L220s loop's function for regulation, but also its contribution to stability.
Finally, to establish an allosteric enzyme, binding sites for allosteric effectors and signaling pathways have evolved, thus destroying two of the active sites to generate two allosteric domains. The resulting binding sites are substantially identical for tyrosine as inhibitor and tryptophan as activator of the yeast wt enzyme. It is now reasonable to ask whether the consequences of effector binding can be changed along the allosteric pathway. In fact, the exchange of the molecular hinge connecting allosteric and catalytic domain presented here showed that loop L220s is one critical part for the transmission of the distinct allosteric signals. Obviously, the difference in size and probably conformation of the loops exchanged caused tertiary structural changes that promoted different rearrangements during signal transduction upon binding of the allosteric effectors. Thus, the signal of tyrosine binding can cause enzyme activation, and tryptophan binding can be a signal for enzyme inhibition. A combination of modified CMs from different organisms leads to a switch of allosteric effects. It would be interesting to identify other residues along the intramolecular signaling pathway, which when substituted also switch between the allosteric signals. Our results presented here prove the hinge region's function for enzyme stability and establish discrimination between allosteric inhibition and activation. Yeast CM is therefore a model enzyme where the detailed understanding of the cooperation of various modules within the molecule allows a specific engineering of a changed regulatory behavior of a protein.
Acknowledgments
We thank Dr. Georg Schnappauf for providing plasmids pME1493 and pME1495, which each contained a gene encoding a CM with a single amino acid substitution. We thank Dr. Nadja Hellmann and Prof. Heinz Decker, Mainz, for help in recording CD spectra, and the members of the laboratory for helpful discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, the Volkswagen-Stiftung, the Niedersächsische Vorab der Volkswagen-Stiftung, and National Institutes of Health Grant GM06920 (to W.N.L.).
Abbreviations
- CM
chorismate mutase
- AnCM
A. nidulans chorismate mutase
- HpCM
H. polymorpha chorismate mutase
- ScCM
S. cerevisiae chorismate mutase
- EcCM
E. coli chorismate mutase
- wt
wild type
References
- 1.Helmstaedt K, Krappmann S, Braus G H. Microbiol Mol Biol Rev. 2001;65:404–421. doi: 10.1128/MMBR.65.3.404-421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacBeath G, Kast P, Hilvert D. Biochemistry. 1998;37:10062–10073. doi: 10.1021/bi980449t. [DOI] [PubMed] [Google Scholar]
- 3.Krappmann S, Lipscomb W N, Braus G H. Proc Natl Acad Sci USA. 2000;97:13585–13590. doi: 10.1073/pnas.240469697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue Y, Lipscomb W N, Graf R, Schnappauf G, Braus G. Proc Natl Acad Sci USA. 1994;91:10814–10818. doi: 10.1073/pnas.91.23.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidheini T, Mösch H U, Evans J N, Braus G. Biochemistry. 1990;29:3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]
- 6.Schmidheini T, Sperisen P, Paravicini G, Hütter R, Braus G. J Bacteriol. 1989;171:1245–1253. doi: 10.1128/jb.171.3.1245-1253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mumberg D, Müller R, Funk M. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinemeyer W, Kleinschmidt J A, Saidowsky J, Escher C, Wolf D H. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnappauf G, Krappmann S, Braus G H. J Biol Chem. 1998;273:17012–17017. doi: 10.1074/jbc.273.27.17012. [DOI] [PubMed] [Google Scholar]
- 10.Giebel L B, Spritz R A. Nucleic Acids Res. 1990;18:4947. doi: 10.1093/nar/18.16.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 12.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito H, Jukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miozzari G, Niederberger P, Hütter R. J Bacteriol. 1978;134:48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krappmann S, Helmstaedt K, Gerstberger T, Eckert S, Hoffmann B, Hoppert M, Schnappauf G, Braus G H. J Biol Chem. 1999;274:22275–22282. doi: 10.1074/jbc.274.32.22275. [DOI] [PubMed] [Google Scholar]
- 16.Schnappauf G, Lipscomb W N, Braus G H. Proc Natl Acad Sci USA. 1998;95:2868–2873. doi: 10.1073/pnas.95.6.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnappauf G, Sträter N, Lipscomb W N, Braus G H. Proc Natl Acad Sci USA. 1997;94:8491–8496. doi: 10.1073/pnas.94.16.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krappmann S, Pries R, Gellissen G, Hiller M, Braus G H. J Bacteriol. 2000;182:4188–4197. doi: 10.1128/jb.182.15.4188-4197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson L O, Borg H, Mikaelsson M. FEBS Lett. 1972;20:199–202. doi: 10.1016/0014-5793(72)80793-2. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Combet C, Blanchet C, Geourjon C, Deléage G. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 22.Sträter N, Schnappauf G, Braus G, Lipscomb W N. Structure (London) 1997;5:1437–1452. doi: 10.1016/s0969-2126(97)00294-3. [DOI] [PubMed] [Google Scholar]
- 23.Peitsch M C. Bio. 1995. /Technology13, 658–660. [Google Scholar]
- 24.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 25.Peitsch M C. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 26.Graf R, Dubaquié Y, Braus G H. J Bacteriol. 1995;177:1645–1648. doi: 10.1128/jb.177.6.1645-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]