Abstract
A G to T mutation has been observed at the third position of codon 249 of the p53 tumor-suppressor gene in over 50% of the hepatocellular carcinoma cases associated with high exposure to aflatoxin B1 (AFB1). Hypotheses have been put forth that AFB1, in concert with hepatitis B virus (HBV), may play a role in the formation of, and/or the selection for, this mutation. The primary DNA adduct of AFB1 is 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 (AFB1-N7-Gua), which is converted naturally to two secondary lesions, an apurinic site and an AFB1-formamidopyrimidine (AFB1-FAPY) adduct. AFB1-FAPY is detected at near maximal levels in rat DNA days to weeks after AFB1 exposure, underscoring its high persistence in vivo. The present study reveals two striking properties of this DNA adduct: (i) AFB1-FAPY was found to cause a G to T mutation frequency in Escherichia coli approximately 6 times higher than that of AFB1-N7-Gua, and (ii) one proposed rotamer of AFB1-FAPY is a block to replication, even when the efficient bypass polymerase MucAB is used by the cell. Taken together, these characteristics make the FAPY adduct the prime candidate for both the genotoxicity of aflatoxin, because mammalian cells also have similar bypass mechanisms for combating DNA damage, and the mutagenicity that ultimately may lead to liver cancer.
Aflatoxin B1 (AFB1), one of the most potent known liver carcinogens, is produced by the common soil fungus Aspergillus flavus. Exposure to this toxin is high in regions of the world where certain foods are improperly stored (1). Hepatitis B virus (HBV) is also common in these regions, and epidemiological evidence indicates that there is a synergistic interaction between AFB1 exposure and HBV infection on the induction of hepatocellular carcinoma (HCC). In over 50% of HCC cases studied in these areas, a characteristic G to T mutation is observed at the third position of codon 249 of the p53 tumor-suppressor gene (2, 3). Whether this specific sequence is an exceptional target for mutations caused by AFB1 or whether the mutation is selected for once it occurs remains to be determined. However, each of these scenarios shares the fundamental early step involving generation of a G to T mutation.
There is substantial evidence that AFB1-induced G to T mutations in cellular ras genes may also be a step in transformation of normal cells to malignant cells (4–6). In humans these mutations occur at the first and second positions of codon 12 in the Ha-ras protooncogene (7) and they are in sequence contexts similar, but not identical, to that of codon 249 in p53.
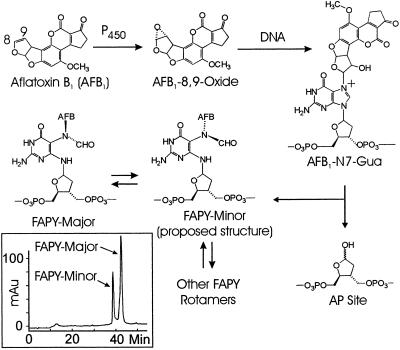
Many studies have defined the mutational spectrum produced after exposure of cells to either the epoxide, which is the toxicologically relevant natural metabolite of AFB1 (8), or to other electrophilic derivatives that serve as models for the epoxide (9). The G to T mutation is predominantly observed (2, 3, 8–19). Studies of mutational landscapes, by their nature, do not elucidate which specific chemical form of AFB1-DNA adduct is responsible for a given mutation (1). The epoxide reacts with DNA to form the primary AFB1-DNA adduct, 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 (AFB1-N7-Gua) (20–26), which can break down into two secondary lesions, the apurinic (AP) site or the AFB1-formamidopyrimidine (FAPY) adduct (Fig. 1) (20). The mutational properties of the AFB1-N7-Gua adduct have been studied in Escherichia coli (16), where it primarily causes G to T mutations, although at a very low frequency (4%). An interesting property of this adduct is that a significant number of the total mutations observed (13%) occurs 5′ to the site at which the adduct forms. NMR data (27) may explain this mutational asymmetry in that the AFB1-position of the adduct is intercalated 5′ to the attached DNA base. The AFB1-FAPY adduct is similar in structure to AFB1-N7-Gua, but data show that these two adducts alter the structure of DNA differently (28, 29). These differences in DNA secondary structure may lead to differences in lethality, mutagenicity, and repair of the two chemical forms of the adduct.
Figure 1.
Aflatoxin biochemistry. AFB1 is activated to the reactive exo-8,9-epoxide form by cytochrome P450 in the liver. The epoxide can react at the N7 position of guanine in DNA to form the primary AFB1-N7-Gua adduct. This product can break down to form two secondary lesions, the AP site and the ring-opened AFB1-FAPY adduct. The FAPY adduct itself consists of two proposed rotameric forms, FAPY major and FAPY minor. FAPY minor is formed first, subsequently equilibrating to a 2:1 ratio of FAPY major:FAPY minor. The FAPY major structure can be found in Stone and coworkers (29), and the FAPY minor structure is yet unresolved, but one possible structure is depicted here. (Inset) Separation of AFB1-FAPY rotamers. HPLC was performed on a 13-base oligonucleotide containing one single AFB1-FAPY adduct. Under the HPLC conditions described, FAPY major can be separated from FAPY minor. The equilibrium mixture shown here is roughly 2:1 major to minor. This mixture was maintained throughout the experiments described in this article and is referred to as FAPY mix. The major and minor peaks were collected and tested as indicated.
The AFB1-FAPY adduct itself consists of an equilibrium mixture of two presumably rotameric forms (FAPY mix) that are separable by HPLC into products termed FAPY major and FAPY minor (Fig. 1 Inset). Over time at 4°C or warmer, the equilibrium of this mixture shifts toward FAPY major. Stone and coworkers (29) attempted to resolve the NMR structure of FAPY and at first obtained complex results, presumably because of the presence of multiple species. After several days at 4°C, the NMR spectrum simplified to reveal presumably the structure referred to as FAPY major in this work. Although the exact chemical nature of FAPY minor remains elusive, a structure for one of the possible rotamers has been proposed (Fig. 1) based on data obtained for the imidazole ring-opened form of 7-methylguanine. This adduct forms two rotamers, indicating that structural duality may be a common characteristic of formamidopyrimidines (30–33). Under the conditions by which the AFB1-FAPY oligonucleotides are made and stored, an equilibrium exists between the major and minor presumed rotamers at a ratio of 2:1, and these oligonucleotides can be used in studies of mutagenicity.
To evaluate the mutagenic properties of AFB1-FAPY, we took advantage of the fact that bacterial cells have mechanisms such as the SOS response for coping with DNA damage. After exposure of E. coli to UV light, the UmuDC operon is derepressed, allowing the error-prone bypass polymerase (polV) to be transcribed (34–38). MucAB, an analogous bypass polymerase in Salmonella typhimurium, is regulated in a similar manner. These bypass polymerases incorporate bases, often incorrectly, opposite the site of DNA damage, allowing for cell survival in a time of crisis. They can also be used as tools to determine which type of base is most often put opposite a specific DNA lesion. Because mammalian cells have similar mechanisms to cope with DNA damage, we used cells induced to express these bypass polymerases in this work.
Materials and Methods
Enzymes and Chemicals.
EcoRI, HaeIII, HinfI, T4 polynucleotide kinase (ATP: 5′-dephosphopolynucleotide 5′phosphotransferase, 2.7.1.78), T4 DNA ligase [poly(deoxyribonucleotide): poly(deoxyribonucleotide) ligase (AMP forming), 6.5.11], exonuclease III (exodeoxyribonuclease III, 3.1.11.2), and uracil DNA glycolsylase (UDG) were from New England Biolabs. [γ-32P]dATP and [α-35S]dATP were from Perkin–Elmer. SfuI and G-50 and G-25 Sephadex columns were from Roche Molecular Biochemicals. Cetricon-3 concentrators were from Amicon. ZipTip C18 pipet tips were from Millipore. Isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were from Gold Biotechnology, St. Louis. N,N′-dimethylformamide (DMF) was from Fisher Scientific.
DNAs and Cell Strains.
M13mp7L2 was obtained from C. W. Lawrence (39). A 13-base oligonucleotide, d(CCTCTTCGAACTC) (boldface G = site of adduct; underscore = SfuI site), and a 53-base uracilated oligonucleotide scaffold, d(AAAACGACGGCCAGUGAAUUGAGUUCGAAGAGGCACUGAAUCAUGGUCAUAGC) (the underlined sequence is complementary to the 13-base insert, and the 20 bases on either side of this sequence are complementary to the flanking regions of the M13mp7L2 genome), as well as mass spectral internal standards, a 12-base oligonucleotide, d(ATTGGTAAGTGC), and a 14-base oligonucleotide d(ATTGGTAAGTCCGA), were obtained from GIBCO/BRL and purified with PAGE. E. coli strains used were DL7 (AB1157; lacΔU169), DL7/pGW16 (DL7; pGW16 mucAB) (this laboratory), GW5100 (JM103, P1−) (G. Walker, Massachusetts Institute of Technology), and NR9050 (F′ prolacIZΔM15, Δprolac, suB) (R. Schaaper, National Institutes of Health).
Construction and Characterization of AFB1-N7-Gua- and AFB1-FAPY-Containing Oligonucleotides.
Oligonucleotides containing single AFB1-N7-Gua and AFB1-FAPY residues in the d(CCTCTTCGAACTC) context (boldface G = site of adduct; underscore = SfuI site) were synthesized based on published procedures (40). First, the oligonucleotide was gel-purified, and 100–250 nmol were resuspended in 90 ml of buffer (10 mM NaHPO4, pH 7/100 mM NaCl/50 mM EDTA). One milligram of AFB1 epoxide (41) in 50 ml of CH2Cl2 was then added and the mixture was shaken for 10 min at 4°C. The same amount of AFB1 epoxide was added once more, again shaking at 4°C for 15 min. The solution was then extracted twice with 100 ml of CH2Cl2 and desalted on a G-25 Sephadex column. Oligonucleotides containing AFB1-N7-Gua adducts were separated from unreacted DNA by HPLC with a Beckman Ultrasphere C18 reverse phase column (part no. 235329). Buffer A was 0.1 M TEAAC (pH 7.5) and buffer B was CH3CN. The gradient used went from 8 to 12% B from 0 to 40 min, and from 12 to 25% B from 40 to 60 min. Fractions were collected and desalted with Centricon-3 concentrators at 4°C. A portion of the AFB1-N7-Gua lesions was converted to AFB1-FAPY through incubation in 0.1 M NaHPO4 (pH 10) at 37°C for 4 h. AFB1-FAPY minor was formed first, subsequently reaching equilibrium with AFB1-FAPY major, at a ratio of 2:1 major:minor. Oligonucleotides containing AFB1-FAPY lesions were separated from those containing AFB1-N7-Gua lesions with the HPLC conditions above. These conditions were also used to separate FAPY major from FAPY minor.
Matrix-Assisted Laser Desorption Ionization–Time-of-Flight (MALDI-TOF) MS.
Thirteen-base oligonucleotides containing FAPY major, FAPY minor, or 2:1 mixture of the major:minor FAPY adducts (FAPY mix) were analyzed on a PerSeptive Biosystems Voyager Elite DE MALDI-TOF mass spectrometer. Twelve-base and 14-base unmodified oligonucleotides were used as internal calibration standards. Ten picomoles of the sample and each standard were mixed in a 10-μl volume and desalted with ZipTips. They were then resuspended in the MALDI matrix (2.5% anthranilic acid/1.1% nicotinic acid/45.5% CH3CN/27.3% 100 mM ammonium citrate) and spotted onto the sample plate. Spectra were obtained in the reflector mode with a laser energy of 2,400, an accelerating voltage of 25,000 V, a grid voltage of 95%, a guide wire voltage of 0.28%, and a delay time of 120 ns. Each spectrum was an average of 128–256 laser shots.
Genome Construction.
Genomes were constructed as in Bailey et al. (16). Four separate ligation reactions were done with each of the inserts: (i) AFB1-N7-Gua, (ii) FAPY mix, (iii) FAPY major, and (iv) unmodified control. These reactions were carried out at 16°C to ensure the integrity of the adducts.
Transformation of AFB1-Modified Genomes into Cell Strains.
Once genomes were constructed and desalted, they were kept on ice and used immediately to transform DL7 (not induced for SOS), UV-irradiated DL7 (expresses UmuDC), DL7/pGW16 (expresses MucAB), or UV-irradiated DL7/pGW16 (expresses both MucAB and UmuDC) by using conditions in Bailey et al. (16). The only procedural difference was that the cells were irradiated in a Stratalinker UV crosslinker at 45 J/m2. Approximately 0.1 pmol of genome in a volume of 10 μl was mixed with 190 μl of the transfection strain for each electroporation. Progeny phage were produced from each transformation and stored at 4°C until needed.
Pooling Progeny Phage.
Between 8 and 40 parallel replicate transformations were carried out per sample per cell strain to generate a progeny phage pool that could be easily examined for mutants. A total of 20,800 infective centers from the eight most successful transformations made up each pool. Replicative form DNA was isolated with the Qiagen (Chatsworth, CA) Midi double-stranded plasmid procedure.
Enrichment for Mutant Genomes.
Approximately 0.5 pmol of DNA isolated from pooled phage was subjected to cleavage by SfuI, a restriction enzyme that cleaves at a unique site within the 13-base region of the inserted oligonucleotide. Digested products, as well as mock-digested controls, were analyzed on agarose gels to determine linearization. Approximately 0.1 pmol of digested or control DNA was desalted on a G-25 Sephadex column and electroporated as above, using DL7 as the transfection strain and NR9050 as the plating bacteria.
Sequencing.
Sequencing was performed on enriched M13 DNA isolated from the cell strain that expressed both UmuDC and MucAB. Two hundred fifty-six blue and 100 clear plaques were sequenced.
Results
MALDI–Time-of-Flight (TOF) MS.
To determine whether the peaks observed in the HPLC trace (Fig. 1 Inset) represented chemically unique structures, MALDI-TOF MS was performed on 13-base oligonucleotides containing FAPY major, FAPY minor, or a 2:1 mixture of the major:minor forms (FAPY mix). Twelve-base and 14-base oligonucleotides were used as internal standards, and in each case, only one peak was observed for the FAPY-containing oligonucleotide. This peak had m/z ratios of 4191.73, 4191.97, and 4191.99, respectively, for each of the three samples. This result indicates that FAPY major and FAPY minor share identical chemical compositions, implying that they are most likely rotamers of one another, as is the case for other formamidopyrimidines (33).
Construction of Genomes Containing AFB1 Adducts.
Single-stranded oligonucleotides containing AFB1-FAPY adducts were analyzed by HPLC after being exposed to the conditions of genome construction. Stability studies were performed on oligonucleotides containing the FAPY major adduct, the FAPY minor adduct, or FAPY mix. Although the FAPY major adduct maintained its integrity through all steps of the procedure, there was, however, some conversion of FAPY minor to the major form. Accordingly, genetic studies were performed with only oligonucleotides containing either FAPY major or FAPY mix. Bailey et al. (40) demonstrated the stability of AFB1-N7-Gua oligonucleotides under these conditions.
After the ligation of the oligonucleotide inserts (either modified or unmodified, as a control), it was noted that the yield of intact modified genomes was low compared with that of unmodified controls, indicating that steric or other factors may inhibit efficient annealing and ligation of the AFB1-FAPY- or AFB1-N7-Gua-containing inserts. The amount of DNA used in each transformation was adjusted based on the percentage of circular ligation product observed with agarose gel analysis.
Cellular Tolerance of AFB1 DNA Adducts.
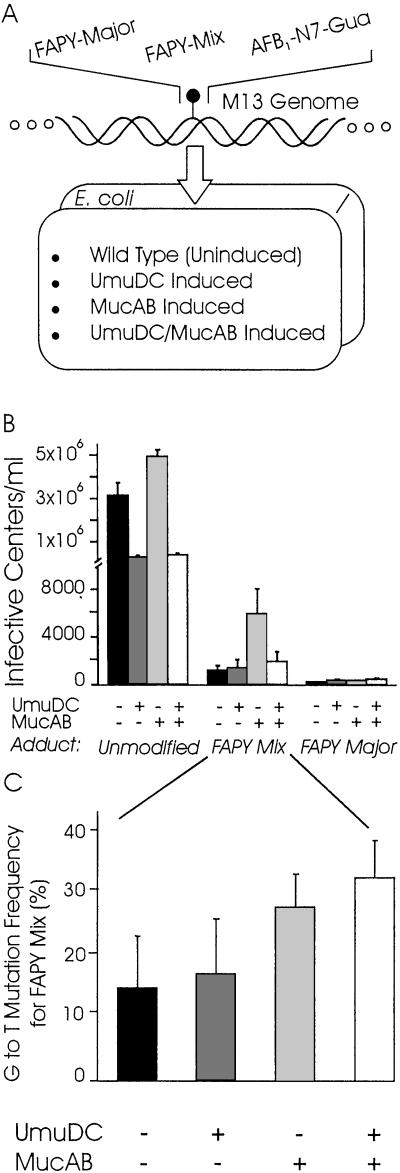
Genomes containing an unmodified insert or an insert containing the FAPY major, FAPY mix, or AFB1-N7-Gua adduct were electroporated into E. coli (Fig. 2A). Several cell strains were used in these experiments to probe the genetic requirements for toxicity. Cells not induced for the SOS response (DL7) or cells that expressed the bypass polymerases UmuDC (UV-irradiated DL7), MucAB (DL7/pGW16), or both UmuDC and MucAB (UV irradiated DL7/pGW16) were used in the electroporations. G to T mutations are scored as light blue plaques, whereas wild-type sequences, other point mutations, and in-frame deletions (provided they are small enough) yield dark blue plaques (16). Clear plaques would result from frame shifts, large deletions, or genomes without inserts.
Figure 2.
Lethality and mutagenicity of AFB1-FAPY. (A) M13 genomes contained a single aflatoxin adduct and were replicated in E. coli expressing different bypass polymerases. (B) Cell strains challenged with FAPY mix, FAPY major, or an unmodified control exhibited different levels of survival. The x axis depicts the adduct with which the cell was challenged and the polymerase status of the cell, whereas the y axis is the number of infective centers per milliliter. (C) The G to T mutation frequency for cell strains challenged with FAPY mix is represented here. Cell strains expressed the bypass polymerases indicated by the x axis. The G to T mutation frequency was calculated by counting the number of light blue plaques and dividing by the total number of blue plaques observed. Error bars for both graphs reflect the SEM for four sets of data.
Each adduct-containing genome was electroporated into each cell strain 8–40 separate times to yield one set of data. Every graph shown represents four sets of data, each set generated from a separate genome construction event. After transformation, the total number of blue plaques was scored as the number of infective centers. Each infective center represents a single transformation event by one M13 genome that was successfully replicated in the host cell, yielding a wild-type or mutant outcome. Thus, we can determine how well or poorly the replication machinery of the cell can handle the challenge of a single AFB1-FAPY adduct. Fig. 2B compares the numbers of infective centers per milliliter of transformed cells for genomes containing an unmodified, FAPY major, or FAPY mix oligonucleotide insert in each of the genetic backgrounds described above. We found that unmodified inserts were tolerated an average of 2–3 orders of magnitude better than those containing a modified insert. When cells were challenged with the FAPY major adduct alone, the genome survived less well than a comparable genome containing FAPY mix. Cells tolerated the AFB1-N7-Gua adduct 1–2 orders of magnitude better than the FAPY mix adduct, as inferred from the number of infective centers obtained (data not shown). These data indicate that the AFB1-N7-Gua adduct is far less effective as a replication block than the AFB1-FAPY adduct. In our hands the FAPY major adduct is the most lethal known aflatoxin-induced replication block in vivo. FAPY mix is not as lethal a block to replication, therefore it is possible that cells that survive the challenge with aflatoxin contain mostly the FAPY minor component of this mixture and, therefore, most of the mutations that arise may be a result of this form of the adduct. It is for this reason that we chose to evaluate the FAPY mix adduct in our mutagenicity studies.
Mutagenicity of the AFB1-FAPY Adduct.
The experimental system allowed the facile scoring of G to T mutations by using the convenient light blue/dark blue color selection described above. Fig. 2C shows the G to T mutation frequency of the FAPY mix adduct in each of the four genetic backgrounds used in this study. Cells not induced for bypass polymerases showed the lowest number of mutations, about 15%. Those expressing UmuDC showed a 17% mutation frequency, whereas the MucAB-expressing cells showed a 27% mutation frequency. Finally, cells expressing both UmuDC and MucAB showed a 32% mutation frequency, which is less than additive, implying that both polymerases may compete to bypass the same adduct. The same trend observed for survival is not observed for mutation frequency and, in fact, the only significant difference discovered here is that cells expressing MucAB, whether or not UmuDC is present, have a slightly higher G to T mutation frequency than the cell strains that do not express the MucAB bypass polymerase. To simplify our analysis, we therefore decided to evaluate the frequency of other types of mutations only in the cell strain expressing both UmuDC and MucAB.
The G to T mutation frequency observed by Bailey et al. (16) for the AFB1-N7-Gua adduct was 4% in the cell strain expressing MucAB. After repeating this experiment for all four cell strains used in these studies, the mutation frequency was found to be between 2–6%, which agrees well with prior results and allows for a direct comparison of the two studies. The modest mutagenicity of AFB1-N7-Gua is striking by contrast with AFB1-FAPY. Indeed, FAPY adducts are at least 6 times as likely to cause G to T mutations as their molecular predecessor, AFB1-N7-Gua.
Enrichment for Non-G to T Mutations.
The non-G to T mutations were expected to be infrequent; therefore, we used a method that would help to enrich the phage pool for mutants by eliminating the majority of wild-type DNA. Progeny phage obtained from the eight transformation events that gave the highest number of transformants in each cell strain were pooled, the population was expanded in cell culture, and the double-stranded (replicative form) M13 DNA was isolated. The oligonucleotide inserts were engineered such that they contained a unique SfuI restriction site that encompassed the target base for evaluation of mutagenesis. If this and the surrounding positions retained their wild-type sequence, the restriction enzyme would successfully cut this DNA, rendering it unable to transform E. coli. However, if a mutation occurred within the restriction site, the DNA would be refractory to digestion and therefore able to transform E. coli successfully.
DNA from progeny phage that were a result of AFB1-FAPY-modified genomes transfected into cells expressing both UmuDC and MucAB was subjected to treatment with SfuI or was mock-treated. The number of dark blue plaques in the digested sample divided by the number of dark blue plaques in the mock-treated sample is termed the restriction-resistant fraction (RRF) and is a first approximation of the mutation frequency. Sequencing the dark blue plaques in the digested samples and multiplying their values by the RRF yields the true frequency of each type of non-G to T mutation present. It should be noted that all of the dark blue plaques did not consist of wild-type or mutant sequences; some were the result of the genetic engineering procedures used in the experiment. For example, some sequences were M13 DNA that had been ligated back together without an insert, containing only a few extra random bases to put the gene back in frame. Others were small deletions that presumably result from errant nuclease action. These species are included in the total used to calculate the fraction of each type of mutation observed. The following overall mutation frequencies were observed: G to A = 1%, total 5′ mutations (C to G and C to T) = 1%, a specific quadruple mutation = 1%, and other multiple mutations = 3%. For comparison, under these genetic conditions the G to T mutation frequency was 32%.
When looking only at the dark blue plaques that were mutants, it can be determined how each specific type of mutation contributed to the total. The following percentages were observed: G to A mutations = 18%, total 5′ mutations (C to T and C to G) = 16%, a specific quadruple mutation = 16%, and other multiple mutations = 49%. Table 1 shows the different multiple mutations and calculates the contribution of each mutation to the total. One surprising result was the presence of a specific quadruple mutation. This sequence was 5′-CCTCTAAAGACTC-3′. The bold bases are mutations, and the underlined base is the originally modified site. The mutation at the target site was a G to A, and these were not included in the total number of G to A mutations. The two 5′ transversions are T to A and C to A, respectively, and the 3′ transition is A to G, none of which were included in the total number of 5′ or 3′ mutations. We believe that the multiple mutations are completely unique to the AFB1-FAPY adduct, specifically the FAPY minor form, because of the fact that FAPY major is a lethal replication block in our system. We also hypothesize that the FAPY minor adduct, before converting to the low energy FAPY major structure, may cause some conformational flexibility of the DNA helix that may interfere with the replication of several bases that are adjacent to the modified site.
Table 1.
Multiple mutations induced by AFB1-FAPY mix
| Mutation type | Sequence (5′–3′) | Contribution*, % |
|---|---|---|
| Wild type | CCTCTTCGAACTC | |
| Quadruple | CCTCTAAAGACTC | 16 |
| Tandem | CTGCTTGAAACTC | 10 |
| CCTCTTGAAACTC | 3 | |
| 5′ or 3′ | CCTCTTTGAACTC | 10 |
| CCTCTTGGAACTC | 3 | |
| CTCCATTGAAGTC | 8 | |
| Deletions | CGACTTC(ΔGAA)CTC | 2 |
| CGTCTTC(ΔGAA)CTC | 8 | |
| Other | CATCTTCTAACTC | 8 |
Underlined base is target site of modification. Boldface indicates mutated bases. Parentheses (Δ) indicate deletions.
Contribution of the type of mutation to the total number of non-G to T mutations observed.
Several small deletions consisting of the codon containing the modified G were also observed. They made up about 10% of the non-G to T mutations and may be a result of the complete bypass of the damaged area by the polymerase. One other occurrence was the point mutation of one or two bases located about five to six positions upstream of the modified G. Most often these mutations were C to T or C to G. It is possible that these mutations are the results of localized infidelity of the error-prone polymerase distal to the damaged site. Such mutations would taper off once the normal replicative DNA polymerase resumed its function.
Discussion
Epidemiological studies indicate that aflatoxin B1 plays a synergistic role with hepatitis B virus in the formation of many human hepatocellular carcinomas (1). It is believed that the carcinogenicity of the toxin is mediated in part by the mutagenic properties of one or more of the AFB1 DNA adducts. Although exposure to HBV alone increases the risk of HCC, the risk is much higher in an individual who is positive for HBV and is at risk for high levels of aflatoxin exposure. Studies indicate that people who live in areas where potential AFB1 exposure is high are 3 times more likely to develop HCC. Those who test positive for HBV are about 7 times more likely to develop HCC, and when both these criteria are met, people are 60 times more likely to develop this disease.
The work presented here evaluated the contributions to lethality and mutagenicity of the aflatoxin formamidopyrimidine adduct, a DNA lesion that is highly persistent in liver in vivo (25). The G to T mutation at the third position of codon 249 of the p53 tumor-suppressor gene has been observed in over 50% of HCC cases as well as in human hepatocytes treated with AFB1 in culture (2, 3). Additionally, activation of human ras genes by specific G to T mutations presumed to be caused by aflatoxin exposure may play a significant role in malignant transformation (4–6). This mutation was observed for the AFB1-N7-Gua adduct in the mutation analysis system used in this study. AFB1-N7-Gua, however, was only weakly mutagenic, with a mutation frequency of 2–6%. By contrast, we have established that AFB1-FAPY produced the same mutation with a frequency of up to 32%; because this adduct is also highly persistent in the genome, it has great potential to play a role in hepatocarcinogenesis.
It would be of interest to know to what extent the structures of the different types of aflatoxin DNA adducts influence their biological properties. The NMR structures of AFB1-DNA adducts provide insight into this issue (27). The AFB1-FAPY adduct stabilizes the DNA duplex by unwinding it by about 15° and increasing stacking interactions between the intercalated aflatoxin moiety and the neighboring bases. In addition there is a possible hydrogen bond between the formyl proton of what is thought to be FAPY major and the exocyclic amino group on a neighboring adenine. The NMR solution structure of AFB1-N7-Gua also reveals an increased melting temperature of the DNA duplex, but only by about 3–5°C (42). Additionally, this form of the adduct bends slightly the DNA helix (28). NMR data to establish the FAPY minor structure remain elusive.
In principle, repair enzymes may recognize AFB1-N7-Gua better than the FAPY analog owing to the greater distortive effect of AFB1-N7-Gua on DNA structure. In support of this view, excision repair systems preferentially remove AFB1-N7-Gua from the DNA of human fibroblasts (43). The FAPY adduct, having a more subtle effect on DNA architecture and a more profound effect on the local melting temperature of the DNA duplex than AFB1-N7-Gua (29, 41, 42), may evade repair, which is in line with its persistence in vivo. The longevity of the FAPY adduct, combined with its high mutagenic potential, makes it a dangerous lesion.
Unfortunately, the sequences used in the two NMR studies of AFB1-N7-Gua and AFB1-FAPY (27, 29) were not identical, which raises the question of whether sequence context may have an effect on the secondary structures these adducts form within DNA. Sequence context may in addition affect whether the adducts are blocks to replication, as has been observed for both the AFB1-N7-Gua and AFB1-FAPY adducts in many in vitro systems (12, 44–48) and/or what frequencies and types of mutations they cause. It is possible that different sequence contexts allow for faster or slower conversion of FAPY minor to FAPY major, if the stability of the adduct depends on hydrogen bonding with neighboring bases.
There are clear differences in the survival of phage DNA containing an AFB1-FAPY adduct when that DNA is replicated in cells expressing different bypass polymerases. UmuDC and MucAB are analogous bypass polymerases (polV) found in E. coli and S. typhimurium, respectively (38, 49–51). Phage DNA harboring the AFB1-FAPY adduct survives best when replicated in cells expressing MucAB, a polymerase that is currently thought to be the most proficient at bypass of bulky lesions (Fig. 2B) (17, 50, 52–58). This study has established that the FAPY major adduct is the strongest block to replication of all of the aflatoxin adducts studied here, irrespective of which bypass polymerase is used by the cell to help it overcome the DNA damage. Thus, this lesion is a prime candidate as a contributor to the extreme toxicity of AFB1. This conclusion may be extrapolated to humans, because mammalian cells have similar lesion bypass mechanisms (59, 60). In contrast, phage DNA containing the AFB1-N7-Gua adduct generates about 2 orders of magnitude more infective centers than that containing the AFB1-FAPY adducts, indicating that the AFB1-N7-Gua adduct is a less formidable threat to the replication machinery of the cell.
AFB1-FAPY induces a G to T mutation frequency that is roughly 6 times that observed for AFB1-N7-Gua. The G to A mutation frequency is similar for the two adducts, the G to C mutation frequency is higher for the AFB1-N7-Gua adduct, and there are also a similar number of 5′ mutations, supporting the notion that it is the bulkiness of the AFB1 adducts that causes adjacent mutations. Additionally, the FAPY minor adduct may induce a zone of conformational flexibility in the vicinity of the adduct. This zone possibly interferes with faithful replication beyond the adduct, causing multiple mistakes to be made by the polymerase, and resulting in several mutations in the immediate area. One particularly interesting case is that of a quadruple mutant that makes up 16% of the non-G to T mutations observed (1% of all mutations caused AFB1-FAPY), demonstrating that multiple mutations are not necessarily random. The lesion itself may determine the type of mutations observed at sites other than the target site, which is consistent with the fact that these mutations are not observed for the AFB1-N7-Gua adduct. One aspect to note regarding the quadruple mutants is that all four of the bases inserted opposite the lesion are pyrimidines, possibly because they are less sterically hindered than their bulkier purine counterparts, and therefore more easily placed opposite a lesion.
In most studies of the mutations observed in DNA globally modified with AFB1, there are several instances in which mutations occur 5′ or 3′ to a G, and sometimes even several bases away. When these mutations occur at a C, they have been commonly attributed to a modified G in the opposite strand. This may or may not be the case, and when the mutations occur at bases other than C, these mutations may be the results of an adduct a few bases away. We have closely evaluated several reports in the literature (61, 62) and have noted that the sequence context used in our studies appears several times. In these sequence contexts there exist mutations that mirror the multiple mutations that we have observed. These mutations could be a result of the adduct itself or a function of a bypass/repair mechanism of the cell. The notion that one DNA adduct can cause more than one mutation, even a few bases away from the adduct site, is supported by the work of others (63), who observe this phenomenon not only for AFB1, but also for benzo(a)pyrene and UV light. All of the aforementioned studies support the observation that mutational hotspots do not always correspond with hotspots for adduct formation (1).
This work, in summary, leads to three main conclusions. The first is that the highly persistent AFB1-FAPY adduct is also highly mutagenic, which poses a long-term threat to the genetic integrity of cells harboring the adduct. Second, the FAPY major form of the adduct is a very strong block to replication, even when cells use bypass polymerases. Similar mammalian replicative and DNA repair systems may have difficulty dealing with this adduct in vivo, leading to toxic effects. Finally, the AFB1-FAPY adduct is responsible for mutations that occur at sites other than the site of initial damage.
Acknowledgments
We thank J. Mendoza, A. Whagmare, D. M. Nekorchuk, and P. DeLeon for technical assistance. We are indebted to Graham Walker for helpful discussions and for providing cell strains. We also thank Gerald Wogan for valuable input. This work was supported by National Institutes of Health Grants CA80024, ES09546, and T32 ES07020.
Abbreviations
- AFB1
aflatoxin B1
- AFB1-N7-Gua
8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1
- FAPY
formamidopyrimidine
- HCC
hepatocellular carcinoma
- HBV
hepatitis B virus
- MALDI
matrix-assisted laser desorption ionization
References
- 1.Smela M E, Currier S S, Bailey E A, Essigmann J M. Carcinogenesis. 2001;22:535–545. doi: 10.1093/carcin/22.4.535. [DOI] [PubMed] [Google Scholar]
- 2.Hsu I C, Metcalf R A, Sun T, Welsh J A, Wang N J, Harris C C. Nature (London) 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 3.Bressac B, Kew M, Wands J, Ozturk M. Nature (London) 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 4.Bos J L, Fearon E R, Hamilton S R, Verlaan-de Vries M, van Boom J H, van der Eb A J, Vogelstein B. Nature (London) 1987;327:293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 5.Vogelstein B, Fearon E R, Hamilton S R, Kern S E, Preisinger A C, Leppert M, Nakamura Y, White R, Smits A M, Bos J L. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 6.McMahon G, Hanson L, Lee J J, Wogan G N. Proc Natl Acad Sci USA. 1986;83:9418–9422. doi: 10.1073/pnas.83.24.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riley J, Mandel H G, Sinha S, Judah D J, Neal G E. Carcinogenesis. 1997;18:905–910. doi: 10.1093/carcin/18.5.905. [DOI] [PubMed] [Google Scholar]
- 8.Benasutti M, Ejadi S, Whitlow M D, Loechler E L. Biochemistry. 1988;27:472–481. doi: 10.1021/bi00401a068. [DOI] [PubMed] [Google Scholar]
- 9.Denissenko M F, Cahill J, Koudriakova T B, Gerber N, Pfeifer G P. Mutat Res. 1999;425:205–211. doi: 10.1016/s0027-5107(99)00038-x. [DOI] [PubMed] [Google Scholar]
- 10.Basu A K, Essigmann J M. Mutat Res. 1990;233:189–201. doi: 10.1016/0027-5107(90)90162-w. [DOI] [PubMed] [Google Scholar]
- 11.Kamiya H, Sakaguchi T, Murata N, Fujimuro M, Miura H, Ishikawa H, Shimizu M, Inoue H, Nishimura S, Matsukage A, et al. Chem Pharm Bull. 1992;40:2792–2795. doi: 10.1248/cpb.40.2792. [DOI] [PubMed] [Google Scholar]
- 12.Levy D D, Groopman J D, Lim S E, Seidman M M, Kraemer K H. Cancer Res. 1992;52:5668–5673. [PubMed] [Google Scholar]
- 13.Puisieux A, Lim S, Groopman J, Ozturk M. Cancer Res. 1991;51:6185–6189. [PubMed] [Google Scholar]
- 14.Foster P L, Eisenstadt E, Miller J H. Proc Natl Acad Sci USA. 1983;80:2695–2698. doi: 10.1073/pnas.80.9.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence C W, Borden A, Banerjee S K, LeClerc J E. Nucleic Acids Res. 1990;18:2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey E A, Iyer R S, Stone M P, Harris T M, Essigmann J M. Proc Natl Acad Sci USA. 1996;93:1535–1539. doi: 10.1073/pnas.93.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster P L, Groopman J D, Eisenstadt E. J Bacteriol. 1988;170:3415–3420. doi: 10.1128/jb.170.8.3415-3420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muench K F, Misra R P, Humayun M Z. Proc Natl Acad Sci USA. 1983;80:6–10. doi: 10.1073/pnas.80.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misra R P, Muench K F, Humayun M Z. Biochemistry. 1983;22:3351–3359. doi: 10.1021/bi00283a008. [DOI] [PubMed] [Google Scholar]
- 20.Busby W F, Jr, Wogan G N. In: Chemical Carcinogens. Searle C, editor. Washington, DC: Am. Chem. Soc.; 1984. pp. 945–1136. [Google Scholar]
- 21.Martin C N, Garner R C. Nature (London) 1977;267:863–865. doi: 10.1038/267863a0. [DOI] [PubMed] [Google Scholar]
- 22.Lin J K, Miller J A, Miller E C. Cancer Res. 1977;37:4430–4438. [PubMed] [Google Scholar]
- 23.Groopman J D, Croy R G, Wogan G N. Proc Natl Acad Sci USA. 1981;78:5445–5449. doi: 10.1073/pnas.78.9.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croy R G, Essigmann J M, Reinhold V N, Wogan G N. Proc Natl Acad Sci USA. 1978;75:1745–1749. doi: 10.1073/pnas.75.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croy R G, Wogan G N. J Natl Cancer Inst. 1981;66:761–768. [PubMed] [Google Scholar]
- 26.Croy R G, Wogan G N. Cancer Res. 1981;41:197–203. [PubMed] [Google Scholar]
- 27.Johnston D S, Stone M P. Biochemistry. 1995;34:14037–14050. doi: 10.1021/bi00043a009. [DOI] [PubMed] [Google Scholar]
- 28.Gopalakrishnan S, Harris T M, Stone M P. Biochemistry. 1990;29:10438–10448. doi: 10.1021/bi00498a002. [DOI] [PubMed] [Google Scholar]
- 29.Mao H, Deng Z, Wang F, Harris T M, Stone M P. Biochemistry. 1998;37:4374–4387. doi: 10.1021/bi9718292. [DOI] [PubMed] [Google Scholar]
- 30.Boiteux S, Belleney J, Roques B P, Laval J. Nucleic Acids Res. 1984;12:5429–5439. doi: 10.1093/nar/12.13.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chetsanga C J, Makaroff C. Chem Biol Interact. 1982;41:235–249. doi: 10.1016/0009-2797(82)90092-8. [DOI] [PubMed] [Google Scholar]
- 32.Chetsanga C J, Bearie B, Makaroff C. Chem Biol Interact. 1982;41:217–233. doi: 10.1016/0009-2797(82)90091-6. [DOI] [PubMed] [Google Scholar]
- 33.Tomasz M, Lipman R, Lee M S, Verdine G L, Nakanishi K. Biochemistry. 1987;26:2010–2027. doi: 10.1021/bi00381a034. [DOI] [PubMed] [Google Scholar]
- 34.Witkin E M. Bacteriol Rev. 1976;40:869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol. Press; 2001. pp. 407–464. [Google Scholar]
- 36.Woodgate R, Levine A S, Koch W H, Cebula T A, Eisenstadt E. Mol Gen Genet. 1991;229:81–85. doi: 10.1007/BF00264216. [DOI] [PubMed] [Google Scholar]
- 37.Sommer S, Bailone A, Devoret R. Mol Microbiol. 1993;10:963–971. doi: 10.1111/j.1365-2958.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- 38.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee S K, Borden A, Christensen R B, LeClerc J E, Lawrence C W. J Bacteriol. 1990;172:2105–2112. doi: 10.1128/jb.172.4.2105-2112.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey E A, Iyer R S, Harris T M, Essigmann J M. Nucleic Acids Res. 1996;24:2821–2828. doi: 10.1093/nar/24.14.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gopalakrishnan S, Byrd S, Stone M P, Harris T M. Biochemistry. 1989;28:726–734. doi: 10.1021/bi00428a047. [DOI] [PubMed] [Google Scholar]
- 42.Jones W R, Johnston D S, Stone M P. Chem Res Toxicol. 1998;11:873–881. doi: 10.1021/tx980047m. [DOI] [PubMed] [Google Scholar]
- 43.Leadon S A, Tyrrell R M, Cerutti P A. Cancer Res. 1981;41:5125–5129. [PubMed] [Google Scholar]
- 44.Yu F L. Carcinogenesis. 1983;4:889–893. doi: 10.1093/carcin/4.7.889. [DOI] [PubMed] [Google Scholar]
- 45.Marien K, Moyer R, Loveland P, Van Holde K, Bailey G. J Biol Chem. 1987;262:7455–7462. [PubMed] [Google Scholar]
- 46.Yu F L, Bender W, Geronimo I H. Carcinogenesis. 1990;11:475–478. doi: 10.1093/carcin/11.3.475. [DOI] [PubMed] [Google Scholar]
- 47.Refolo L M, Conley M P, Sambamurti K, Jacobsen J S, Humayun M Z. Proc Natl Acad Sci USA. 1985;82:3096–3100. doi: 10.1073/pnas.82.10.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnston D S, Stone M P. Chem Res Toxicol. 2000;13:1158–1164. doi: 10.1021/tx000129m. [DOI] [PubMed] [Google Scholar]
- 49.Sutton M D, Smith B T, Godoy V G, Walker G C. Annu Rev Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- 50.Perry K L, Elledge S J, Mitchell B B, Marsh L, Walker G C. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldsmith M, Sarov-Blat L, Livneh Z. Proc Natl Acad Sci USA. 2000;97:11227–11231. doi: 10.1073/pnas.200361997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulaeva O I, Wootton J C, Levine A S, Woodgate R. J Bacteriol. 1995;177:2737–2743. doi: 10.1128/jb.177.10.2737-2743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hauser J, Levine A S, Ennis D G, Chumakov K M, Woodgate R. J Bacteriol. 1992;174:6844–6851. doi: 10.1128/jb.174.21.6844-6851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woodgate R. Mutat Res. 1992;281:221–225. doi: 10.1016/0165-7992(92)90012-7. [DOI] [PubMed] [Google Scholar]
- 55.O'Grady P I, Borden A, Vandewiele D, Ozgenc A, Woodgate R, Lawrence C W. J Bacteriol. 2000;182:2285–2291. doi: 10.1128/jb.182.8.2285-2291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe M, Nohmi T, Ohta T. Mutat Res. 1994;314:27–37. doi: 10.1016/0921-8777(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 57.Bennett C B, Luo X, Refolo L M, Humayun M Z. Mutat Res. 1988;202:223–234. doi: 10.1016/0027-5107(88)90186-8. [DOI] [PubMed] [Google Scholar]
- 58.Szekeres E S, Jr, Woodgate R, Lawrence C W. J Bacteriol. 1996;178:2559–2563. doi: 10.1128/jb.178.9.2559-2563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 60.Gerlach V L, Aravind L, Gotway G, Schultz R A, Koonin E V, Friedberg E C. Proc Natl Acad Sci USA. 1999;96:11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aguilar F, Hussain S P, Cerutti P. Proc Natl Acad Sci USA. 1993;90:8586–8590. doi: 10.1073/pnas.90.18.8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambamurti K, Callahan J, Luo X, Perkins C P, Jacobsen J S, Humayun M Z. Genetics. 1988;120:863–873. doi: 10.1093/genetics/120.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Courtemanche C, Anderson A. Mutat Res. 1999;430:23–36. doi: 10.1016/s0027-5107(99)00113-x. [DOI] [PubMed] [Google Scholar]