Abstract
In Dictyostelium, a transient increase in intracellular cGMP is important for cytoskeletal rearrangements during chemotaxis. There must be cGMP-binding proteins in Dictyostelium that regulate key cytoskeletal components after treatment with chemoattractants, but to date, no such proteins have been identified. Using a bioinformatics approach, we have found four candidate cGMP-binding proteins (GbpA–D). GbpA and -B have two tandem cGMP-binding sites downstream of a metallo β-lactamase domain, a superfamily that includes cAMP phosphodiesterases. GbpC contains the following nine domains (in order): leucine-rich repeats, Ras, MEK kinase, Ras guanine nucleotide exchange factor N-terminal (RasGEF-N), DEP, RasGEF, cGMP-binding, GRAM, and a second cGMP-binding domain. GbpD is related to GbpC, but is much shorter; it begins with the RasGEF-N domain, and lacks the DEP domain. Disruption of the gbpC gene results in loss of all high-affinity cGMP-binding activity present in the soluble cellular fraction. GbpC mRNA levels increase dramatically 8 h after starvation is initiated. GbpA, -B, and -D mRNA levels show less dramatic changes, with gbpA mRNA levels highest 4 h into starvation, gbpB mRNA levels highest in vegetative cells, and gbpD levels highest at 8 h. The identification of these genes is the first step in a molecular approach to studying downstream effects of cGMP signaling in Dictyostelium.
Dictyostelium discoideum is a small, motile eukaryote that is remarkably similar to mammalian cells in morphology and cell biology. Dictyostelium cells have a highly developed chemotactic response. During vegetative growth they respond to bacterial compounds as a mechanism for finding food; upon starvation they secrete and chemotax toward cAMP, thus using cAMP as a beacon for aggregation so that multicellular fruiting bodies are formed. In mammals, chemotaxis plays key roles in defense against infection, wound healing, angiogenesis, embryogenesis, and metastasis of cancer cells. Overall, the signal transduction pathways associated with chemotaxis are conserved between Dictyostelium and higher organisms (1). With its relatively small genome (34 MB; ≈10,000 genes), Dictyostelium offers the virtue of a simple setting in which the cellular processes and signal transduction pathways associated with chemotaxis can be understood in detail.
Migration of cells up a chemotactic gradient involves asymmetric remodeling of the cytoskeleton during the first minute after stimulation, and therefore requires that cytoskeletal components be tightly regulated. In Dictyostelium, cGMP transiently increases when cells are exposed to chemoattractants (2, 3). Cells with two guanylyl cyclase genes disrupted do not increase cGMP in response to chemoattractant, and have a partial chemotaxis defect, indicating that cGMP signaling is important for the chemotactic response (4).
cGMP signaling in mammals is important in physiological processes such as smooth muscle tone, epithelial electrolyte transport, neuronal excitability, and phototransduction (5). Because cGMP signaling is not well documented in yeasts, Dictyostelium is the simplest model organism in which cGMP signaling pathways can be studied. For cGMP to function, it must bind to and regulate the activity of key proteins. In metazoans cGMP signaling is mediated by cGMP-dependent protein kinases (PKGs), cGMP-gated ion channels, a cAMP/cGMP-regulated RasGEF (6), and cGMP-regulated phosphodiesterases (PDEs). The cGMP-binding sites in these proteins are homologous to the Escherichia coli catabolite gene activator protein (CAP), except for the PDEs, which bind cGMP by using GAF domains (cGMP-specific and stimulated PDEs, Anabaena adenylate cyclases, and E. coli FhlA; ref. 7). Remarkably, no cGMP-binding proteins have been cloned from Dictyostelium. Biochemical evidence for such proteins comes from studies quantitating cGMP-binding sites in cellular lysates (8), and the observations that myosin light chain kinase-A (MLCK-A) and cGMP PDE are activated by cGMP in crude lysates (9, 10).
We have taken a bioinformatics approach to identifying such proteins. The Dictyostelium genome is well advanced and partial or complete sequence for ≥95% of genes is publicly available. (L. Eichinger, personal communication). We have mined this data resource and discovered four proteins predicted to bind cGMP. These proteins have novel domain structures compared with cGMP targets that have been identified in metazoans.
Methods
Sequence Databases.
The Dictyostelium genome is being sequenced at the following centers: genome.imb-jena.de/dictyostelium (Genome Sequencing Center Jena); www.uni-koeln.de/dictyastelium (University of Cologne); dictygenome.bcm.tmc.edu (Baylor College of Medicine); and www.sanger.ac.uk/Projects/D_discoideum (Sanger Centre). Data from a large-scale cDNA Project in Tsukuba, Japan can be obtained at www.csm.biol.tsukuba.ac.jp.
Identification and Sequencing of the gbp Genes.
We searched the genomic and cDNA databases of individual sequence reads by using the blast search engines available at each of the sequencing centers' web sites, as well as a central repository of sequences available at the San Diego Supercomputer Center (www.sdsc.edu/mpr/dicty/). Query sequences included all known cGMP-binding sites from ion channels, PKGs, and CNrasGEF, as well as cAMP-binding sites from CAP, Rap1-GEF (11, 12), and PKA regulatory subunits (PKA-Rs). In addition, we searched using 11 different GAF domain sequences.
For each gene, sequences were assembled from individual reads downloaded from the sequencing centers, using DNASTAR (Lasergene, Madison, WI). For uncertain portions, additional data were obtained by sequencing cDNAs and genomic PCR products. All introns were verified from cDNAs or by reverse transcription (RT)-PCR. For gbpB and -D, the 3′ ends of the ORF were deduced based on sequences from the Japanese cDNA project. For gbpC, the 3′ end was clear based on a TAA stop codon, followed by an A-rich sequence typical of Dictyostelium terminators. A long ORF on the opposite strand ending 567 bp downstream of the stop codon (data not shown) makes an additional downstream exon unlikely. For gbpA, the 3′ end was determined by 3′RACE (Invitrogen). The 5′ ends of the gbpA and -B ORFs were determined from cDNA clones. The 5′ ends of the gbpC and -D ORFs were assigned based on the fact that they are immediately preceded by at least 470 bp (gbpC) and 220 bp (gbpD) of AT-rich sequences typical of intergenic regions.
Total RNA Preparation and Northern Analysis.
One hundred million log-phase cells were allowed to develop on 42.5-mm Whatman No. 50 filters, as described (13). At 4 h intervals, a filter was placed in 10 ml of TRIzol Reagent (Invitrogen) and gently agitated for 5 min to dissolve the cells. The filter was removed, and the RNA was isolated according to the manufacturer's protocol. The yield decreased during development; 1.12 mg was recovered at 0 h, and 0.45 mg at 24 h. For Northern blots, 6.2 μg of total RNA was fractionated by agarose-formaldehyde gel electrophoresis, transferred to nitrocellulose, and probed as described (14). As probes, cloned DNA fragments (0.7–1.2 kb) from each gene were radioactively labeled using a rediprime II kit (Amersham Pharmacia).
Gene Disruption of gbpC.
A 874-bp portion of gbpC was amplified from genomic DNA using the primers ATCATTGATTTCGGTACAAG and TCTTCCTCTGTGAAAATATC. This fragment was cloned, and the Bsr selection cassette (15) was inserted in the same orientation as the gbpC gene into the BglI site between the kinase and the RasGEF-N domains. The resulting insert was amplified with the primers described above, and 4 μg of the PCR product was used to transform DH1 (“wild type”) cells by electroporation. Transformants were selected in medium with 10 μg/ml blasticidin. Knockouts were screened by PCR and confirmed by Southern analysis. Ninety-five percent of the transformants had a disrupted gbpC locus.
Cyclic Nucleotide Binding Assays.
Cells were starved for 4 h in 10 mM sodium phosphate (pH 6.5), washed two times with lysis buffer (40 mM Hepes/NaOH (pH 7.0)/0.5 mM EDTA), resuspended at 108 cells per ml in lysis buffer containing 0.25 M sucrose, and passed through a 0.45-μm Nuclepore filter. The resulting lysate was precleared by centrifugation for 2 min at 14,000 × g, followed by centrifugation for 1 h at 48,000 × g. Binding reactions (200 μl) contained 100 μl of high-speed supernatant, 0.5× lysis buffer, 0.125 M sucrose, 50 mM phosphate (pH 6.1), 3 mM MgCl2, 5 mM DTT, and 10 nM [3H]cGMP (with or without 250 nM cold cGMP) or 10 nM [3H]cAMP. Nonspecific binding was determined by including 30 μM cGMP or cAMP. Reactions were incubated for 15 min at 0°C and filtered through 0.45 μm of nitrocellulose, and the filters then washed two times with 50 mM phosphate (pH 6.1), dried, and analyzed by liquid scintillation counting.
Results
Identification of the gbp Genes.
We took a bioinformatics approach to identify cyclic nucleotide (cNMP)-binding proteins in the Dictyostelium genome, using low-threshold blast searches with known cAMP- and cGMP-binding proteins. We have identified four novel cNMP-binding proteins. We have designated these genes gbpA–D, and the proteins GbpA–D, for cGMP-binding proteins, because, as discussed below, they have a higher likelihood of binding cGMP than cAMP. Their domain structures are presented in Fig. 1A, and an annotated version of their genomic sequences is shown in Figs. 5–8, which are published as supporting information on the PNAS web site, www.pnas.org. Each protein has two potential cGMP-binding sites with homology to CAP, as well as one or more other domains that are discussed in more detail below. We also found one GAF domain protein (GafA), but based on the crystal structure and mutagenesis studies of a yeast GAF domain (16), this Dictyostelium protein is not predicted to bind cGMP.
Figure 1.
Four candidate cGMP-binding proteins. (A) The domain structures of the GbpA–D proteins are shown approximately to scale. The putative cGMP-binding sites are labeled “cG.” The amino acid present at a position known to correlate with cNMP specificity is indicated; this position is also shaded pink and marked with an asterisk in B. (B) The putative cGMP-binding sequences from the Gbp proteins are aligned with known cAMP-binding sites from type I bovine PKA-R (GenBank accession no. P05987) and CAP (P03020), the cGMP-binding domains from mouse PKGII (L12460), bovine cGMP-gated ion channel (X51604), and a domain from human CNrasGEF, which binds both cAMP and cGMP (KIAA0313). The two cNMP sites found in most of the proteins are distinguished by a numerical suffix. Residues are shaded black if they are identical to the consensus for each position in the alignment, and shaded gray if they are similar to the consensus. The residues thought to be involved in cNMP specificity in CAP are shaded yellow and indicated with open circles. The E-values for a Pfam cNMP domain were obtained from a Conserved Domain Search (NCBI). For GbpD.1, the E-value was obtained after manually removing the 63-aa insert indicated in the alignment.
An alignment of the putative cGMP-binding sites from the Dictyostelium Gbp proteins with known cAMP and cGMP-binding sites is presented in Fig. 1B. Overall, the Dictyostelium sites are quite divergent from those previously described, although all except the first site in GbpD are readily recognized as cNMP-binding sites in an NCBI Conserved Domain (CD) search against the Pfam protein domain collection (17). The E-values from these CD searches are reported in Fig. 1B. Lower values indicate a better match to the consensus sequence; by this measure the first site in GbpA and the second site in GbpB are the best fits to the cNMP-binding motif, but the other scores also indicate significant similarity. The first site in GbpD was identified by eye, and this sequence represents a good match to the consensus for approximately the first half of the domain; the second half is more degenerate. The second site in GbpD has a large insertion, but inspection of the PKA-R crystal structures reveals that a large loop can be accommodated at this position (18, 19).
For eukaryotic cAMP and cGMP-binding proteins, specificity is thought to be determined by the highly conserved “FGE . . . R(T/A)A” motif, where typically an “RTA” or occasionally an “RSA” is present in cGMP-binding sites, and an “RAA” is present in cAMP-binding sites (20). Crystal structures for type I and II PKA-Rs have been determined (18, 19); these structures, as well as earlier modeling studies (20), provide a structural basis for cyclic nucleotide specificity. When cGMP is superimposed on cAMP, and the RAA is replaced by RTA or RSA, the Ser or Thr side chain can accept a hydrogen bond from the amino nitrogen of the C2 carbon of cGMP. In the CAP structure, the conformation of the bound cAMP is anti, rather than syn as in PKA (21). As a result, the residues likely to be important in specificity differ in these two evolutionarily related classes of proteins. CAP is a dimer, and T127 from one subunit, together with S128 from the other are implicated in defining specificity. The regions corresponding to T127 and S128 are not conserved in GbpA–D, or in the other eukaryotic proteins (Fig. 1B), suggesting that for them this region in not involved in cNMP recognition. Also, 8-Br-cGMP, which strongly favors the syn conformation, activates cGMP targets in Dictyostelium (9, 22).
Based on these criteria, we expect that the cGMP binds to the Dictyostelium proteins in a syn conformation, and have chosen PKA, rather than CAP, in evaluating specificity in our proteins. We predict that GbpA.1, both sites in GbpB and -C, and GbpD.2 bind cGMP preferentially to cAMP, because they have a Ser or Thr at the position marked with an asterisk in Fig. 1B. GbpA.2 and GbpD.1 have very degenerate FGE . . . RTA motifs, and thus may not bind either cyclic nucleotide with high affinity.
GbpA and -B.
GbpA and -B have a metal hydrolase domain upstream of two tandem cGMP-binding sites (Fig. 2). These metal hydrolase domains are in the metallo β-lactamase (MBL) superfamily, which includes a broad range of enzymes that use metal catalytically in hydrolysis reactions (23). MBL itself has two bound Zn2+ ions that are coordinated by His and Asp residues (24). These residues are conserved in GbpA and -B, presumably reflecting shared metal-binding properties. In MBL the Zn2+ ions, together with two serines and a tyrosine, are involved in catalysis. These residues are not conserved in GbpA or -B, suggesting that these proteins have a different catalytic function.
Figure 2.
The metal hydrolase domain from GbpA and -B. The domains from GbpA and -B are aligned with DPDE, the Dictyostelium cAMP PDE (J02628), and MBL, the L1 MBL from Stenotrophomonas maltophilia (CAB63489). Residues are shaded black if they are identical to the consensus for each position in the alignment, and shaded gray if they are similar to the consensus. The conserved His and Asp residues that bind Zn2+ in the MBL structure are shaded pink and marked with an asterisk, and the Ser and Tyr residues involved in β-lactam hydrolysis by MBL are shaded yellow and indicated by a plus sign.
GbpA and -B do not show particularly strong similarity to any characterized member of the MBL superfamily, so it is not possible to predict substrate specificity on this basis. However, several cAMP PDEs are in the MBL superfamily, including one from Dictyostelium (25). Because a cGMP PDE activity that is stimulated by cGMP has been identified in Dictyostelium lysates (9), we suggest that GbpA and/or -B are cGMP-activated cGMP PDEs. The metal hydrolase domains in GbpA and -B are only 39% identical, leaving open the possibility that they catalyze different reactions.
N-Terminal Half of GbpC.
GbpC is a large (2631 aa), multidomain protein. The first three domains—i.e., the leucine-rich repeats (LRRs), Ras, and MEK kinase domains—are unique to GbpC, whereas the C-terminal half, containing the RasGEF domain and cGMP-binding sites, is very similar to GbpD. The six 22–23-aa-long LRRs in GbpC are aligned in Fig. 7B. LRRs occur in a wide variety of bacterial and eukaryotic proteins, and function by forming complexes with other proteins (26, 27). Immediately downstream of the LRRs, GbpC has a domain with homology to Ras (Fig. 3A). This domain has highest homology to a region of the Drosophila CG5483 gene product (28). The second best match in a blast search is KIAA1790, a protein deduced from a human partial cDNA clone (29). An alignment of the Ras domains from these proteins, together with the human H-Ras sequence, is shown in Fig. 3A. The catalytic residues responsible for the basal GTPase activity of Ras are present in the Dictyostelium protein, suggesting that this domain functions as a GTPase.
Figure 3.
Sequence analysis of GbpC and -D. Residues are shaded black if they are identical to the consensus for each position in the alignment, and shaded gray if they are similar to the consensus. (A) Alignment of the Ras sequences from GbpC, the Drosophila CG5483 protein (D.m., GenBank accession no. AAF55793), a human protein encoded by the KIAA1790 cDNA (H.s., BAB47419), and human H-Ras (P01112). Key residues involved in GTP hydrolysis are highlighted and marked with an asterisk. (B) Alignment of the protein kinase sequences from GbpC, transforming growth factor β-activated protein kinase 1 (Tak1, O43318), a RAF homolog from Arabidopsis thaliana (Ctr1, A45178), and mixed lineage kinase 1 (Mlk1, AAG44591). The twelve nearly invariant protein kinase residues (41) are highlighted and indicated with an asterisk. For positions that show a conserved residue in >90% of Tyr kinases, but not Ser/Thr kinases (42), the preferred amino acid is indicated above the alignment. (C) Alignment of the RasGEF catalytic domains from GbpC and -D with those from human Sos-1 (Q07889) and Saccharomyces cerevisiae cdc25 (P04821). The bar above the alignment in C indicates the helical hairpin involved in nucleotide exchange. (D) Alignment of the DEP domain from GbpC with mouse dishevelled protein (Dv1, P51141), mouse pleckstrin (Plk., AAG29513), and C. elegans Egl-10 (Egl10, P49809). Residues implicated in forming a β-hairpin arm and electric dipole are highlighted and indicated by asterisks.
For GbpC's protein kinase domain, all 12 of the nearly invariant protein kinase residues are conserved (asterisks in Fig. 3B), strongly suggesting that it has catalytic activity. Overall the GbpC kinase domain is most similar (31–34% identity) to Arabidopsis CTR1, mammalian transforming growth factor β (TGF-β)-activated kinase 1, and mixed lineage kinase 1, which are all MEKKs (mitogen-activated/extracellular signal-regulated protein kinase kinase kinase). MEKKs activate the MEK family of protein kinases, which in turn activates ERKs. Although GbpC and the other sequences in Fig. 3B are hybrid between those of Tyr and Ser/Thr kinases, MEKKs are specific for Ser/Thr residues, and GbpC presumably shares this feature.
GbpC appears to be unique to Dictyostelium; no protein with a similar domain architecture exists in the current sequence database. However, proteins are found in other organisms that are similar to the N-terminal half of GbpC ending after the kinase domain. The Drosophila CG5483 gene product, the GenBank entry most similar to GbpC's Ras domain, has a series of LRRs upstream of the Ras domain, and a protein kinase domain downstream, making its layout very similar to the N-terminal half of GbpC, and suggesting evolutionary conservation. This suggestion is strengthened by the fact that CG5483's kinase domain, like that of GbpC, is most similar to mixed lineage kinase. The human protein KIAA1790 also has a Ras domain very similar to that of GbpC with a mixed lineage protein kinase domain downstream. KIAA1790 is a partial cDNA clone that begins just upstream of the Ras domain, so despite the fact that LRRs are not found in the reported sequence, they could be present in the uncharacterized 5′ end of the gene. It appears that GbpC arose as a duplication of GbpD, followed by fusion with a CG5483/KIAA1790-like gene.
GbpD and the C-Terminal Half of GbpC.
GbpD and the C-terminal half of GbpC have similar domain structures and are predicted to have a cGMP-regulated RasGEF activity. RasGEFs activate Ras by promoting the exchange of GDP for GTP. This catalytic activity is carried out by the C-terminal RasGEF domain, whereas the RasGEF-N domain is thought to play a structural role (30). GbpC and -D both have RasGEF domains (aligned in Fig. 3C) and RasGEF-N domains (aligned in Fig. 7C). The domains found in GbpD are better matches to the consensus than are those found in GbpC. In particular, it is noteworthy that the GbpC RasGEF domain does not appear to have the helical hairpin (indicated above the alignment in Fig. 3C) that is thought to play an important role in catalytic activity (30). However, GbpC's RasGEF domain could potentially act on GbpC's own Ras domain, and this missing helical hairpin may reflect an adaptation for intramolecular signaling.
The DEP domain found immediately upstream of the RasGEF catalytic domain in GbpC is not present in GbpD, even though it is in the portion of GbpC that is similar to GbpD (Fig. 1A). DEP domains are found in a wide variety of proteins, and are named for dishevelled, Egl-10, and pleckstrin—three well studied proteins in which they are found (31). DEP domains target proteins to the membrane, but the residues involved in this function are not well defined. DEP domains also mediate protein–protein interactions, and for the dishevelled DEP domain this occurs via an electric dipole that is formed in part by a β-hairpin arm (32). The two glycines that flank the β-hairpin arm, and the one basic and two acidic residues that form the electric dipole, are conserved the GbpC DEP domain (Fig. 3D).
GbpC and -D both have a GRAM domain between their two cGMP-binding sites (highlighted in Figs. 7 and 8). This motif was named after glucosyltransferases, Rab-like GTPase activators, and myotubularins—the most well characterized proteins that contain this domain (33). The function of GRAM domains is unknown, but they are proposed to mediate protein–protein interactions (33).
GbpD and the C-terminal half of GbpC do not appear to have a direct homolog in metazoans. Although Ras and Rap1 GEFs containing one or two cNMP-binding sites have been found in mammals, Caenorhabditis elegans and Drosophila melanogaster, in each of these cases the cNMP-binding site(s) are upstream of the RasGEF domain, rather than downstream as in GbpC and -D. Each of the metazoan proteins also has a DEP domain, as is the case for GbpC, but its position in the primary sequence differs as well. The metazoan proteins also lack the GRAM domain found in GbpC and -D. Thus it appears that cyclic nucleotide-regulated GEFs evolved separately in these lineages.
gbpC-Nulls Lack a cGMP-Binding Protein.
GbpC binding to cyclic nucleotides was evaluated by assaying the soluble fraction of wild-type and gbpC− cells. Disruption of gbpC had no effect on binding of 10 nM cAMP: 242 ± 12 fmol/mg protein for wild-type, and 222 ± 20 fmol/mg protein for gbpC−. In contrast, binding of 10 nM cGMP was reduced from 221 ± 12 fmol/mg for wild type to 17.7 ± 1.8 fmol/mg for gbpC−, indicating that GbpC binds cGMP, as predicted from the sequence. The residual binding activity in gbpC− cytosol indicates that one or more additional cGMP-binding proteins are present. At 260 nM cGMP, binding increased 4-fold to 853 ± 88 fmol/mg for wild type and 22-fold to 419 ± 60 fmol/mg for gbpC−. The greater relative increase in binding in the null extract suggests that one or both GbpC-binding sites becomes saturated at approximately 10 nM, while the remaining Gbps have higher Kd values.
Developmental Expression of the gbp Genes.
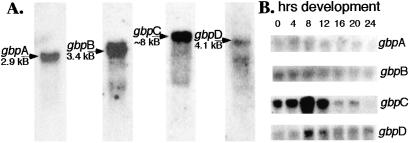
Dictyostelium development is a process rich in signaling events and cytoskeletal changes, and changes in mRNA levels for the gbp genes may reflect specific roles for these proteins in vegetative growth or during development. We used Northern blotting to determine changes in expression during development. In addition, the mRNA size determined by this analysis provides an independent measure for evaluating the gbp gene sequences presented in Fig. 5–8. gbpA is maximally expressed at 4 h (Fig. 4B). The mRNA is 2.9 kb (Fig. 4A), consistent with our mapping of the transcript by using cDNAs and 3′ RACE—a 2,607-bp ORF and 300 bp of untranslated sequence. gbpB is maximally expressed in vegetative cells as a 3.4-kb mRNA (Fig. 4), consistent with the 3,291-bp ORF and 122 bp of untranslated sequence that we observed from sequencing cDNA clones. For both gbpA and -B, the changes in gene expression levels are fairly small. In contrast, gbpC expression increases dramatically by 8 h development. The size of the gbpC mRNA is ≈8 kb, consistent with the 7,896-bp ORF that we have determined. The level of gbpD mRNA is also maximal at 8 h, although the increase is less dramatic than is observed with gbpC. The mRNA is ≈4.1 kB (Fig. 4A), consistent with a 3,929-bp ORF for gbpD and 24–68 bp of 3′ untranslated sequence, as indicated by two clones from the cDNA project.
Figure 4.
Northern analysis. (A) Total RNA from vegetative wild-type cells was probed with radioactive DNA fragments from the indicated genes. The indicated size of the transcript was calculated from RNA markers (Promega). (B) Total RNA prepared at 4-h intervals during development was probed with the same DNA fragments as in A. The number of hours of development is indicated for each lane.
Discussion
Relationship of the Gbp Proteins to cGMP Targets in Higher Eukaryotes.
We are confident that we have identified all of the candidate cGMP-binding proteins (both in the CAP and GAF families) that are represented in the current Dictyostelium sequence databases. Given the advanced stage of the genome sequencing effort, all cGMP signaling in Dictyostelium may well be mediated by GbpA–D. Remarkably, we did not find direct homologs of any of the cGMP targets that have been identified in higher eukaryotes. Rather, GbpA–D represent four previously uncharacterized proteins that in turn do not have clear homologs in higher eukaryotes. Our findings indicate that cGMP signaling evolved independently in these two lineages.
Proposed Regulatory Mechanism for GbpC and -D.
GbpC and -D are implicated in Ras signaling, because they each contain RasGEF domains and GbpC contains a Ras domain. Ras signaling in Dictyostelium is complex, as evidenced by the fact that Dictyostelium contain at least ten Ras genes (34)—a disproportionately large number compared with its genome size. Dictyostelium appears to have a similarly high number of RasGEFs; two have been described (35, 36), GbpC and -D are described here, and our searches of the Dictyostelium genomic sequence databases readily revealed at least 18 more (data not shown).
The juxtaposition of RasGEF and cGMP-binding domains in GbpD implies that the RasGEF activity is regulated by cGMP; and given that GbpC and -D likely have a common origin, our working model is that the RasGEF activity of GbpC is also regulated by cGMP. We propose that the MEKK activity of GbpC is directly regulated by its upstream Ras domain; this suggestion is based on the fact that Raf, another MEKK, is well known to be directly activated by Ras. Assuming that GbpC's own RasGEF domain activates its Ras domain, then the MEKK activity of GbpC would require cGMP, where this dependence is transmitted from the cGMP-binding domains through the RasGEF and Ras domains on the same polypeptide chain. However, in light of the multiplicity of Ras and RasGEF proteins in Dictyostelium, it is important to consider the possibility that GbpC is involved in a network of Ras signaling, rather than functioning as a signal transduction pathway within one polypeptide.
Potential Roles for GbpA–D in Chemotaxis.
cGMP signaling plays an important role in chemotaxis in Dictyostelium. We propose, based on sequence analysis, that GbpA and/or -B are cGMP PDEs, and preliminary data indicates that gbpA nulls do indeed have reduced cGMP PDE activity (L.B. and P.J.M.V.H., unpublished results). Thus GbpA, and possibly GbpB, may be involved in temporally and spatially controlling the cGMP signal in response to chemoattractants, whereas GbpC and -D would be responsible for relaying the cGMP signal downstream so that the cytoskeletal changes responsible for chemotactic movements occur. GbpC and -D likely activate a Ras during chemotaxis; perhaps GbpC's own Ras domain, or one of the other Dictyostelium Ras proteins. Given the homology of GbpC's kinase domain to MEKKs, it is likely that GbpC phosphorylates a MEK in a MAP kinase cascade. Dictyostelium appears to have at least three MAP kinase pathways (37–39). The Dictyostelium proteins ERK2 and Mek1 are both important for chemotaxis, and are in separate pathways. Given the role of cGMP signaling in chemotaxis, it follows that GbpC might activate ERK2 through an unidentified MEK, or it might phosphorylate Mek1. GbpC may also phosphorylate other types of targets. For example, myosin II activity is tightly regulated during chemotaxis, and the activation of myosin light chain kinase-A (MLCK-A) is cGMP-dependent in crude lysates (10, 40). Identification of GbpA–D as candidate downstream players in cGMP signaling paves the way to a molecular approach to studying cGMP signaling during chemotaxis in this important model organism.
Supplementary Material
Acknowledgments
We are indebted to all of the teams involved in the Dictyostelium sequencing projects. We thank Ana C. Urbin-Reyes for technical assistance, Eugene Koonin for helpful discussions, and L. Aravind for help in identifying GAF proteins. This work was supported by grants from the National Science Foundation and the American Heart Association–New England Affiliate (to J.L.S.), and a grant from the Netherlands Organization of Scientific Research (to P.J.M.V.H.).
Abbreviations
- CAP
E. coli catabolite gene activator protein
- MBL
metallo β-lactamase
- MEKK
mitogen-activated/extracellular-signal regulated protein kinase kinase kinase
- PDE
phosphodiesterase
- PKA-R
cAMP-dependent protein kinase regulatory subunit
- RasGEF
Ras guanine nucleotide exchange factor
Footnotes
References
- 1.Parent C A, Devreotes P N. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 2.Mato J M, Van Haastert P J M, Krens F A, Rhijnsburger E H, Dobbe F C P M, Konijn T M. FEBS Lett. 1977;79:331–336. doi: 10.1016/0014-5793(77)80814-4. [DOI] [PubMed] [Google Scholar]
- 3.Wurster B, Schubiger K, Wick U, Gerisch G. FEBS Lett. 1977;76:141–144. doi: 10.1016/0014-5793(77)80139-7. [DOI] [PubMed] [Google Scholar]
- 4.Roelofs J, Meima M, Schaap P, Van Haastert P J M. EMBO J. 2001;20:4341–4348. doi: 10.1093/emboj/20.16.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruth P. Pharmacol Ther. 1999;82:355–372. doi: 10.1016/s0163-7258(98)00067-9. [DOI] [PubMed] [Google Scholar]
- 6.Pham N, Cheglakov I, Koch C A, de Hoog C L, Moran M F, Rotin D. Curr Biol. 2000;10:555–558. doi: 10.1016/s0960-9822(00)00473-5. [DOI] [PubMed] [Google Scholar]
- 7.Aravind L, Ponting C P. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 8.Kuwayama H, Van Haastert P J M. J Biol Chem. 1996;271:23718–23724. doi: 10.1074/jbc.271.39.23718. [DOI] [PubMed] [Google Scholar]
- 9.Van Haastert P J M, Van Lookeren Campagne M M. J Cell Biol. 1984;98:709–716. doi: 10.1083/jcb.98.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silveira L A, Smith J L, Tan J L, Spudich J A. Proc Natl Acad Sci USA. 1998;95:13000–13005. doi: 10.1073/pnas.95.22.13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Rooij J, Zwartkruis F J, Verheijen M H, Cool R H, Nijman S M, Wittinghofer A, Bos J L. Nature (London) 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki H, Springett G M, Mochizuki N, Toki S, Nakaya M, Matsuda M, Housman D E, Graybiel A M. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 13.Sussman M. In: Dictyostelium discoideum: Molecular Approaches to Cell Biology. Spudich J A, editor. Vol. 28. Orlando: Academic; 1987. pp. 9–30. [Google Scholar]
- 14.Brown T, Mackey K. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1997. [Google Scholar]
- 15.Sutoh K. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- 16.Ho Y-S J, Burden L M, Hurley J H. EMBO J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diller T C, Madhusudan, Xuong N, Taylor S S. Structure (London) 2001;9:73–82. doi: 10.1016/s0969-2126(00)00556-6. [DOI] [PubMed] [Google Scholar]
- 19.Su Y, Dostmann W R, Herberg F W, Durick K, Xuong N H, Ten Eyck L, Taylor S S, Varughese K I. Science. 1995;269:807–813. doi: 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- 20.Shabb J B, Corbin J D. J Biol Chem. 1992;267:5723–5726. [PubMed] [Google Scholar]
- 21.Weber I T, Steitz T A. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- 22.Kuwayama H, Ecke M, Gerisch G, Van Haastert P J M. Science. 1996;271:207–209. doi: 10.1126/science.271.5246.207. [DOI] [PubMed] [Google Scholar]
- 23.Aravind L. In Silico Biol. 1999;1:69–91. [PubMed] [Google Scholar]
- 24.Ullah J H, Walsh T R, Taylor I A, Emery D C, Verma C S, Gamblin S J, Spencer J. J Mol Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 25.Lacombe M L, Podgorski G J, Franke J, Kessin R H. J Biol Chem. 1986;261:16811–16817. [PubMed] [Google Scholar]
- 26.Andrade M A, Perez-Iratxeta C, Ponting C P. J Struct Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 27.Kobe B, Deisenhofer J. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 28.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 29.Nagase T, Nakayama M, Nakajima D, Kikuno R, Ohara O. DNA Res. 2001;8:85–95. doi: 10.1093/dnares/8.2.85. [DOI] [PubMed] [Google Scholar]
- 30.Boriack-Sjodin P A, Margarit S M, Bar-Sagi D, Kuriyan J. Nature (London) 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 31.Ponting C P, Bork P. Trends Biochem Sci. 1996;21:245–246. [PubMed] [Google Scholar]
- 32.Wong H C, Mao J, Nguyen J T, Srinivas S, Zhang W, Liu B, Li L, Wu D, Zheng J. Nat Struct Biol. 2000;7:1178–1184. doi: 10.1038/82047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doerks T, Strauss M, Brendel M, Bork P. Trends Biochem Sci. 2000;25:483–485. doi: 10.1016/s0968-0004(00)01664-9. [DOI] [PubMed] [Google Scholar]
- 34.Wilkins A, Insall R H. Trends Genet. 2001;17:41–48. doi: 10.1016/s0168-9525(00)02181-8. [DOI] [PubMed] [Google Scholar]
- 35.Insall R H, Borleis J, Devreotes P N. Curr Biol. 1996;6:719–729. doi: 10.1016/s0960-9822(09)00453-9. [DOI] [PubMed] [Google Scholar]
- 36.Wilkins A, Chubb J R, Insall R H. Curr Biol. 2000;10:1427–1437. doi: 10.1016/s0960-9822(00)00797-1. [DOI] [PubMed] [Google Scholar]
- 37.Gaskins C, Clark A M, Aubry L, Segall J E, Firtel R A. Genes Dev. 1996;10:118–128. doi: 10.1101/gad.10.1.118. [DOI] [PubMed] [Google Scholar]
- 38.Ma H, Gamper M, Parent C, Firtel R A. EMBO J. 1997;16:4317–4332. doi: 10.1093/emboj/16.14.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung C Y, Reddy T B, Zhou K, Firtel R A. Genes Dev. 1998;12:3564–3578. doi: 10.1101/gad.12.22.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith J L, Silveira L A, Spudich J A. EMBO J. 1996;15:6075–6083. [PMC free article] [PubMed] [Google Scholar]
- 41.Hanks S K, Hunter T. In: The Protein Kinase FactsBook: Protein-Serine Kinases. Hardie G, Hanks S, editors. San Diego: Academic; 1995. pp. 7–47. [Google Scholar]
- 42.Hubbard S R, Wei L, Ellis L, Hendrickson W A. Nature (London) 1994;372:746–753. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.