Abstract
Selection of the division site in Escherichia coli is regulated by the min system and requires the rapid oscillation of MinD between the two halves of the cell under the control of MinE. In this study we have further investigated the molecular basis for this oscillation by examining the interaction of MinD with phospholipid vesicles. We found that MinD bound to phospholipid vesicles in the presence of ATP and, upon binding, assembled into a well-ordered helical array that deformed the vesicles into tubes. Stimulation of the MinD ATPase by addition of MinE led to disassembly of the tubes and the release of MinD from the vesicles. It is proposed that this MinE-regulated dynamic assembly of MinD underlies MinD oscillation.
A cell must identify specific locations in intracellular space to undergo morphogenesis. During the cell cycle the cytoskeletal machinery for cytokinesis must be placed precisely at midcell to generate two equal-sized progeny cells. How does the cell obtain positional information to spatially regulate the placement of the cytokinetic ring? In bacteria such as Escherichia coli at least two negative mechanisms cooperate to provide positional information to localize the cytokinetic Z ring. One mechanism involves inhibition of Z ring assembly in the vicinity of the nucleoid (1). The second mechanism consists of the min system, which prevents Z ring assembly away from midcell (2, 3). In the absence of min spatial regulation is relaxed and Z rings frequently form near the cell poles, resulting in minicells.
Spatial regulation by the min system occurs through the positioning of MinC, an antagonist of FtsZ assembly (4–6), on the membrane away from midcell (7, 8). MinC is not static, however, but oscillates between the two halves of the cell under the direction of the other two components of the min system, MinD and MinE (7, 8). Through this oscillation, MinC alternately occupies the membrane in polar zones without occupying the membrane at midcell. Thus, the midcell region is free of MinC, allowing the Z ring to assemble there.
In the absence of MinE, MinD is at the membrane all along the periphery of the cell (9). In the presence of MinE, however, MinD rapidly oscillates between polar zones (10, 11). During an oscillation cycle a polar zone of MinD recedes from midcell toward the pole as a new polar zone is formed in the other half of the cell, emanating from the pole. MinE is present as a ring that appears to be at the receding edge of the MinD polar zone during the shrinking phase although some MinE also colocalizes to the MinD polar zone (12, 13).
MinD is a peripheral membrane ATPase and a member of the large ParA family of ATPases, many of which are involved in plasmid partitioning (9). Its activity is stimulated by MinE but only in the presence of phospholipids (14). This dependency on phospholipid suggests that MinD binds directly to phospholipids and undergoes a conformational change so that it is susceptible to MinE stimulation. MinE mutants that have reduced capacity to stimulate the MinD ATPase induce a slower oscillation, providing further support for a direct coupling between MinE stimulation of MinD ATPase and MinD oscillation (14). These results have led to a model in which: (i) the ATP form of MinD binds to the membrane, (ii) membrane binding results in a conformational change in MinD such that it is susceptible to MinE-induced ATP hydrolysis, and (iii) MinD is released from the membrane after ATP hydrolysis. In the present study we have tested this model.
Several observations suggest that MinD assembles into a dynamic structure during the oscillation. First, the movement of MinD appears cooperative (10), and second, the specific activity of the MinD ATPase displays a strong dependence on the MinD concentration, implying oligomerization of MinD (14). In this study we find that ATP promotes MinD assembly on phospholipid vesicles that is reversed by MinE-stimulated ATP hydrolysis.
Experimental Procedures
Strain and Plasmids.
The E. coli K12 strain JS964 (MC1061 malP∷lacIqΔmin∷kan) was used in this study (15). The plasmids pZH106 (gfp-minD) and pZH115 (minD) have been described (14). The plasmid pZH115–16 (minDK16Q) was constructed similarly to pZH115 except that the 5′ primer was designed to alter amino acid 16 of MinD from K to Q. PCR was also used to place minD K16Q downstream of gfp under the control of the arabinose promoter in plasmid pJC106 (14) to give pZH106–16.
Fluorescence Microscopy and Cell Growth.
The method is similar to that used by Hu and Lutkenhaus (7). Cells were grown in LB at 30°C and the gfp-minD fusions were induced with 0.01% arabinose. At 1–2 h after induction a sample was placed on a microscope slide and photographed with a Nikon Optiphot fluorescence microscope with a MagnaFire charge-coupled device camera from Optronics International, Chelmsford, MA. Images were imported into Adobe photoshop for assembly.
Protein Purification and Preparation of E. coli Phospholipid Vesicles.
The purification of MinD and MinE has been described (14). MinDK16Q was purified by the same procedure used for the wild-type protein, and it behaved very similarly during purification. E. coli phospholipids were purchased from Avanti Polar Lipids, and phospholipid vesicles (small unilamellar vesicles) were prepared by sonication. The E. coli phospholipids were suspended on ice in buffer A (25 mM Tris⋅HCl, pH 7.5/40% glycerol), followed by rapid 50-fold dilution in buffer B (25 mM Tris⋅HCl, pH 7.5/50 mM KCl). After a 30-min incubation on ice, phospholipid vesicles were collected by centrifugation for 20 min at 36,500 rpm. The vesicles were suspended at 2 mg/ml, sonicated six times for 10 min each, and stored at −80°C until used. The diameters of the phospholipid vesicles ranged from 0.1 to 2 μm as determined by electron microscopy (EM) and negative staining (see Fig. 2A).
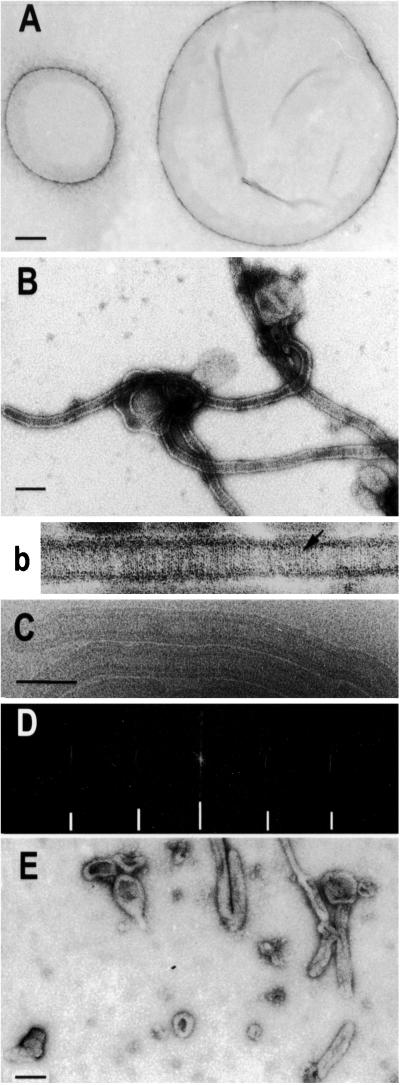
Figure 2.
MinD tubulates vesicles in the presence of ATP. (A) Negative stain image of phospholipid vesicles. The diameter of the vesicles ranged from 0.1 to 2 μm. (B and b) Negative stain image of MinD tubes. A reaction mixture containing MinD (6 μM), phospholipid vesicles (40 μg/ml), and 1 mM ATP was incubated for 10 min at 30°C and samples were taken for EM and negative staining. The tube in b is at a higher magnification and striations are visible (see arrow). (C) Cryo-EM of MinD tubes. Tubes prepared as in B were visualized by cryo-EM. (D) The computed diffraction pattern of a tube visualized by cryo-EM indicates the tubes are well ordered and helical. The layer lines are indicated by hatch marks and are at 1/59 and 1/28.5 Å−1. Similar layer lines were obtained for tubes visualized by negative staining. (E) MinE causes disassembly of MinD tubes. MinE (6 μM) was added to MinD tubes prepared as in B and samples were taken 15 min later for EM. (Scale bars: 100 nm.)
MinD ATPase and ATP Binding Assays.
The MinD ATPase assay was essentially as described (14). MinD (6 μM), phospholipid vesicles (40 μg/ml), and 1 mM (γ-32P) ATP (all are final concentrations) were mixed in 50 μl of buffer C (25 mM Tris⋅HCl, pH 7.5/50 mM KCl/5 mM MgCl2) and incubated at 30°C for 15 min. MinE was added to the reaction mixture at various concentrations. To assess ATP binding to MinD and MinD K16Q we followed the procedure described for ATP binding to FtsA (16).
EM of the Interaction of MinD with Phospholipid Vesicles.
In the standard reaction MinD (6 μM), phospholipid vesicles (40 μg/ml), and nucleotide (1 mM ATP) were mixed at room temperature in 25 μl of buffer C. The reaction was incubated at 30°C for 5–10 min. Ten-microliter samples were taken for EM and negative staining. To assay MinE's effect on MinD tubes, the above MinD mixture, but containing 0.5 mM ATP, was incubated at 30°C for 5–10 min before addition of MinE. Incubation was continued at 30°C for 30 min and a sample was taken for EM.
Negatively stained samples were prepared by adsorption of ≈5–10 μl of specimen to glow-discharged carbon-coated grids, followed by brief washes with water and unbuffered 2% uranyl acetate, and then dried. For cryo-EM, aliquots were applied to grids coated with perforated carbon films (Quantifoil) and blotted immediately before plunging into liquified ethane cooled with liquid nitrogen. These samples were stored in liquid nitrogen until examined. Minimal-dose recording of both negatively stained and unstained frozen-hydrated specimens was done with a JEOL 1200EX microscope, using a Gatan cryo-transfer system for cryo-EM. In this case, images were recorded at magnifications of 40,000–60,000 at doses of 10–20 e−/Å2 on Kodak SO163 film developed for maximal electron speed.
Images were selected for analysis by optical diffractometry and digitized with a HiScan drum scanner (San Marcos, TX) by using a primary resolution of 30 μm on the film, for a pixel of 5–7.5 Å on the specimen. Areas exhibiting straight tubes and clear striations were windowed, minimizing background carbon or surrounding ice, and power spectra (squared Fourier transforms) were calculated by using the spider suite of image processing programs (17).
Sedimentation Assay for MinD Binding to Phospholipid Vesicles.
For the sedimentation assay the reaction conditions were the same as for the EM assay except that the reaction volume was increased from 25 to 100 μl. In various assays the MinD concentration varied from 1.5 to 24 μM and the phospholipid concentration was varied from 20 to 160 μg/ml. After incubation at 30°C for 10 min samples were centrifuged at 80,000 rpm at 30°C in a Beckman TLA 100.2 rotor. After centrifugation the supernatants were carefully removed and the pellets were resuspended in 100 μl of SDS-sample buffer. A 20-μl aliquot of the sample was electrophoresed on 12.5% or 15% SDS/PAGE and stained with Coomassie brilliant blue, and the bands were quantitated with digital imaging equipment from Alpha Innotech (San Leandro, CA).
Light Scattering Assay for Disassembly of MinD Tubes.
Disassembly of MinD tubes was monitored by 90° angle light scattering in a Hitachi fluorometer (model F-3010) with both the excitation and emission wavelengths set at 350 nm and a slit width of 1.5 nm. MinD (6 μM), phospholipid (120 μg), and ATP (1 mM) were mixed in buffer C in a final volume of 300 μl. The mixture was incubated at 30°C for 15 min. MinE was then added and the change in light scattering was plotted.
Results
MinD Binds to Phospholipid Vesicles in the Presence of ATP.
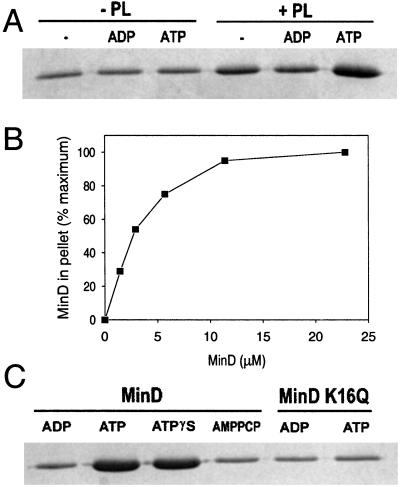
To investigate the possibility that MinD interacts directly with phospholipid we used a sedimentation assay in which proteins associated with phospholipid vesicles are pelleted. Purified MinD (6 μM) was incubated with or without phospholipid vesicles (40 μg/ml) and nucleotides (1 mM) and centrifuged. Fig. 1A contains the gel analysis of the pellets from a typical assay. In the absence of phospholipid vesicles some MinD was in the pellet, but the amount was the same whether ADP, ATP, or no nucleotide was added. The pellets represent the background in this assay (the pellets are not washed after centrifugation). In the presence of phospholipid vesicles significantly more MinD was pelleted with ATP than ADP. In the absence of nucleotide some MinD was associated with the vesicles, presumably because of nonspecific association. The binding of MinD to phospholipid vesicles was saturable at a molar ratio of MinD to phospholipid of ≈1:25 (Fig. 1B).
Figure 1.
ATP regulates MinD binding to phospholipid vesicles. (A) MinD cosediments with phospholipid vesicles in the presence of ATP. Phospholipid vesicles (small unilamellar vesicles) were assembled from purified E. coli phospholipids by sonication. MinD (6 μM) was incubated with or without phospholipid vesicles (40 μg/ml) in the absence of nucleotide or with the addition of ADP or ATP. After 5 min incubation at 30°C the samples were centrifuged and the pellets were analyzed by SDS/PAGE on a 12.5% gel. (B) Saturable binding of MinD to phospholipid vesicles. MinD at various concentrations was incubated with phospholipid vesicles (40 μg/ml) and ATP and centrifuged, and the amount in the pellet was determined. The maximum amount of MinD in the pellet was set at 100%, which corresponded to 2.6 μM. (C) Analysis of MinD binding to membranes in the presence of the nonhydrolyzable analogues of ATP, ATPγS, and AMPCP. Also, the binding of MinD K16Q to phospholipid vesicles was determined. After incubation of MinD or MinD K16Q with phospholipid vesicles and nucleotide for 5 min at 30°C samples were centrifuged and the pellets were analyzed by SDS/PAGE.
The nonhydrolyzable analogue ATPγS [adenosine-5-O-(3-thiotriphosphate)] also supported MinD binding to phospholipid vesicles, almost to the same extent as ATP (Fig. 1C). We infer that MinD binding does not require ATP hydrolysis. Another ATP analogue, AMPPCP (adenylyl [β,γ-methylene]diphosphonate), did not promote MinD binding to the membrane (Fig. 1C). We infer that only ATPγS is capable of inducing a conformational change in MinD that has a high affinity for phospholipid vesicles.
MinD Assembles on Phospholipid Vesicles Forming Tubes.
The interaction of MinD with phospholipid vesicles was also analyzed by EM. Incubation of MinD with phospholipid vesicles and ATP resulted in the dramatic conversion of vesicles to tubes with a diameter of 50–100 nm (Fig. 2 A and B). Often several tubes were observed emanating from the same vesicle and occasional branching was observed. In higher magnification (Fig. 2B) striations running perpendicular to the long axis of the tubes were visible, suggesting that MinD assembled into a polymer on the vesicles causing constriction into tubes. Images were also obtained by cryo-EM (Fig. 2C). In these images the phospholipid bilayer surrounding the lumen of the tube is clearly visible. The analysis of computed diffraction patterns of images from both negative staining and cryo-EM revealed that MinD formed a well-ordered helical lattice with a lateral spacing of 59 Å (Fig. 2D).
Tube formation occurred at phospholipid concentrations ranging from 20 to 160 μg/ml. In contrast, tube formation was very sensitive to the MinD concentration. Tube formation was readily observed at MinD concentrations of 6 μM or more; however, at 3 μM tubes were only rarely observed. At 1.5 μM no tubes were observed. At these lower MinD concentrations MinD bound to the vesicles (Fig. 1B) but was insufficient to drive tube formation. Thus, tube formation required two steps. The first step involved MinD-ATP binding to phospholipid vesicles, followed by MinD-ATP self-assembly. MinD has to be at or above a critical concentration (3.0 μM) to drive the self-association necessary to form tubes.
Evidence for two distinct steps in tube formation also comes from analysis of the effects of ATP analogues. The formation of MinD tubes strictly depended on the addition of ATP; no tubes were observed with ADP, AMPPCP, or in the absence of nucleotide, which do not support MinD binding to vesicles. Also, no tubes were formed in the presence of ATPγS, although this analogue promoted MinD binding to phospholipid vesicles. This finding raised the possibility that tube formation required a basal level of ATP hydrolysis. However, tube formation occurred just as readily at 0°C (on ice) as at 30°C, suggesting ATP hydrolysis was not required (data not shown). We suggest that ATPγS does not fully mimic ATP as a ligand for MinD (see Discussion).
MinD K16Q Is Deficient in Tube Formation and Binding to Phospholipid Vesicles.
Several mutations have been described that alter residues within the ATP binding pocket of MinD and inactivate MinD function (9, 18). One such mutation results in a K16Q substitution in the Walker A motif. Our initial characterization of MinD K16Q revealed that it had less that 50% of the wild-type basal ATPase activity and that this was not stimulated by MinE (data not shown). However, this mutant protein bound ATP as well as wild-type MinD, demonstrating that the deficiency in hydrolysis was not caused by a lack of ATP binding (data not shown). Incubation of MinD K16Q with phospholipid vesicles and ATP, however, did not lead to tube formation. We therefore investigated binding to phospholipid vesicles. As shown in Fig. 1C MinD K16Q did not bind to phospholipid vesicles in the presence of ATP, thus explaining its inability to form tubes. We infer that this mutant is unable to adopt the conformational change in response to ATP binding, which results in high affinity for phospholipid vesicles. Thus, this mutant behaves similarly to wild-type MinD in the presence of AMPPCP or ADP.
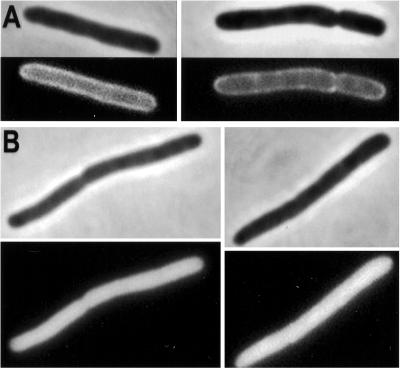
The results of the in vitro assays indicated that the MinD K16Q protein is unable to bind to the membrane. To see whether this was the case in vivo green fluorescent protein (GFP)-MinD K16Q was expressed in a Δmin strain. Cells expressing GFP-MinD had a typical halo appearance in fluorescence microscopy, indicating that GFP-MinD was at the membrane (Fig. 3A). In contrast, in cells expressing GFP-MinD K16Q the fluorescence was uniformly distributed throughout the cell (Fig. 3B). These results confirm that MinD K16Q is deficient in interaction with the membrane in vivo.
Figure 3.
Localization of MinD K16Q in vivo. The localization of GFP-MinD was compared with GFP-MinD K16Q. (A) JS964 (Δmin) containing pZH106 (gfp-minD) was induced for 2 h with 0.01% arabinose and photographed. Two cells are displayed. (B) JS964 (Δmin) containing pZH106–16 (gfp-minD K16Q) was induced with 0.01% arabinose and examined by fluorescence microscopy 2 h later. Two cells are displayed. Immunoblot analysis demonstrated that the level of GFP-MinD and GFP-MinD K16Q were similar at 2 h after induction (data not shown).
MinD Tubes Disassemble in the Presence of MinE.
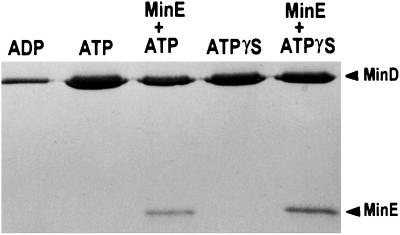
The effect of stimulating MinD ATPase by MinE on the interaction of MinD with phospholipid vesicles was examined by sedimentation. Addition of MinE decreased the amount of MinD recovered in the pellet by more than 50% (Fig. 4). In contrast, the addition of MinE had little effect on the amount of MinD in the pellet when the nonhydrolyzable analogue, ATPγS, was used. These results argue that stimulating MinD's ATPase activity results in release of MinD from the membrane. Interestingly, MinE was also present in the pellet in proportion to the amount of MinD. Only a background level of MinE was in the pellet in the absence of MinD (data not shown). These results confirm that MinE binds to MinD as previously inferred from genetic studies and yeast two-hybrid results (2, 19).
Figure 4.
MinE induces release of MinD from phospholipid vesicles. MinD (6 μM) was mixed with phospholipid (40 μg/ml) and 0.5 mM ATP to assemble tubes. MinE was added and incubated for 30 min. Samples were taken and the amount of MinD bound to membrane was determined by sedimentation and SDS/PAGE. In a second reaction ATPγS was used in place of ATP.
Because MinE stimulates MinD's ATPase activity and causes its release from the membrane we would also expect MinE to cause disassembly of MinD tubes. EM revealed that preformed MinD tubes were largely disassembled by 15 min after MinE addition (Fig. 2E). Instead of structured tubes, mostly vesicles were observed, indicating that the structured tubes mostly disassembled upon ATP hydrolysis. Some elongated vesicles, which may be tubes in the process of disassembly, are still visible.
The kinetics of MinD tube disassembly were followed by light scattering as was previously done with tubes formed by dynamin (20). Dynamin assembles on phospholipid vesicles to form tubes. Upon the addition of GTP the tubes vesiculate, which is accompanied by a decrease in the light scattering signal. Fig. 5A shows that light scattering signal obtained from preassembled tubes is stable in the absence of MinE, consistent with the tubes being stable. The stability of tubes was also verified by EM (data not shown).
Figure 5.
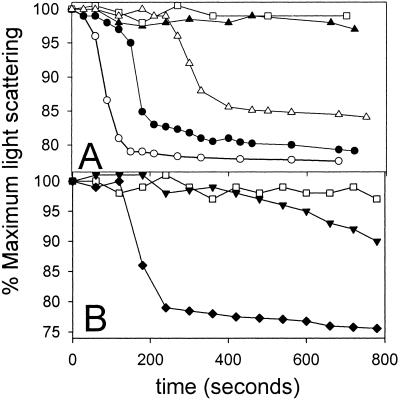
MinE induces tube disassembly as monitored by light scattering. Tubes were prepared as in Fig. 2B. A sample (300 μl) of the preassembled tubes was placed in a Hitachi fluorometer (model F-3010) and light scattering was monitored at 90°. (A) At 0 time MinE was added and the decrease in light scattering was measured. The MinE concentration was as follows: □, no MinE; ▴, 0.44 μM; ▵, 1.1 μM; ●, 2.2 μM; and ○, 4.4 μM. (B) □, no MinE; ♦, MinE (2.2 μM); ▾, MinE4 (2.2 μM) was added at 0 time to tubes assembled as in A and the change in light scattering was monitored.
The addition of MinE led to a cooperative decrease in the light scattering signal after a lag. The length of the lag was inversely proportional to the MinE concentration. At 4.4 μM MinE the tubes disassembled within 3 min whereas at 0.44 μM MinE the light scattering did not start to decrease for >10 min. However, once the signal started to decrease the rate appeared independent of the MinE concentration. These results suggest that ATP hydrolysis leads to cooperative disassembly of the tubes. Control experiments demonstrated that addition of MinE to phospholipid vesicles did not display any change in light scattering. To further examine the role of MinE-stimulated ATPase we used a MinE mutant that we previously isolated (14). This mutant, MinE4, is unable to stimulate MinD oscillation in vivo. In vitro it is deficient in stimulation of the MinD ATPase and therefore would not be expected to affect the light scattering. Indeed, the light scattering assay revealed that MinE4 was less efficient that the wild type at inducing a decrease in the light scattering (Fig. 5B). This result indicates that MinE4 is deficient in inducing tube disassembly, which was confirmed by EM (data not shown). This result further supports the conclusion that disassembly of MinD tubes requires MinE-induced ATP hydrolysis.
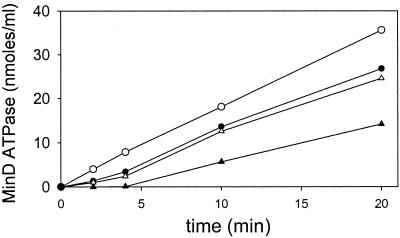
We also examined the kinetics of the ATPase activity of MinD under these conditions (Fig. 6). We observed that the MinD ATPase displayed a lag that was inversely proportional to the MinE concentration. The lag was 4.5 min when the molar ratio of MinE to MinD was 0.08. However, there was no perceptible lag when the molar ratio of MinE to MinD was increased to 0.8.
Figure 6.
MinE stimulation of MinD ATPase activity displays a lag. MinD tubes were assembled as in Fig. 5A. At 0 time MinE was added and the ATPase activity was determined by measuring the release of 32Pi. The MinE concentrations are as follows: ▴, 0.44 μM; ▵, 1.1 μM; ●, 2.2 μM; and ○, 4.4 μM. The basal activity observed in the absence of MinE was subtracted from the values before plotting.
Discussion
MinD undergoes a dramatic oscillation between the two poles of the cell in the presence of MinE; however, the molecular mechanism of this oscillation is unknown (10). Previously, we reported that MinE stimulated MinD ATPase in the presence of phospholipids and provided evidence that this stimulation was required for the oscillation (14). In this study we report that in the presence of ATP MinD binds to phospholipid vesicles and undergoes self-assembly, constricting the vesicles into tubes. Furthermore, these tubes are dynamic, disassembling upon stimulation of the MinD ATPase by MinE. This reversible assembly of MinD on vesicles, regulated by ATP and MinE, is likely to be the mechanism that underlies the oscillation of MinD.
Assembly of MinD on Phospholipid Vesicles: Role of ATP.
Previously (14), we argued that the ATP form of MinD underwent self-assembly at the membrane. The argument was based on the observations that: (i) MinD is at the membrane in the absence of MinE, the activator of the MinD ATPase (9, 14); (ii) the specific activity of the MinD ATPase shows a strong dependence on the MinD concentration, typical of self-interacting systems (14); and (iii) the movement of MinD during an oscillation cycle appears cooperative (10). The present results confirm this suggestion as we demonstrate that MinD undergoes a concentration- and ATP-dependent assembly on phospholipid vesicles to form tubes with a well-ordered helical lattice. Furthermore, the dependence of tube formation on the MinD concentration is the same as previously observed for stimulation of the MinD ATPase (14), arguing that assembly of MinD into an oligomeric species is required for ATPase activity.
Assembly of MinD into a polymer depended on a phospholipid surface, as no assembly of MinD was observed in the absence of phospholipid vesicles. The pitch of the MinD helix on the tubes was determined at 59 Å, a distance that can accommodate the long axis of a MinD monomer [a MinD monomer can be approximated as a rectangular box with a height of 57 Å and a length and depth of 35 Å; determined from Hayashi et al. (18)]. Such a packed array of MinD cannot occur in vivo as there is insufficient MinD to coat half of the inner surface of the cell membrane (21). However, we suggest that the formation of MinD polar zones observed in vivo is caused by MinD assembly into polymers on the membrane. The existence of these polymers and how they are arranged within these polar zones remains to be determined. We also do not know why MinD assembly into a polar zone during the oscillation appears to initiate from the pole.
The results of this study clearly indicate two distinct steps in the formation of MinD tubes (Fig. 7). The first step involves MinD binding to phospholipid vesicles. Both ATP and ATPγS are able to support this step although another ATP analogue, AMPPCP, does not. Interestingly, the structures of MinD with ADP or AMPPCP bound are identical, arguing that for MinD AMPPCP is a poor analogue of ATP (18). Also, we observed that MinD K16Q-bound ATP but was unable to bind to phospholipid vesicles. We suggest that this amino acid substitution alters MinD such that it does not respond to ATP binding.
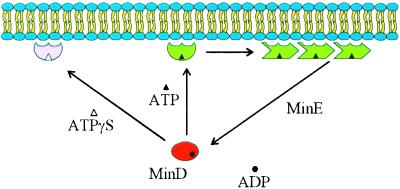
Figure 7.
Model for dynamic association of MinD with the membrane. In this model MinD undergoes a conformational change upon binding ATP, causing it to associate with the membrane. Binding to the membrane causes an additional conformational change that favors the self-assembly of MinD into polymers. Interaction of MinD polymers with MinE leads to ATP hydrolysis and release of MinD. ATPγS is able to induce the first conformational change but MinD-ATPγS is unable to undergo the second change that would lead to assembly. MinD K16Q in the presence of ATP behaves similarly to MinD in the presence of ADP or AMPPCP (not diagrammed).
The second step in tube formation is the self-assembly of the vesicle-associated MinD into a polymer that constricts the vesicle. This step is very dependent on the MinD concentration and displays a critical concentration around 3 μM, which is similar to the estimated in vivo concentration (9). We suggest a model in which ATP induces a conformational change in MinD, leading to a higher affinity for phospholipid vesicles. The binding of MinD to phospholipid vesicles then leads to a second conformational change, favoring MinD assembly (Fig. 7). MinD self-association has also been observed in the yeast two-hybrid system (22). We suggest that MinD ATPγS undergoes the first conformational change leading to higher affinity for phospholipid vesicles, but is unable to undergo the second conformational change in response to association with vesicles. Structures of MinD with ATPγS and ATP should be quite revealing as to the conformational changes necessary for MinD to bind to membranes and undergo self-assembly.
Role of MinE Stimulation of MinD ATPase and Implications for MinD Oscillation.
From our previous study (14) we suggested that MinE stimulation of MinD ATPase resulted in dissociation of MinD from the membrane. This possibility would provide a mechanism for MinE's role in stimulating the oscillation of MinD (10). In this study we observed that MinE induced disassembly of MinD tubes and reduced the amount of MinD associated with phospholipid vesicles. The release of MinD from the membrane after MinE-induced ATP hydrolysis is consistent with the different affinities we observed for the ADP and ATP forms of MinD for phospholipid vesicles.
Interestingly, both the stimulation of MinD ATPase and disassembly of the tubes displayed a lag that was inversely proportional to the MinE concentration. The existence of this lag suggests that MinE has to overcome a kinetic barrier for activation of the MinD ATPase. Such a barrier may result from self association of MinE [MinE is a dimer (23)] or MinE having to nucleate assembly on MinD filaments. However, the cosedimentation of some MinE with MinD, ATP and phospholipid vesicles suggests that stimulating ATP hydrolysis may be the rate-limiting step. Regardless of the mechanism, it is likely that this lag contributes to the timing of MinD oscillation in vivo. The estimated in vivo concentration for MinE is just below 1 μM, indicating that the lag could be significant (24).
With other nucleotide-dependent assembly systems, such as tubulin and actin, nucleotide hydrolysis is coupled to assembly. For example, with tubulin and FtsZ, the catalytic site is comprised of a sandwich of two subunits (25–27). In contrast, with MinD nucleotide hydrolysis is not coupled to assembly, but instead depends on MinE. This dependency on a second protein provides a mechanism for MinD assembly to be spatially separated from MinD disassembly.
Our model for MinD oscillation is based on the ability of MinE to stimulate the MinD ATPase in the presence of phospholipids (14) and the reported behavior of GFP fusions to MinD and MinE (12, 13). In the model we suggested that MinD oligomerizes and binds to the membrane in the presence of ATP (14). In this study we have confirmed that MinD binds to phospholipid vesicles and is in an oligomerized state. This oligomerization is cooperative and consistent with the cooperative behavior of MinD movement in vivo.
In our oscillation model (14) we also suggested that MinD movement was initiated by MinE stimulation of MinD ATPase, releasing MinD from the membrane. In this study we demonstrated that MinD release from phospholipid vesicles required stimulation of ATP hydrolysis by MinE. In the model the oscillation results, in part, from the different concentrations of MinD and MinE. With MinD in excess, MinE is occupied at one pole for some time in removing MinD. As MinD is released it diffuses and assembles in the other cell half. Only when MinE reaches the pole and has dislodged all of the MinD is it released. It then reassembles at the growing end of the new MinD polar zone. Why it reassembles at the end of the MinD zone is not clear, although it was suggested that MinD assembled into polymers and that MinE primarily assembled at the ends (14).
Recently, computer simulations have been done to mimic MinD oscillation (28–30). Interestingly, in all simulations oscillation requires only MinD, MinE, and the membrane; no prelocalized components are necessary. In these simulations it is assumed that MinD binds to the membrane and is displaced by MinE, assumptions that we have confirmed in this study. It was also assumed that MinD recruits MinE to the membrane for which there is good in vivo evidence (31). Here we show that MinE can be cosedimented with MinD and phospholipids, demonstrating that MinE is bound to MinD in the presence of phospholipids. In those simulations, as well as our model, oscillation results from the membrane release of MinE lagging the release of MinD.
MinD Family.
MinD is a member of the large ParA family of ATPases (32, 33). The prototype of this family is ParA, which is required for partitioning of plasmid P1 (34). An additional member, Soj is involved in regulating the initiation of sporulation in Bacillus subtilis (35). Interestingly, at least two of these members, Soj and ParA of pB171, undergo an oscillation on the time scale of minutes (36–38). It is likely that reversible polymerization also underlies the cooperative movement of these proteins. Soj is of particular interest as it oscillates between the poles of the cell and therefore may resemble MinD in undergoing reversible assembly on the membrane regulated by ATP.
The min System.
The min system is composed of three components that spatially regulate cell division in E. coli (2). With this work the biochemical activities of these three components can now be summarized. MinC is a division inhibitor that antagonizes FtsZ assembly (4–6). It is composed of two domains: an N-terminal domain that interacts with FtsZ and a C-terminal domain responsible for dimerization and binding to MinD (19, 39, 40). MinD and MinE function to spatially regulate MinC by causing it to rapidly oscillate between polar zones without occupying midcell (7, 8). As we show in this study, the formation of polar zones of MinD is likely to involve the ATP-dependent assembly of MinD at the membrane. MinE stimulates MinD ATPase activity, dislodging MinD, and therefore MinC, from the membrane in such a way as to ensure oscillation of MinD.
Acknowledgments
We thank Amit Mukherjee for helpful discussions. This work was supported by Public Health Service Grant GM 29764.
Abbreviations
- EM
electron microscopy
- GFP
green fluorescent protein
- ATPγS
adenosine 5-O-(3-thiotriphosphate)
- AMPPCP
adenylyl [β,γ-methylene]diphosphonate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Yu X C, Margolin W. Mol Microbiol. 1999;32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]
- 2.de Boer P A, Crossley R E, Rothfield L I. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 3.Bi E, Lutkenhaus J. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. Proc Natl Acad Sci USA. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Z, Lutkenhaus J. J Bacteriol. 2000;182:3965–3971. doi: 10.1128/jb.182.14.3965-3971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichoff S, Lutkenhaus J. J Bacteriol. 2001;183:6630–6635. doi: 10.1128/JB.183.22.6630-6635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Lutkenhaus J. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 8.Raskin D M, de Boer P A. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer P A, Crossley R E, Hand A R, Rothfield L I. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raskin D M, de Boer P A. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowland S L, Fu X, Sayed M A, Zhang Y, Cook W R, Rothfield L I. J Bacteriol. 2000;182:613–619. doi: 10.1128/jb.182.3.613-619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale C A, Meinhardt H, de Boer P A. EMBO J. 2001;20:1563–1572. doi: 10.1093/emboj/20.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu X, Shih Y L, Zhang Y, Rothfield L I. Proc Natl Acad Sci USA. 2001;98:980–985. doi: 10.1073/pnas.031549298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z, Lutkenhaus J. Mol Cell. 2001;7:1337–1343. doi: 10.1016/s1097-2765(01)00273-8. [DOI] [PubMed] [Google Scholar]
- 15.Pichoff S, Vollrath B, Touriol C, Bouche J P. Mol Microbiol. 1995;18:321–329. doi: 10.1111/j.1365-2958.1995.mmi_18020321.x. [DOI] [PubMed] [Google Scholar]
- 16.Feucht A, Lucet I, Yudkin M D, Errington J. Mol Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- 17.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi I, Oyama T, Morikawa K. EMBO J. 2001;20:1819–1828. doi: 10.1093/emboj/20.8.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Cao C, Lutkenhaus J. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sweitzer S M, Hinshaw J E. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 21.Rothfield L I, Shih Y L, King G. Cell. 2001;106:13–16. doi: 10.1016/s0092-8674(01)00432-9. [DOI] [PubMed] [Google Scholar]
- 22.Szeto J, Ramirez-Arcos S, Raymond C, Hicks L D, Kay C M, Dillon J A. J Bacteriol. 2001;183:6253–6264. doi: 10.1128/JB.183.21.6253-6264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King G F, Rowland S L, Pan B, Mackay J P, Mullen G P, Rothfield L I. Mol Microbiol. 1999;31:1161–1169. doi: 10.1046/j.1365-2958.1999.01256.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Rowland S, King G, Braswell E, Rothfield L I. Mol Microbiol. 1998;30:265–273. doi: 10.1046/j.1365-2958.1998.01059.x. [DOI] [PubMed] [Google Scholar]
- 25.Nogales E, Downing K H, Amos L A, Lowe J. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee A, Saez C, Lutkenhaus J. J Bacteriol. 2001;183:1–8. doi: 10.1128/JB.183.24.7190-7197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheffers D J, de Wit J G, Den Blaauwen T, Driessen A J. Biochemistry. 2002;41:521–529. doi: 10.1021/bi011370i. [DOI] [PubMed] [Google Scholar]
- 28.Meinhardt H, de Boer P A. Proc Natl Acad Sci USA. 2001;98:14202–14207. doi: 10.1073/pnas.251216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howard M, Rutenberg A D, de Vet S. Phys Rev Lett. 2001;87:278102-1–278102-4. doi: 10.1103/PhysRevLett.87.278102. [DOI] [PubMed] [Google Scholar]
- 30.Kruse K. Biophys J. 2002;82:618–627. doi: 10.1016/S0006-3495(02)75426-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raskin D M, de Boer P A. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 32.Yamaichi Y, Niki H. Proc Natl Acad Sci USA. 2000;97:14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayes F. Mol Microbiol. 2000;37:528–541. doi: 10.1046/j.1365-2958.2000.02030.x. [DOI] [PubMed] [Google Scholar]
- 34.Abeles A L, Friedman S A, Austin S J. J Mol Biol. 1985;185:261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- 35.Grossman A D. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 36.Quisel J D, Lin D C, Grossman A D. Mol Cell. 1999;4:665–672. doi: 10.1016/s1097-2765(00)80377-9. [DOI] [PubMed] [Google Scholar]
- 37.Marston A L, Errington J. Mol Cell. 1999;4:673–682. doi: 10.1016/s1097-2765(00)80378-0. [DOI] [PubMed] [Google Scholar]
- 38.Ebersbach G, Gerdes K. Proc Natl Acad Sci USA. 2001;98:15078–15083. doi: 10.1073/pnas.261569598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szeto T H, Rowland S L, King G F. J Bacteriol. 2001;183:6684–6687. doi: 10.1128/JB.183.22.6684-6687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordell S C, Anderson R E, Lowe J. EMBO J. 2001;20:2454–2461. doi: 10.1093/emboj/20.10.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]