Abstract
The essential, rapamycin-sensitive TOR kinases regulate a diverse set of cell growth-related readouts in response to nutrients. Thus, the yeast TOR proteins function as nutrient sensors, in particular as sensors of nitrogen and possibly carbon. However, the nutrient metabolite(s) that acts upstream of TOR is unknown. We investigated the role of glutamine, a preferred nitrogen source and a key intermediate in yeast nitrogen metabolism, as a possible regulator of TOR. We show that the glutamine synthetase inhibitor l-methionine sulfoximine (MSX) specifically provokes glutamine depletion in yeast cells. MSX-induced glutamine starvation caused nuclear localization and activation of the TOR-inhibited transcription factors GLN3, RTG1, and RTG3, all of which mediate glutamine synthesis. The MSX-induced nuclear localization of GLN3 required the TOR-controlled, type 2A-related phosphatase SIT4. Other TOR-controlled transcription factors, GAT1/NIL1, MSN2, MSN4, and an unknown factor involved in the expression of ribosomal protein genes, were not affected by glutamine starvation. These findings suggest that the TOR pathway senses glutamine. Furthermore, as glutamine starvation affects only a subset of TOR-controlled transcription factors, TOR appears to discriminate between different nutrient conditions to elicit a response appropriate to a given condition.
The Saccharomyces cerevisiae targets of rapamycin TOR1 and TOR2 are phosphatidylinositol kinase-related protein kinases that activate cell growth in response to nutrient availability (1). A TOR deficiency or rapamycin treatment causes a nutrient stress response, including inhibition of translation initiation, arrest in the G1 phase of the cell cycle, glycogen accumulation, autophagy, down-regulation of glycolysis, rRNA, tRNA, and r-protein genes, and up-regulation of tricarboxylic acid (TCA) cycle genes (1–8). Nitrogen is a particularly important nutrient in TOR signaling (7–10). Of the several hundred genes affected when TOR function is inhibited by rapamycin, the most affected are the nitrogen-regulated genes involved in the assimilation of alternative nitrogen sources (7, 8, 10). Furthermore, TOR controls the phosphorylation and presumably the activity of the nitrogen-regulated kinase NPR1 (11).
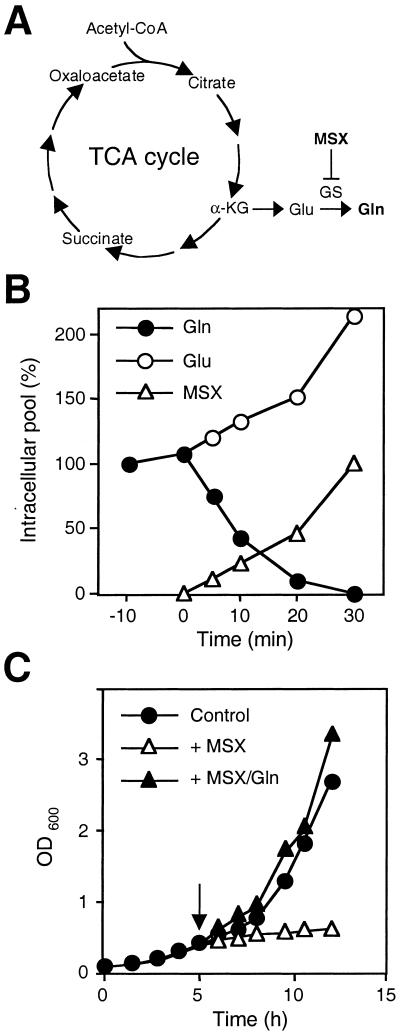
Among the wide variety of nitrogenous compounds used by yeast cells, glutamine is the preferred nitrogen source (reviewed in ref. 12). Glutamine is a key intermediate in nitrogen metabolism, required for the biosynthesis of purine and pyrimidine nucleotides and other nitrogen-containing molecules such as NAD+ (12). In yeast cells, synthesis of glutamine is catalyzed exclusively by glutamine synthetase (GS) (Fig. 1A), encoded by the GLN1 gene (13). Cells grown in the absence of glutamine up-regulate expression of GLN1. How yeast cells sense glutamine is unknown.
Figure 1.
(A) Schematic drawing of the TCA cycle and metabolic reactions for the synthesis of glutamine. α-KG, α-ketoglutarate. (B) Effect of MSX on the size of the intracellular pools of glutamine and glutamate. Wild-type (JK9–3da) cells were grown in SD and treated with MSX. Time refers to MSX treatment. MSX was added at 0 min. A 100% glutamine pool corresponds to 13.1 μmol/g of protein; a 100% glutamate pool corresponds to 42.4 μmol/g of protein. Open triangles refer to the intracellular accumulation of MSX. A 100% MSX pool corresponds to 19.9 μmol/g of protein. (C) Effect of the GS inhibitor MSX on cell growth. Wild-type (JK9–3da) cells were grown in SD and treated with MSX (2 mM) (▵) or simultaneously with MSX and glutamine (5 mM) (▴). Control refers to nontreated cells (●). The arrow indicates time of treatment.
The nitrogen-regulated genes are expressed upon nitrogen limitation or rapamycin treatment. TOR inhibits expression of nitrogen-regulated genes by sequestering the GATA-binding transcription factors GLN3 and GAT1 (also called NIL1) in the cytoplasm (9). In the presence of a good nitrogen source, GLN3 is phosphorylated in a TOR-dependent manner and thereby tethered to the cytoplasmic URE2 protein. Upon nitrogen limitation or rapamycin treatment, GLN3 is dephosphorylated by the type 2A-related phosphatase SIT4, released from URE2, and translocated into the nucleus where it activates target genes (9). The nature of the nitrogenous compound(s) TOR senses to inhibit GLN3 and GAT1 is unknown. Magasanik and colleagues (12–14) have proposed that GLN3 is inhibited by glutamine, which, in turn, suggests that TOR may sense glutamine.
In addition to GLN3 and GAT1, TOR inhibits the nuclear localization and activation of several other nutrient-responsive transcription factors, such as MSN2, MSN4, RTG1, and RTG3 (8, 9). The partially redundant Zn2+-finger transcription factors MSN2 and MSN4 activate a large number of stress-related genes in response to several types of stress, including carbon limitation (15–17). The basic helix–loop–helix/Zip transcription factors RTG1 and RTG3 and the cytoplasmic protein RTG2 comprise an inter-organelle communication pathway termed retrograde regulation (18, 19). Retrograde regulation activates RTG-target genes in the nucleus in response to a mitochondrial defect. More specifically, RTG1 and RTG3 form a heterodimer (19) that activates expression of the genes encoding the TCA and glyoxylate cycle enzymes, leading to the production of α-ketoglutarate, a precursor of glutamate and ultimately glutamine (Fig. 1A). External glutamate (20) or glutamine (8) represses RTG-dependent expression, suggesting that TOR may sense one or both of these amino acids to inhibit RTG1/3. It is not known whether glutamate or glutamine mediates retrograde regulation (and possibly TOR signaling) as these two amino acids can be interconverted in the cell. Furthermore, the nature of the link between TOR-mediated control of RTG1/3 possibly in response to glutamine or glutamate and retrograde regulation in response to a mitochondrial defect is also unknown.
Here we establish a link between TOR signaling and glutamine. We show that the TOR-controlled transcription factors GLN3, RTG1, and RTG3 are activated in glutamine-starved cells, even in the presence of glutamate. In contrast, no activation of the TOR-regulated transcription factors GAT1, MSN2, or MSN4 was observed upon glutamine starvation. We also found that glutamine depletion has no effect on r-protein mRNA levels, another TOR-dependent readout. Our results suggest that the TOR pathway regulates a subset of its targets in response to intracellular glutamine. Our results also suggest that glutamine mediates retrograde regulation via TOR.
Materials and Methods
Strains, Media, and Reagents.
The complete genotypes of yeast strains used in this study are listed in Table 1. The composition of rich medium (yeast extract/peptone/dextrose) and synthetic minimal medium (SD) supplemented with the appropriate nutrients was as described (21). SD contains ammonium as a nitrogen source, unless otherwise indicted. All cultures were incubated at 30°C. Rapamycin (provided by Sandoz Pharmaceutical) was used at a final concentration of 200 ng/ml. A 1 mg/ml stock solution of rapamycin was prepared in the drug vehicle 90% ethanol, 10% Tween-20. Control cells were treated with drug vehicle alone. l-methionine sulfoximine (MSX) was obtained from Sigma and used at a final concentration of 2 mM. Cells treated with MSX were grown in SD.
Table 1.
Strains of S. cerevisiae used in this study
| Strain | Genotype |
|---|---|
| JK9-3da | MATa leu2-3,112 ura3-52 trp1 his4 rme1 HMLa |
| TB50a | MATa leu2-3,112 ura3-52 trp1 his3 rme1 HMLa |
| TB102-1a | JK9-3da gat1∷kanMX |
| TB103-1d | TB50a gln3∷HIS3MX |
| TB105-3b | TB50a gln3∷kanMX gat1∷HIS3MX |
| TB106-2a | JK9-3da GAT1-HA3-kanMX |
| TB123 | JK9-3da GLN3-myc13-kanMX |
| TB136-2a | JK9-3da GLN3-myc13-kanMX sit4∷kanMX |
| JC33-6c | JK9-3da GLN3-myc13-kanMX gln1∷kanMX |
| JC18-2b | JK9-3da GLN3-myc13-kanMX tor1∷LEU2 |
| JC37-1a | TB50a RTG1-GFP-kanMX |
| K699 | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL+ |
| EY0733 | K699 rtg1∷TRP1 |
| EY0734 | K699 rtg2∷TRP1 |
| EY0735 | K699 rtg3∷TRP1 |
Determination of Intracellular Amino Acid Pools.
Wild-type (JK9–3da) cells were grown in SD to logarithmic phase and treated with 2 mM MSX. Samples equivalent to 15 OD600 units were taken at different times and placed immediately on ice. Cells were washed twice with 50 ml of ice-cold water, resuspended in 500 μl of freshly prepared 5% trichloroacetic acid, and incubated on ice for 30 min with gentle agitation. The samples were then centrifuged to remove cellular debris and precipitated proteins. The size of intracellular amino acid pools and the intracellular MSX pool were determined with a Biochrom 20 plus amino acid analyzer (Amersham Pharmacia).
GS Assay.
The transferase activity of GS was measured as described (22).
Indirect Immunofluorescence.
To localize GLN3-myc, GAT1-HA, MSN2-myc, and MSN4-HA, the appropriate cells were grown at 30°C in SD and treated with rapamycin, drug vehicle alone, or MSX for the indicated times. GLN3-myc, GAT1-HA, MSN2-myc, and MSN4-HA were visualized by indirect immunofluorescence on whole fixed cells, as described (9). DNA was stained with 4′,6-diamidino-2-phenylindole (Sigma) at a concentration of 1 μg/ml. Cells were visualized with a Zeiss Axiophot microscope (×100 objective).
Fluorescence Microscopy.
Strain JC37–1a, expressing a functional C-terminally green fluorescent protein-tagged RTG1, was constructed by using a PCR-derived cassette (23). Cells were grown to logarithmic phase (OD600, 0.4–0.6) in minimal medium containing 5 mM glutamate as the only nitrogen source. Samples were observed by using a Zeiss microscope (model Axioplan 2 imaging) equipped with an XBO 75 W/2 xenon short-arc lamp.
RNA Analysis.
Reverse transcriptase (RT)-PCR.
Total yeast cell RNA was prepared by using the RNeasy MiniKit (Qiagen, Chatsworth, CA) as recommended by the manufacturer. RNA preparations were treated with DNase (New England Biolabs) for 15 min at room temperature and washed with RNeasy mini spin columns as described by the manufacturer (Qiagen). RT-PCR was performed by using the Titan One Tube RT-PCR Kit (Roche Diagnostics) and a Biometra thermocycler. The samples were first incubated at 55°C for 30 min (for reverse transcription) and then as follows for thermocycling: 94°C for 2 min (one time); then 94°C for 50 s, 52°C for 1 min, and 68°C for 75 s (24 times); and then 68°C for 5 min (one time). The sequence of the PCR primers is as follows: GLN1, 5′-CGTTTGGATCGATGGTACTG-3′ and 5′-CGCAAACAGTTTCACACATG-3′; MEP2, 5′-ACGGGTGGTAACTCGTTGAC-3′ and 5′-TACAACCACCCACACCATGG-3′; ACT1, 5′-ATGGATTCTGAGGTTGCTGC-3′ and 5′-ACCTTCATGGAAGATGGAGC-3′.
Northern blot.
Northern blot analysis was performed as described (6). DNA probes were generated as described (8).
Results
The GS Inhibitor MSX Induces Glutamine Starvation.
To investigate the role of glutamine in TOR signaling, we sought to specifically deplete cells of glutamine. MSX is an irreversible and highly specific inhibitor of GS activity (24). We examined whether MSX inhibits GS activity and induces glutamine depletion in yeast cells. MSX (2 mM) was added to cells growing exponentially in defined minimal medium, and aliquots were removed at various time points and assayed for GS activity and intracellular amino acid pool sizes. GS activity was 90% inhibited within 10 min of MSX addition and was undetectable within 30 min (data not shown). In agreement with the observed inhibition of GS, the intracellular glutamine pool was reduced to an undetectable level and the glutamate pool was increased 2-fold, within 30 min of MSX addition (Fig. 1B). MSX had no significant effect on the size of the intracellular pool of any other amino acid (data not shown). Furthermore, MSX inhibited growth within 30 min of treatment, and this growth inhibition was completely suppressed by the presence of glutamine in the culture medium (Fig. 1C). Glutamine also restored growth in cells that had previously been arrested with MSX (data not shown). Other amino acids, including glutamate, did not suppress the effect of MSX on growth (data not shown). Thus, MSX inhibits glutamine synthesis, and MSX treatment is an effective and rapid method to induce conveniently and specifically glutamine depletion in yeast cells. As a result of GS inactivation and glutamine depletion, MSX treatment rapidly inhibits cell growth.
Intracellular Glutamine Inhibits GLN3 via the TOR Signaling Pathway.
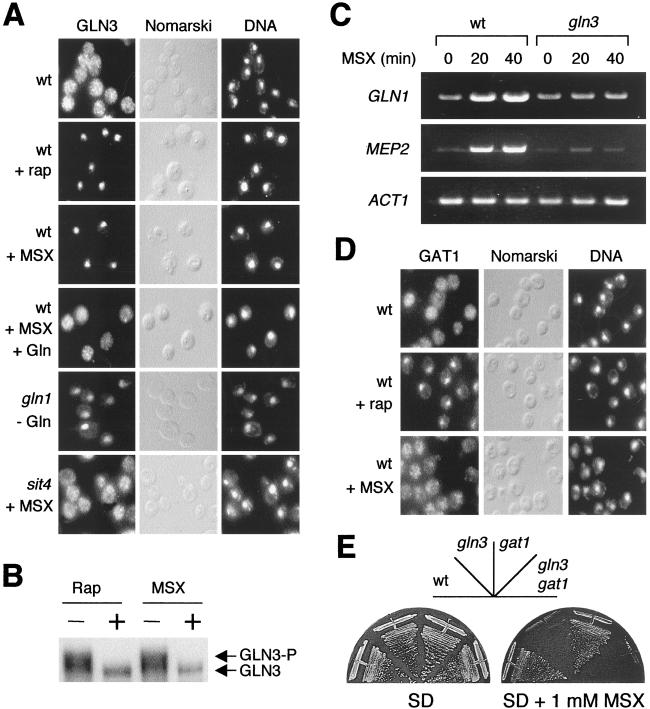
To investigate whether the TOR signaling pathway responds to intracellular levels of glutamine, we treated cells with MSX and examined the cellular location of the TOR-controlled transcription factor GLN3 (Fig. 2). TOR maintains GLN3 in the cytoplasm (9). Similarly to rapamycin, MSX induced strong nuclear accumulation of GLN3, after 15–20 min of treatment (Fig. 2A). To confirm that MSX-induced nuclear localization of GLN3 is caused by glutamine depletion, glutamine was added to MSX-treated cells. Addition of glutamine rapidly countered the nuclear localization of GLN3 caused by MSX; within 5 min, glutamine induced cytoplasmic accumulation of GLN3 in cells previously treated with MSX for 20 min (Fig. 2A). To further confirm the specificity of MSX, the cellular location of GLN3 was also examined in gln1 mutant cells. As expected, GLN3 was cytoplasmic in gln1 mutant cells grown in the presence of external glutamine (data not shown). Within 20 min of shifting gln1 cells to glutamine-free medium, GLN3 accumulated in the nucleus, as observed in wild-type cells treated with MSX (Fig. 2A). Deprivation for tryptophan or leucine, by shifting trp1 leu2 cells to tryptophan- or leucine-free medium, did not affect the cellular localization of GLN3 (data not shown). Thus, glutamine controls the cellular localization of GLN3; more precisely, glutamine prevents nuclear accumulation of GLN3. It is not known whether glutamine induces nuclear export of GLN3 or prevents GLN3 from shuttling back into the nucleus.
Figure 2.
Glutamine starvation specifically activates the transcription factor GLN3 but not GAT1. (A) Localization of GLN3-myc (GLN3) in a wild-type (wt) (TB123) and gln1 (JC33–6c) and sit4 (TB136–2a) mutant cells. Wild-type cells were grown in SD and treated either with rapamycin (+ rap) or MSX (+ MSX) for 20 min. Wild-type cells also were treated with MSX for 20 min and then with glutamine (5 mM final concentration) for 5 min (+ MSX + Gln). Untreated wild-type cells were used as a control. gln1 mutant cells were grown in SD supplemented with 0.3% glutamine and shifted to glutamine-free medium for 20 min. sit4 mutant cells were treated with MSX for 20 min. All strains were grown in SD at 30°C. Cells and DNA were visualized by Nomarski optics and 4′,6-diamidino-2-phenylindole staining (DNA). (B) GLN3 is dephosphorylated in glutamine-starved cells. Wild-type cells (TB123) were grown in SD and treated with either rapamycin or MSX for 25 min. GLN3-myc was detected by immunoblotting. (C) Reverse transcriptase–PCR analysis of total RNA from wild-type (JK9–3da) and gln3 (TB103–1d) mutant cells with oligonucleotide primers specific to the actin gene (ACT1) and two nitrogen-regulated genes, GLN1 and MEP2. Cells were grown in SD to an OD600 of 0.5 and treated with MSX for either 20 or 40 min. Nontreated cells (time 0) were used as a control. (D) GAT1-HA (GAT1) was visualized by indirect immunofluorescence in wild-type (TB106–2a) cells untreated (Control) or treated with rapamycin (+ rap) or MSX (+ MSX) for 20 min. (E) Growth of wild-type (JK9–3da), gln3 (TB103–1d), gat1 (TB102–1a), and gln3 gat1 (TB105–3b) mutant cells in SD or SD supplemented with a sublethal concentration (1 mM) of MSX. Plates were incubated at 30°C for 3 days.
To confirm that glutamine prevents GLN3 activation, we examined the GLN3 phosphorylation state and the expression of GLN3 target genes in glutamine-depleted cells. Similar to rapamycin treatment, MSX treatment caused dephosphorylation of GLN3 (Fig. 2B). Expression of the GLN3 target genes GLN1 and MEP2 was induced in wild-type cells treated with MSX for 20 min (Fig. 2C). GLN1 and MEP2 were not induced in MSX-treated gln3 mutant cells (Fig. 2C), indicating that MSX-induced expression of GLN1 and MEP2 is via GLN3. Thus, glutamine inhibits activation of GLN3.
The type 2A-related phosphatase SIT4 is required for the dephosphorylation and nuclear localization of GLN3 (9). TOR inhibits the nuclear localization of GLN3 by inactivating SIT4. To determine whether glutamine inhibits GLN3 via the TOR pathway, we examined the location of GLN3 in MSX-treated sit4 mutant cells. In contrast to wild-type cells, GLN3 remained cytoplasmic in sit4 cells treated with MSX (Fig. 2A), indicating that SIT4 mediates the nuclear localization of GLN3 in response to glutamine starvation. Thus, glutamine inhibits GLN3 via the TOR pathway.
To further investigate the role of TOR in GLN3 inhibition, we examined the cellular location of GLN3 in tor1 mutant cells treated with MSX. Surprisingly, nuclear accumulation of GLN3 in glutamine-starved tor1 mutant cells was significantly delayed (30–40 min) compared with GLN3 localization in similarly treated wild-type cells (data not shown). A possible explanation for the slow activation of GLN3 in tor1 mutant cells is that the basal intracellular level of glutamine is higher in cells lacking TOR1, and thus MSX-induced glutamine depletion occurs later. Indeed, we found that the intracellular level of glutamine in tor1 cells was 2.8-fold higher than in wild-type cells (data not shown).
Intracellular Glutamine, but Not Glutamate, Inhibits RTG1 and RTG3.
RTG1 and RTG3, as a heterodimeric transcription complex, activate transcription of genes involved in the TCA and glyoxylate cycles (19, 20). TOR and the presence of glutamine or glutamate (see Introduction) in the growth medium inhibit nuclear localization of RTG1 and RTG3 (8, 25). To determine whether RTG1 is regulated by intracellular glutamine, cells expressing RTG1-green fluorescent protein were treated with MSX in the presence of glutamate. As reported previously (8, 25), RTG1 was found mainly in the cytoplasm in MSX-untreated, glutamate-grown cells (Fig. 3A). Similar to rapamycin treatment, MSX treatment induced nuclear localization of RTG1 within 30 min, even in the presence of glutamate (Fig. 3A). As expected, the presence of glutamine in the medium suppressed the effect of MSX (data not shown). Thus, RTG1 and likely RTG3 are inhibited by intracellular glutamine, and not by glutamate.
Figure 3.
Glutamine starvation activates the transcription factors RTG1 and RTG3. (A) RTG1 localized in the nucleus in glutamine-depleted cells. Wild-type (JC37–1a) cells expressing a functional RTG1-green fluorescent protein (GFP) fusion protein were grown in SD containing 5 mM glutamate and treated either with rapamycin (+ rap) for 15 min or MSX (+ MSX) for 30 min. (B) Induction of the RTG-target gene CIT2 by rapamycin and MSX. Wild-type (K699) cells grown in glutamate-containing SD were treated with rapamycin and/or MSX for 30 min in the presence or the absence of glutamine. Cells were then harvested and total RNA was prepared and analyzed by Northern blotting, blotting for the specified mRNAs (Top). (C) RTG genes mediate the induction of CIT2 in MSX-treated cells. Wild-type (K699) cells and rtg1 (EY0733), rtg2 (EY0734), or rtg3 (EY0735) mutant cells were grown in glutamate-containing SD and treated with MSX for 30 min. Cells were then harvested and total RNA was isolated. Expression of CIT2 and ACT1 was determined by Northern blotting as described in Materials and Methods.
Expression of the CIT2 gene, encoding the peroxisomal form of citrate synthase, is positively regulated by the transcription factors RTG1 and RTG3 and the cytoplasmic protein RTG2 (8, 18–20). To confirm that glutamine inhibits RTG1 and RTG3, we measured CIT2 expression in MSX-treated cells. Cells were treated with MSX in the presence of glutamate, and CIT2 mRNA levels were assayed by Northern analysis. Like rapamycin (8), MSX strongly induced CIT2 expression (Fig. 3B). The presence of glutamine in the growth medium inhibited the induction of CIT2 by MSX, indicating that CIT2 is up-regulated by glutamine starvation. To confirm that expression of CIT2 is inhibited by glutamine, CIT2 mRNA levels were examined in wild-type and gln1 mutant cells shifted to glutamine-free (but glutamate-containing) medium. CIT2 expression was induced only in the gln1 mutant cells, confirming that CIT2 is inhibited by glutamine (data not shown). The induction of CIT2 in glutamine-depleted cells is mediated by the RTG proteins, as deletion of RTG1, RTG2, or RTG3 abrogated the induction of CIT2 upon MSX treatment (Fig. 3C); induction of CIT2 is independent of GLN3, as deletion of GLN3 had no effect on CIT2 expression (data not shown). These results suggest that the RTG branch of TOR signaling is regulated (inhibited) by glutamine. Furthermore, the findings that MSX-treated cells accumulate glutamate and that exogenously added glutamate cannot suppress the effect of MSX or a gln1 mutation on the RTGs suggest that the RTGs are not regulated by glutamate, or at least not by glutamate alone. Glutamate must be converted to glutamine (via GS) to inhibit the RTGs.
Not All TOR-Controlled Transcription Factors Are Affected by Glutamine.
GAT1, like GLN3, is a TOR-controlled GATA transcription factor (8) that activates the expression of nitrogen-regulated genes (14, 26). To investigate whether glutamine controls GAT1, we examined the cellular location of GAT1 in MSX-treated cells. In contrast to our findings on GLN3, MSX did not induce nuclear accumulation of GAT1 (Fig. 2D), indicating that not all TOR-controlled transcription factors respond to glutamine. In agreement with the findings that MSX activates GLN3 but not GAT1, gln3 but not gat1 mutant cells are hypersensitive to MSX (Fig. 2E). gln3 mutant cells are hypersensitive to glutamine depletion presumably because they are unable to induce expression of the GLN1 gene encoding GS (see above).
TOR also inhibits nuclear localization of MSN2 and MSN4 (8). As observed for GAT1, MSX did not affect the localization of MSN2 and MSN4. MSN2 and MSN4 remained in the cytoplasm even after prolonged treatment with MSX (data not shown), indicating that these transcription factors do not respond to glutamine.
TOR positively controls expression of the ribosomal protein genes via one or more unknown transcription factors (6). As observed previously (6), transcription of the ribosomal protein gene RPL30 was down-regulated upon rapamycin treatment (Fig. 3B). However, no effect was observed in response to MSX (Fig. 3B). Thus, the TOR-controlled transcription factor mediating ribosomal protein gene expression is not affected by glutamine levels, again indicating that glutamine affects only a subset of TOR-controlled readouts.
Discussion
TOR positively controls cell growth in response to nutrients. However, the specific nutrients sensed by TOR are unknown. We demonstrate that glutamine, the preferred nitrogen source of yeast cells, controls TOR readouts. Glutamine, like TOR, inhibits the nuclear localization and activation of the transcription factors GLN3, RTG1, and RTG3. Furthermore, upon glutamine depletion, the TOR-controlled phosphatase SIT4 mediates the nuclear localization of at least GLN3. These findings suggest that glutamine inhibits GLN3, RTG1, and RTG3 via the TOR pathway, and that the TOR pathway senses glutamine. In mammalian cells, TOR responds to essential amino acids used for protein synthesis (27, 28). In yeast, TOR may respond to glutamine both as an amino acid required for protein synthesis and as a nitrogen source. Thus, TOR in yeast may sense glutamine, among other nutrients (see below), because it is a particularly critical nutrient indicator (see below). It is not known whether the TOR pathway senses glutamine itself or a glutamine metabolite. However, the finding that 6-diazo-5-oxo-l-norleucine, a highly toxic analog of glutamine and an inhibitor of glutamine metabolism, does not induce GLN3 nuclear localization (data not shown) suggests that the TOR pathway senses glutamine itself.
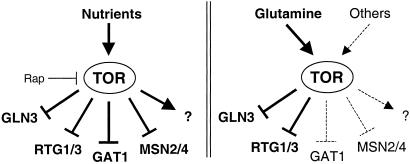
Although TOR may respond to glutamine, not all TOR readouts are affected upon glutamine starvation. The TOR-controlled transcription factors GAT1, MSN2, MSN4, and an unknown factor involved in the regulation of ribosomal protein genes are not affected by glutamine depletion. This finding suggests that the TOR pathway senses a variety of nutrients to elicit a cellular response appropriate to a given nutrient condition (Fig. 4). Indeed, as suggested by Schreiber and colleagues (29), TOR may act as a multichannel processor to differentially regulate gene expression in response to different nutrient conditions. TOR may inhibit only GLN3, RTG1, and RTG3 in response to glutamine because these transcription factors, but not GAT1, MSN2, and MSN4, mediate glutamine synthesis. It is not known in response to which nutrients TOR inhibits GAT1, MSN2, and MSN4, but at least GAT1 has been proposed to respond to a nutrient other than glutamine (14). Rapamycin treatment presumably mimics general starvation, rather than starvation for a single nutrient, and thus affects all of the above transcription factors (Fig. 4).
Figure 4.
TOR regulates a specific subset of proteins in response to glutamine. (Left) In the presence of nutrients, TOR keeps the transcription factors GLN3, GAT1, RTG1/3, and MSN2/4 inactive. (Right) Glutamine is the specific signal that regulates GLN3, RTG1, and RTG3, whereas GAT1, MSN2, and MSN4 are regulated by other metabolites. The question mark refers to an unknown transcription factor that regulates r-protein genes.
Is there a link between TOR-mediated control of RTG1/3 in response to glutamine and retrograde regulation in response to a mitochondrial defect? α-Ketoglutarate is produced in the mitochondria by the TCA cycle and is a precursor in the synthesis of glutamate and ultimately of glutamine (Fig. 1A). Thus, a mitochondrial defect may reduce indirectly the intracellular pool of glutamine, which may, in turn, relieve TOR-mediated inhibition of RTG1 and RTG3. In other words, retrograde regulation of RTG1 and RTG3 in response to a mitochondrial defect is via TOR. However, at present we cannot distinguish this possibility from an alternative model in which TOR signaling and retrograde regulation constitute distinct pathways that converge on RTG1 and RTG3. Furthermore, although a mitochondrial defect may also reduce the intracellular level of glutamate, contrary to the suggestion of Liu and Butow (20), our findings suggest that glutamate (or at least glutamate alone) is not the signal that links mitochondrial integrity to expression of RTG-target genes.
The finding that glutamine controls TOR signaling underscores the importance of glutamine in nitrogen metabolism (reviewed in ref. 12). Our finding that glutamine also controls the RTGs and thereby the TCA cycle suggests that glutamine also has an important function in carbon metabolism. Indeed, glutamine may play a key role in the integration of nitrogen and carbon metabolism by TOR. According to this notion, TOR may sense only the quality of the nitrogen source rather than the quality of the nitrogen and the carbon source.
Acknowledgments
We thank members of the laboratory for helpful discussions. J.L.C. is a recipient of a Federation of European Biochemical Societies Long-Term Fellowship. This work was supported by grants from the Canton of Basel and the Swiss National Science Foundation (to M.N.H.).
Abbreviations
- GS
glutamine synthetase
- MSX
l-methionine sulfoximine
- SD
synthetic minimal medium
- TCA
tricarboxylic acid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Schmelzle T, Hall M N. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 2.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 3.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noda T, Ohsumi Y. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 5.Zaragoza D, Ghavidel A, Heitman J, Schultz M C. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powers T, Walter P. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardwick J S, Kuruvilla F G, Tong J K, Shamji A F, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komeili A, Wedaman K P, O'Shea E K, Powers T. J Cell Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck T, Hall M N. Nature (London) 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 10.Cardenas M E, Cutler N S, Lorenz M C, Di Como J C, Heitman J. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. EMBO J. 1998;17:6924–2931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magasanik B. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 283–317. [Google Scholar]
- 13.Benjamin P M, Wu J-I, Mitchell A P, Magasanik B. Mol Gen Genet. 1989;217:370–377. doi: 10.1007/BF02464906. [DOI] [PubMed] [Google Scholar]
- 14.Stanbrough M, Rowen D W, Magasanik B. Proc Natl Acad Sci USA. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith A, Ward M P, Garrett S. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorner W, Durchschlag E, Martinez-Pastor M T, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M. J Bacteriol. 1998;180:1044–1052. doi: 10.1128/jb.180.5.1044-1052.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao X, Butow R A. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 19.Jia Y, Rothermel B, Thornton J, Butow R A. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Butow R A. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman F. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. San Diego: Academic; 1991. pp. 3–21. [Google Scholar]
- 22.Mitchell A P, Magasanik B. J Biol Chem. 1983;258:119–124. [PubMed] [Google Scholar]
- 23.Longtine M S, McKenzie A, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 24.Manning J M, Moore S, Rowe W B, Meister A. Biochemistry. 1969;8:2681–2685. doi: 10.1021/bi00834a066. [DOI] [PubMed] [Google Scholar]
- 25.Sekito T, Thornton J, Butow R. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coffman J A, Rai R, Cooper T G. J Bacteriol. 1996;177:6910–6918. doi: 10.1128/jb.177.23.6910-6918.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara K, Yonezawa K, Weng Q-P, Kozlowski M T, Belham C, Avruch J. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Campbell L E, Miller C M, Proud C G. Biochem J. 1998;334:261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shamji A F, Kuruvilla F G, Schreiber S L. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]