Abstract
The nuclear translocation of peptide hormones, such as the somatolactogenic hormone prolactin, after receptor internalization has been widely reported. Prolactin has been demonstrated to interact with cyclophilin B, a member of the immunophilin family of proteins. Cyclophilin B interaction with prolactin potentiated prolactin-induced proliferation, cell growth, and the nuclear retrotransport of prolactin. These effects could be abrogated by the removal of the peptidyl-prolyl isomerase activity of cyclophilin B. Our findings indicate that the intranuclear prolactin/cyclophilin B complex acts as a transcriptional inducer by interacting directly with Stat5, resulting in the removal of the Stat-repressor protein inhibitor of activated Stat 3 (PIAS3), thereby enhancing Stat5 DNA-binding activity and prolactin-induced, Stat5-mediated gene expression.
The pleiotropic actions of the somatolactogenic hormone prolactin (PRL) are mediated by signaling through the prolactin receptor, a member of the type I family of cytokine receptors (1–3). Upon ligand binding and dimerization of the receptor, several signaling cascades are activated, including Ras–Raf, Fyn–Vav and Jak2–Stat5 (4–7). Activated Jak2 phosphorylates Stat5 on tyrosine residues, inducing Stat5 dimerization and translocation to the nucleus (8). Intranuclear Stat5 binds to consensus Stat5 response elements, resulting in the transactivation of numerous PRL-specific genes, of which βcasein is the most extensively studied (9). This Stat5 transcriptional activation can be cooperatively enhanced by the glucocorticoid receptor (GR) and C/EBPβ (10–12).
While the above-mentioned pathways are all associated with PRL-induced signaling, activation of the PRL receptor is also associated with ligand internalization via an endosomal-like pathway across the endoplasmic reticulum (ER) and nuclear envelopes (13, 14). The phenomenon of protein retrotransport was initially characterized through the study of retrotranslocated viral and bacterial proteins and peptides destined for presentation on the major histocompatibility complex (15–18). These studies revealed that protein retrotransport depends on transport through the protein-conducting channel formed by the Sec61 complex in the ER membrane. Electron microscopy studies employing colloidal gold-labeled PRL demonstrated that upon ligand internalization into endosomes, approximately 90% of the internalized PRL either remained within the endosome or was degraded. However, the remaining 10% was detected as passing from the endosome through multivesicular bodies and the Golgi/ER to arrive in the nucleus within 2 h poststimulation (13). The functional significance of nuclear PRL was demonstrated by the finding that a nuclear-targeted construct of PRL containing the simian virus 40 large T antigen nuclear localization sequence provided a necessary comitogenic stimulus for IL-2-driven growth (19). The nuclear retrotransport and potential action of peptide hormones and growth factors is widespread, as epidermal growth factor, insulin, growth hormone, platelet-derived growth factor, IL-5, and others have been noted within the nucleus (20–23). However, the mechanism of nuclear function has remained uncertain.
By yeast two-hybrid analysis, we recently identified cyclophilin B (CypB) as a PRL-interacting protein (24). CypB is a member of the larger immunophilin family of cis–trans peptidyl-prolyl isomerases, which also includes the FK506 binding proteins and target of rapamycin (Tor) (25). CypB has been observed in the ER and nucleus and can be found in appreciable levels in blood (150 ng/ml) and breast milk (26, 27). Whereas its role as a chaperone in protein transport through the ER (28) or as a costimulatory molecule for T lymphocytes (29) has been speculated upon, the exact function of CypB has remained uncertain. We demonstrated CypB could potentiate PRL-driven proliferation up to 18-fold with a concurrent 4.5-fold increase in viable cell number over PRL stimulation alone (24). This increased proliferation was not due to an alteration of Jak2–Stat5 activation, but did correspond to a marked increase in intranuclear PRL. Moreover, the removal of a putative nuclear localization sequence in the N terminus of CypB abrogated the potentiation in PRL-induced proliferation and the enhancement of PRL nuclear transport, indicating a necessary role for CypB in these functions.
Herein we describe a direct interaction between the PRL/CypB complex and Stat5 that is dependent on the enzymatic activity of CypB. This interaction occurs after PRL/CypB nuclear internalization and is terminated upon Stat5 binding to DNA. This interaction of PRL/CypB results in an increase in Stat5-mediated gene transcription. The enhancement of Stat5-mediated gene transcription is associated with increased Stat5 DNA-binding activity that results from the release of a repressor of Stat activity, protein inhibitor of activated Stat 3 (PIAS3). These findings demonstrate mechanistically how an intranuclear polypeptide hormone can potentiate its own signal, and perhaps contribute to its own specificity.
Materials and Methods
Generation and Expression of Recombinant Proteins.
PRL-myc, CypB, and CypB-NT were expressed as described (24). CypB-PPI was generated by overlapping mutagenesis. The forward CypB primer containing an EcoRI site 5′-CGGAATTCGCCGATGAGAAGAAGAAGGGG-3′ was combined with the reverse primer 5′-GCCCTGGATCATGGCGTCCTTGATTACGGCATGGAATTTGCT-3′ that mutated residues R63 and F68 to alanines (underlined), whereas the reverse CypB primer containing a XhoI site 5′-CGCTCGAGCTCCTT-GGCGATGCCAAAGGG-3′ was combined with the forward primer 5′-AGCAAATTCCATGCCGTAATCAAGGACGCCATGATCCAGGGC-3′ (mutated residues underlined). The glutathione S-transferase (GST)-Stat5a fragments were generated by PCR as follows: Stat5aN (amino acids 1–327) primer pairs: 5′-CGGAATTCATGGCGGGCTGGATTCAGGCCCAG-3′ and 5′-CGGCGGCCGCTCAGGACAGGGCTGAGATGATGTCTGT-3′; Stat5aDBD (amino acids 327–515) primer pairs 5′-CGGAATTCCTGGTCACCAGCACGTTCATCGAG-3′ and 5′-CGGCGGCCGCTCAGGAGAACTTCATGTTGAGCGCCTCACA-3′; Stat5aC (amino acids 515–793) primer pairs 5′-CGGAATTCTTCAAGGCTGAGGTGCAG-3′ and 5′-CGGCGGCCGCTCAGGACAAGGAGCTTCTGGCAGAAGT-3′. The resulting PCR products were purified, cloned into pGEX (CLONTECH), and expressed as described (30).
Cell Culture.
Nb2 (PRL-dependent, rat T cell lymphoma) cells were maintained and rested in serum-free medium as described (24). The human breast cancer cell line T47D was maintained in DMEM with 10% FBS while Chinese hamster ovary (CHO) cells were maintained in F12 with 10% FBS.
In Vitro Binding Assays and Immunoprecipitations.
In vitro binding assays were performed as described (24). Lysates from 5 × 106 resting Nb2 cells stimulated with PRL (150 ng/ml) with or without CypB (500 ng/ml) or CypB-PPI (500 ng/ml) for 20 min at 37°C were immunoprecipitated with either anti-PRL or one of the following antibodies (all from Santa Cruz Biotechnology), anti-Stat5a, anti-PIAS1/3, anti-PIAS1, or anti-PIAS3, followed by immunoblot analysis with these respective antibodies. For unlabeled oligonucleotide competition analysis, lysates from unstimulated or stimulated Nb2 cells were incubated with 10, 100, or 1000 ng of double-stranded DNA. To assess the in vitro interaction between PRL, CypB and Stat5a, 35S-labeled Stat5a was generated by the TnT Quick coupled Transcription/Translation system (Promega). Stat5a generated in this system was subsequently demonstrated to be tyrosine phosphorylated and active in gel-shift assays (data not shown). One microliter of labeled Stat5a was admixed in 200 μl of in vitro binding buffer with 100 ng of recombinant PRL ± 200 ng of recombinant CypB or CypB-PPI before immunoprecipitation with anti-PRL antibody as above. Anti-PRL immunoprecipitates were visualized by autoradiography. For GST pull-downs, 1 μg of each of the above-described GST-Stat fusions was coupled to glutathione-Sepharose and admixed in binding buffer with 500 ng of myc-tagged PRL, V5-tagged CypB, or a combination of the two. Bound proteins were separated by SDS/PAGE 12% and subjected to sequential immunoblot analysis with anti-myc, anti-V5-horseradish peroxidase (HRP) (Invitrogen) and anti-GST-HRP (Santa Cruz Biotechnology).
Nuclear Extract Preparation and Electrophoretic Mobility-Shift Assays.
To assess Stat5 DNA-binding ability, nuclear extracts (3 μg) from unstimulated and PRL-stimulated Nb2 cells were incubated overnight at 4°C with the addition of 100 ng of PRL, 100 ng of CypB, or 100 ng of PRL/100 ng of CypB along with 1 μg of polyclonal anti-His (Santa Cruz Biotechnology). As controls, stimulated extracts were also incubated with 1 μg of polyclonal anti-Stat5a (Zymed, South San Francisco, CA), 100 ng of BSA, or 1 μg of polyclonal anti-His. These extracts were subjected to mobility shift assays as described (24) with the addition of 32P-labeled double-stranded 25-mer oligonucleotide containing a Stat5 consensus binding site from the β-casein promoter: 5′-TTTTAGATTTCTAGGAATCAAATC-3′ (consensus binding site in bold). Variability in probe loaded from sample to sample was less than 1%.
Plasmid Constructs, Transient Transfections, and Reporter Gene Assays.
pEF-hPRLr-long encoding for the long isoform of the human PRL receptor has been described (31). pcDNA3.1 containing both full-length rat Stat5a and Stat5b was a kind gift of Li Yu-Lee (Baylor University, Houston, TX). The reporter construct LHRE-TK-Luc containing the Stat5 DNA-binding sites from the promoter region of β-casein 5′ to a luciferase reporter was a kind gift of R. J. M. Ross (Sheffield University, Sheffield, England). pCMV5 containing full-length murine PIAS3 was a kind gift of Dr. Ke Shuai (University of California, Los Angeles). CHO cells were transfected with 2 μg of human PRLr-long, LHRE-TK-Luc, and 0.5–2 μg of PIAS3 by using FuGene6 (Roche Diagnostics). Twenty-four hours after transfection, cells were washed and maintained in DMEM-ITS+, while treated with 150 ng/ml PRL with or without 500 ng/ml CypB or CypB-PPI for 24 h. Where indicated, dexamethasone was used at 10−7 M. Luciferase assays were performed by standard methods by using the Luciferase Assay System (Promega) and the Monolight 2010 (Analytical Luminescence Labs, San Diego). All statistical analysis was generated by standard t test, using prism software (GraphPad, San Diego).
Results
Enzymatically Inactive Form of CypB Is Unable to Potentiate PRL-Driven Proliferation.
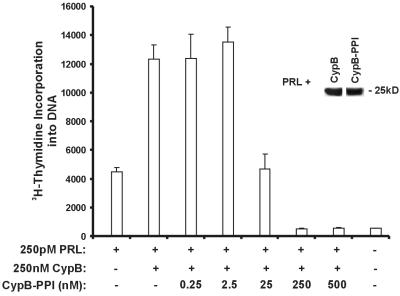
We previously showed that wild-type CypB potentiated PRL-driven proliferation and facilitated PRL nuclear retrotransport (24). In contrast, an N-terminal deletion mutant lacking a putative nuclear localization signal (NLS) neither potentiated proliferation nor facilitated nuclear retrotransport. To assess the role of the peptidyl-prolyl isomerase (PPIase) activity of CypB in this phenomenon, an enzymatically inactive form was generated by conventional PCR-based mutagenesis in which amino acids arginine-63 and phenylalanine-68 were mutated to alanines, and termed CypB-PPI. It has previously been demonstrated that mutation of these residues removes >90% of the enzymatic activity (32). After expression and purification of CypB-PPI from Escherichia coli, a lack of enzyme activity was confirmed by a standard PPIase assay (data not shown). Further evaluation of the CypB-PPI mutant revealed that its interaction with PRL was not affected by deletion of the enzymatic activity (Fig. 1 Inset). The function of the CypB-PPI mutant was then tested on the PRL-dependent rat T lymphoma cell line, Nb2. CypB-PPI was found to competitively inhibit the potentiation of PRL-driven proliferation induced by CypB at concentrations only 10-fold above physiologic concentrations (Fig. 1). Similar results were obtained when the human breast carcinoma cell line T47D was used, whereas anti-PRL immunofluorescence studies indicated comparable levels of PRL nuclear retrotransport when either CypB-PPI or wild-type CypB was used (data not shown). Therefore, while the enzymatic activity of CypB is required for the potentiation of PRL-driven proliferation, it does not appear to be needed for PRL nuclear retrotransport.
Figure 1.
CypB-PPI competitively inhibits PRL/CypB-driven proliferation. Nb2 cells were stimulated with 250 pM PRL with or without 250 nM CypB in the presence of 0.25 to 500 nM CypB-PPI (n = 3). Mean absolute [3H]thymidine incorporation (±SEM) observed in the unstimulated Nb2 cells used here = 5.2 × 102 cpm, for the Nb2 cells stimulated with 250 pM PRL alone = 4.4 × 103 cpm. (Inset) In vitro interaction between PRL and CypB or CypB-PPI was assessed by admixing 100 ng of PRL and 500 ng of each of the CypB proteins. Anti-PRL immunoprecipitates were subjected to immunoblot analysis with anti-CypB.
CypB Enhances PRL-Induced β-Casein Gene Transcription.
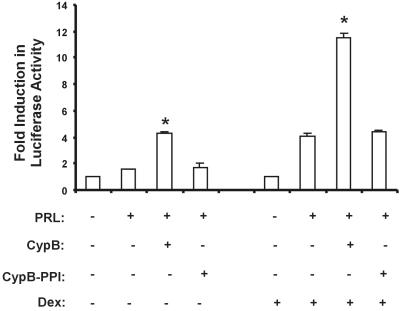
To analyze the role of the intranuclear PRL/CypB complex, luciferase reporter studies using the promoter region of β-casein, a PRL-responsive gene, were undertaken. Upon PRL stimulation of CHO transfectants in the absence of dexamethasone, a small induction in luciferase activity was noted, which increased 2-fold with coadministration of CypB (Fig. 2). The coaddition of CypB-PPI, an enzymatically inactive form of CypB that has been demonstrated to inhibit the potentiation of PRL/CypB-induced proliferation (data not shown), with PRL did not enhance the PRL-driven luciferase activity over PRL alone. With the addition of 10−7 M dexamethasone, there was a 4-fold increase in PRL-induced activity. This increase was potentiated 12-fold with the coadministration of CypB (Fig. 2 Right), whereas the coaddition of CypB-PPI with PRL did not enhance PRL-driven luciferase activity. This steroid-mediated enhancement of PRL-induced activity has been described (11, 33). However, the enhancement of Stat5 activity by the exogenous coadministration of both PRL and CypB, which depends on the PPIase activity of CypB, has not been described.
Figure 2.
PRL/CypB enhances Stat5 transcriptional activity. CHO cells were transfected with 2 μg of plasmids encoding for long human PRLr isoform and a β-casein luciferase reporter construct. After transfection, the cells were washed, placed in defined medium, and stimulated for 24 h in the presence or absence of dexamethasone. Total cell lysates were analyzed for luciferase activity. Data are representative of one of four experiments. Error bars represent SEM; * denotes P < 0.05 as compared with PRL stimulation alone.
Functional Consequences of PRL/CypB Interaction with Stat5.
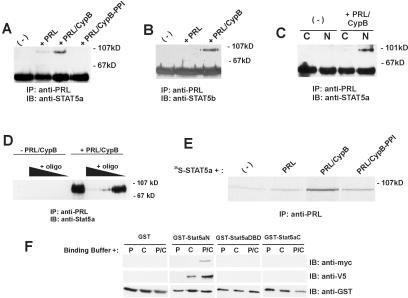
To examine whether the up-regulation of Stat5 activity could be due to a direct interaction between Stat5 and the PRL/CypB complex, coimmunoprecipitation studies were used. The Nb2 cell line stimulated with exogenous epitope-tagged PRL in the presence or absence of CypB demonstrated a direct interaction between Stat5a and PRL in the presence of CypB (Fig. 3A) that occurred within the nucleus (Fig. 3C). Subsequent studies have quantitated that the amount of Stat5 bound to the PRL/CypB complex at 30 min poststimulation is 37% of the total amount of Stat5 present within the cell (data not shown). This interaction was only very weakly observed with the addition of PRL alone and was not present with the coadministration of CypB-PPI. Similar results were obtained when immunoblotting with an antibody specific for Stat5b (Fig. 3B). Stat1 and Stat3 have also been demonstrated to be involved in PRL signal transduction. To test whether these Stats could interact with the PRL/CypB complex, Nb2 and T47D cells were stimulated with PRL in the presence or absence of CypB followed by immunoprecipitation with anti-PRL and then followed by immunoblot analysis with either Stat1 or Stat3. These analyses demonstrated no interaction between PRL/CypB and either Stat1 or Stat3 (data not shown). To confirm a direct interaction between Stat5a and PRL in the presence of CypB, 35S-labeled Stat5a was used in in vitro binding assays with recombinant PRL in the presence or absence of CypB or CypB-PPI. Upon immunoprecipitation with anti-PRL, a direct interaction between PRL and Stat5a was observed in the presence of CypB (Fig. 3E). The nature of this interaction was further investigated by adding double-stranded DNA containing a consensus Stat5-binding site to cell extracts before immunoprecipitating with anti-PRL. As demonstrated in Fig. 3D, the interaction between Stat5a and the PRL/CypB complex can be inhibited by the addition of the double-stranded DNA in a dose-dependent manner. This indicates that once bound to DNA, Stat5 no longer interacts with the PRL/CypB complex. Given the transient nature of the PRL/CypB interaction with Stat5, it was reasoned as unlikely that the PRL/CypB complex was acting directly as a coactivator. To determine which region of Stat5a was involved in the interaction with the PRL/CypB complex, the following GST-Stat5a fusion proteins were generated for use in in vitro binding assays: an N-terminal fragment, Stat5N (amino acids 1–327), a DNA-binding domain fragment (amino acids 327–515), and a C-terminal fragment consisting of the activation domain (AD) (amino acids 515–793). As demonstrated in Fig. 3F, CypB is capable of interacting with the N-terminal fragment of Stat5a and this interaction is enhanced with the inclusion of PRL.
Figure 3.
Analysis of Stat5/PRL/CypB interaction. (A) PRL/CypB complex interacts with Stat5a. Total cell lysates from stimulated or unstimulated Nb2 cells were immunoprecipitated (IP) overnight with polyclonal anti-PRL antibody and were subjected to immunoblot (IB) analysis with anti-Stat5a. Data are representative of one of three experiments. (B) PRL/CypB complex interacts with Stat5b. As in A, but subjected to immunoblot analysis with anti-Stat5b. Data are representative of one of two experiments. (C) PRL/CypB/Stat5 interaction occurs within in the nucleus. Cytoplasmic (C) or nuclear (N) extracts from unstimulated or PRL/CypB-stimulated Nb2 cells were immunoprecipitated and subjected to immunoblot analysis as in A. (D) Stat5 binding-site oligonucleotide inhibits the interaction of Stat5 with PRL/CypB. Nb2 cells (5 × 106) were stimulated for 30 min or left unstimulated. Unlabeled oligonucleotide (1 μg on the left; 10 ng, 100 ng, or 1 μg on the right) was added to lysates before immunoprecipitation and immunoblotting as in A. Data are representative of one of two experiments. (E) In vitro interaction between Stat5a, PRL, and CypB. One microliter of 35S-labeled Stat5a was mixed with 100 ng of recombinant PRL in the presence or absence of 200 ng of recombinant CypB or CypB-PPI. Data are representative of one of two experiments. (F) Mapping of PRL/CypB-binding site on Stat5a. One microgram of GST or GST-Stat5a fusions was coupled to glutathione Sepharose and incubated overnight with 500 ng of PRL-myc (P) in the presence or absence of 500 ng of CypB-V5 (C). Bound proteins were detected by sequential immunoblot analysis with anti-myc, anti-V5-HRP, and anti-GST-HRP (HRP, horseradish peroxidase). Data are representative of one of two experiments.
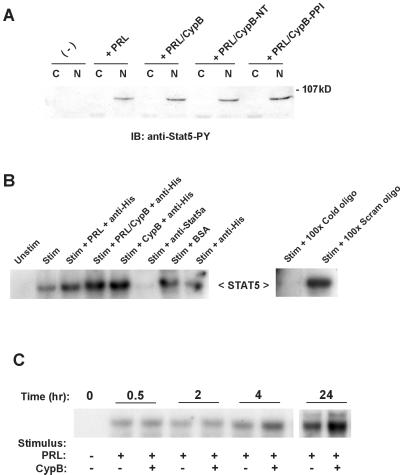
To determine whether PRL stimulation resulted in an increase of phosphorylated Stat5 in the nucleus, which could explain increased Stat5 transcriptional activity, Nb2 cells were stimulated with PRL alone or in the presence of CypB, CypB-PPI, or the previously described CypB-NT (24). After immunoblotting cytoplasmic (C) and nuclear (N) extracts with an anti-phosphotyrosine-Stat5 antibody, we found no difference in the levels or localization of activated Stat5 among any of the treatments (Fig. 4A). To test whether the PRL/CypB complex might alter the ability of Stat5 to bind DNA, electrophoretic mobility-shift assays were performed with nuclear extracts from PRL-stimulated Nb2 cells and a 32P-labeled probe containing a consensus Stat5 binding site. As indicated in Fig. 4B, the addition of PRL alone increased Stat5-binding activity to the labeled probe, and this binding was further enhanced by the coaddition of CypB. The presence of Stat5 within the DNA-binding complex was confirmed by the addition of anti-Stat5a antibody, which inhibited Stat5 from interacting with the probe. The addition of anti-His along with the PRL and CypB did not supershift the complex, confirming the observations in Fig. 3D that indicated a transient interaction between the PRL/CypB complex, terminated by DNA binding. This observation was confirmed in intact cells as Nb2 cells stimulated with PRL in the presence or absence of CypB for up to 24 h demonstrated a marked increase in Stat5-binding activity in those samples receiving the coaddition of exogenous PRL/CypB in comparison to those receiving PRL alone (Fig. 4C). This gradual increase in Stat5 activity coincides both with the known kinetics for PRL nuclear retrotransport (24) and with the kinetics previously reported for β-casein gene transcription (34). These data suggest that the enhancement of Stat5-binding activity and transcriptional activity might be due to the removal of a transcriptional repressor from Stat5 by the PRL/CypB complex.
Figure 4.
Analysis of intranuclear activated Stat5. (A) CypB mutants have no effect on Stat5 activation or nuclear transport. Cytoplasmic and nuclear extracts from stimulated or unstimulated Nb2 cells were subject to immunoblot analysis with a monospecific anti-phosphotyrosine-Stat5 antibody. Data are representative of one of three experiments. (B) Exogenous PRL and CypB enhance Stat5 DNA-binding ability in vitro. Nuclear extracts obtained from stimulated and unstimulated Nb2 cells were incubated with 100 ng of PRL, CypB, PRL/CypB, 1 μg of anti-His, 1 μg of anti-Stat5a, 100 ng of BSA, 100-fold excess of unlabeled oligonucleotide or 100-fold excess of scrambled oligonucleotide before the addition of 1 μl of 32P-labeled probe. Complexes were separated on a 6% gel and analyzed by autoradiography. Data are representative of one of four experiments. (C) Exogenous PRL and CypB enhance Stat5 DNA-binding ability in vivo. Three microliters of nuclear extracts from stimulated and unstimulated Nb2 cells was subjected to gel-shift analysis as stated above. Data are representative of one of two experiments.
The PRL/CypB Complex Disrupts the Interaction of Stat5 and PIAS3.
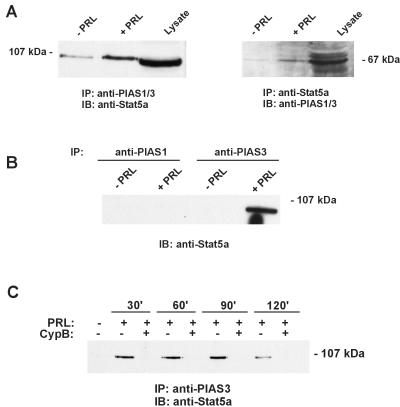
A known family of Stat repressors are the PIAS proteins (35, 36). To date, an interaction between Stat5 and a member of the PIAS family has not been reported. To determine whether these repressors could interact with Stat5, coimmunoprecipitation studies were performed and demonstrated a hormone-inducible interaction between Stat5 and PIAS3 (Fig. 5). This interaction was further assessed in the presence of PRL and CypB. As demonstrated in Fig. 5, after stimulation with PRL alone, there was once again an inducible interaction between Stat5a and PRL. However, the inducible PIAS3/Stat5 interaction observed with PRL stimulation was abrogated after the coaddition of CypB, suggesting a possible mechanism for the enhanced Stat5 DNA-binding activity after treatment with exogenous PRL and CypB.
Figure 5.
PIAS3 interacts with Stat5a after PRL stimulation. (A) PRL stimulation induces an interaction between Stat5 and PIAS1/3. Total cell lysates from stimulated and unstimulated Nb2 cells were immunoprecipitated with either polyclonal anti-PIAS1/3 or anti-Stat5a, followed by immunoblot analysis with either anti-Stat5a or anti-PIAS1/3, respectively. Data are representative of one of two experiments. (B) Stat5 interacts specifically with PIAS3. Total cell lysates from stimulated and unstimulated Nb2 cells were immunoprecipitated with either anti-PIAS1 or anti-PIAS3-specific antibodies, and subjected to immunoblot analysis with anti-Stat5a. Data are representative of one of two experiments. (C) Exogenous addition of CypB inhibits Stat5/PIAS3 interaction. Total cell lysates from stimulated and unstimulated Nb2 cells were immunoprecipitated with anti-PIAS3 followed by immunoblot analysis with anti-Stat5a. Data are representative of one of two experiments.
PIAS3 Repression of Stat5 Activity Can Be Overcome by Exogenous PRL/CypB.
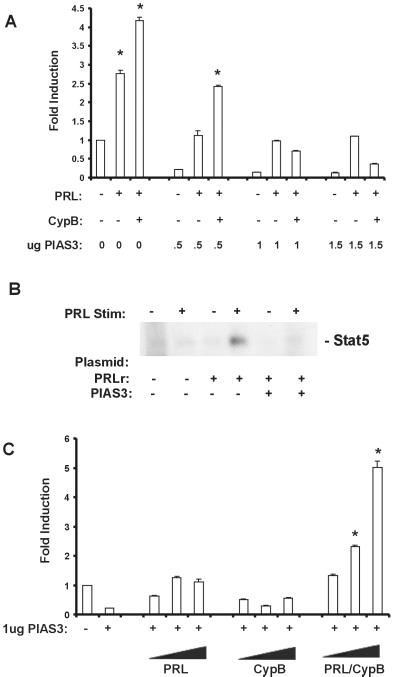
To formally demonstrate that PIAS3 can repress Stat5 transcriptional activity, luciferase reporter assays were undertaken with increasing amounts of PIAS3 cDNA. As demonstrated in Fig. 6A, a dose-dependent repression of both PRL- and PRL/CypB-induced Stat5 transcriptional activity was observed that required twice as much PIAS3 cDNA to repress reporter levels stimulated by PRL/CypB vs. those stimulated with PRL alone. This repression of Stat5 transcriptional activity was also observed by an inhibition in Stat5 DNA-binding ability in gel-shift experiments (Fig. 6B). To further confirm a role for PRL/CypB in altering Stat5/PIAS3 interaction, the ability of increasing amounts of PRL/CypB to overcome PIAS3-mediated repression of Stat5 was tested. PRL or CypB stimulation alone was insufficient to overcome PIAS3-mediated repression; however, increasing amounts of PRL/CypB restored Stat5 transcriptional activity to levels comparable to those observed in the absence of excess PIAS3 (Fig. 6C vs. Fig. 6A). These data demonstrate that PIAS3 serves as a repressor of Stat5 transcriptional activity, and its repressor functions can be disrupted by exogenous addition of PRL/CypB resulting in the release of PIAS3, increased Stat5 DNA-binding activity, and enhanced Stat5-mediated gene expression.
Figure 6.
PIAS3-mediated repression of Stat5 transcriptional activity. (A) Overexpression of PIAS3 represses Stat5 transcriptional activity. CHO cells were transfected as described in Fig. 2 with the addition of the indicated amounts of PIAS3 cDNA. Twenty-four hours after transfection, cells were washed, placed into defined medium, and stimulated for 24 h, and lysates were analyzed for luciferase activity. Data are representative of one of three experiments. Error bars represent SEM; * denotes P < 0.05 as compared with untransfected, unstimulated control. (B) Overexpression of PIAS3 inhibits Stat5 DNA-binding ability. CHO cells were cotransfected with plasmids expressing the long form of the PRLr and PIAS3 before gel-shift analysis as described in Materials and Methods. (C) PIAS3 repression of Stat5 activity can be overcome by exogenous addition of PRL/CypB. CHO cells were transfected as described in Fig. 2 with the addition of 1 μg of PIAS3 cDNA. After transfection, cells were placed into defined medium and stimulated with increasing amounts (PRL = 100, 250, and 500 ng/ml; CypB = 250, 500, and 1000 ng/ml; and PRL/CypB = 100/250, 250/500, and 500/1000 ng/ml) for 24 h. Total cell lysates were analyzed for luciferase activity. Data are representative of one of three experiments. Error bars represent SEM; * denotes P < 0.05 as compared with untransfected, unstimulated control.
Discussion
Herein we report a role for the intranuclear PRL/CypB complex as a transcriptional inducer of Stat5 activity by its nuclear interaction with Stat5 and release of the repressor PIAS3. This interaction and induction of transcriptional activity depended on the PPIase activity of CypB. As such, these phenomena represent a mechanism through which a surface receptor ligand functions directly within the nucleus to modulate transcriptional activity.
After ligand-induced dimerization of the PRLr, numerous signaling pathways are activated, one of which is the Jak–Stat pathway, with Stat5a serving as the primary Stat family member involved in PRL signaling. Upon activation by Jak2-mediated tyrosine phosphorylation, Stat5 peptides dimerize and translocate to the nucleus, where they serve as transcriptional activators of several PRL-responsive genes, most notably whey acidic protein and β-casein (for review, see ref. 37). As for most transcription factors, several coactivators and repressors that interact with Stat5 have been identified. The most notable coactivator of PRL-induced Stat5 activity on the β-casein promoter is the glucocorticoid receptor. It has been demonstrated that the optimal response at the β-casein promoter is achieved upon costimulation with PRL and glucocorticoids (9). We also observed this synergistic enhancement as evidenced by an increase in luciferase activity upon the inclusion of dexamethasone in our PRL-stimulated CHO transfectants using endogenous glucocorticoid receptors (Fig. 2). The further enhancement observed with the inclusion of dexamethasone and the addition of exogenous PRL and CypB can be attributed to the increased pool of activated Stat5 upon which the dexamethasone/glucocorticoid receptor complex can act. Furthermore, it is important to point out that the results reported here were obtained by using physiologic concentrations of PRL/CypB and endogenous proteins where possible.
Two other known enhancers of Stat5 activity are the p300/CBP complex and N-myc-interacting protein (Nmi). p300/CBP are histone acetyltransferases involved in chromatin remodeling that associate with Stat5 upon PRL stimulation (38). This interaction appears to enhance Stat transcriptional activity by linking Stat to the basal transcriptional machinery. Nmi has been identified (39) as a Stat5-interacting protein by yeast two-hybrid analysis and found to enhance Stat5-mediated gene transcription in IL-2-responsive T cells and also to increase the formation of Stat5-p300/CBP complexes in these cells. We have examined whether the PRL/CypB complex interacts with either of these complexes and have found no evidence for such an interaction (data not shown). Therefore, the enhancement in Stat5 activity in the presence of PRL/CypB does not appear to occur through any of the previously identified mechanisms.
Whereas the above mechanisms are all involved in enhancing Stat5 activity, a growing body of evidence exists delineating the pathways through which Stat5 activity is down-regulated or inhibited. One direct mechanism of Stat inhibition is provided by the PIAS family of proteins. This family is comprised of five members: PIAS1, PIAS3, PIASxα, PIASxβ, and PIASy. These proteins bind to activated Stat family members upon cytokine stimulation and inhibit DNA-binding ability without directly dephosphorylating the Stat proteins. Therefore, given their constitutive expression, they appear to function by titrating the amount of activated Stat available after stimulation. Although PIAS1 has been demonstrated to bind to Stat1 (36) and PIAS3 to bind to Stat3 (35), it is not apparent from the literature that these PIAS family members were tested for their ability to bind other Stat family members, notably Stat5. By using polyclonal antibodies against both PIAS1 and PIAS3, we have demonstrated a PRL-inducible interaction between Stat5a and PIAS3 (Figs. 5 A and B). This interaction occurred only when cells were stimulated with PRL alone; upon stimulation with PRL and CypB, the association between PIAS3 and Stat5a was inhibited (Fig. 5C). Moreover, we have also shown by transient transfection studies that increasing amounts of PIAS3 inhibited PRL-induced Stat5-mediated transactivation of the β-casein promoter (Fig. 6A) and that increasing amounts of exogenous PRL/CypB overcame this PIAS3-mediated repression, whereas PRL or CypB by themselves could not (Fig. 6C). Therefore, not only was there a PRL-inducible interaction between PIAS3 and Stat5a, but this interaction served the function of inhibiting Stat5 activity. This inhibition was overcome by the coaddition of exogenous PRL and CypB. This occurs by the direct interaction of the PRL/CypB complex with the N terminus of Stat5a, the same region the aforementioned coactivators of Stat5 activity have been demonstrated to bind. Therefore, it is speculated that the PRL/CypB interaction with the N terminus of Stat5 induces a conformational change within Stat5, by the PPIase activity of CypB that mediates the release of PIAS3.
The release of the inhibitory effect of PIAS3 on Stat5 appears to be dependent on the PPIase activity of CypB. In this context, a regulatory role for the members of the immunophilin family, to which CypB belongs, in signal transduction and transcriptional activation is increasingly apparent (40). FK506-binding protein 12 has been demonstrated to serve as a negative regulator of transforming growth factor-β receptor endocytosis, thereby enhancing signaling through this receptor (41). Matrin CYP is an 88-kDa nuclear located PPIase that associates with the nuclear matrix and the C-terminal domain of RNA polymerase II (42), whereas cyclophilin 40 (Cyp40) within the nucleus has been demonstrated to interact with c-Myb and inhibit its binding to DNA (43). FK506-binding protein 52 has been shown to interact with interferon regulatory factor (IRF)-4 and inhibit its activity by conferring a structural modification on IRF-4 by its PPIase activity (44). Like the other immunophilin family members, CypB modulates transcriptional activity through its enzymatic activity. CypB significantly enhanced the PRL/CypB/Stat5 interaction within the nucleus (Fig. 3 A and C) and potentiated Stat5-mediated gene transcription (Fig. 2) by the release of PIAS3 (Fig. 5C). In contrast, the exogenous addition of the CypB-PPI mutant abrogated both of these effects. Therefore, in addition to their well-characterized roles as chaperones facilitating protein folding, the immunophilins are also important in the transcriptional activity of numerous genes, including Stat5.
In conclusion, this model of retrotransport of the PRL/CypB complex into the nucleus, where it serves as transcriptional inducer of Stat5 action, has considerable parallels with the genomic and nongenomic actions of steroid hormones (45). Collectively, these data support the argument that the summation of signals triggered by ligand at the cell surface and within the nucleus results in the spectrum of recognized steroid and peptide hormone actions. Modulation of these actions, such as those pharmacologically induced by steroid receptor antagonists, have had considerable clinical utility. Given the increasingly appreciated role of PRL in human breast cancer (46, 47) and prostate cancers (48), our findings with CypB-PPI suggest that similar clinical utility may be obtained through the modulation of the genomic actions of peptide hormones.
Acknowledgments
We thank Drs. Li Yu-Lee, Ke Shuai, and R. J. M. Ross for plasmid constructs, Drs. Rob Wilson and Steve Liebhaber for their review of the manuscript, and Dr. Dave Ross for his help with the luciferase assays. This study was supported in part by National Institutes of Health Grants F32DK10043 (to M.A.R.) and R01CA69294 (to C.V.C.) and American Cancer Society Grant RPG00307TBE (to C.V.C.)
Abbreviations
- GST
glutathione S-transferase
- PRL
prolactin
- PPIase
peptidyl-prolyl isomerase
- CypB
Cyclophilin B
- PIAS
protein inhibitor of activated Stat
- PRLr
prolactin receptor
- ER
endoplasmic reticulum
- CHO
Chinese hamster ovary
References
- 1.Goffin V, Kelly P A. Clin Endocrinol. 1996;45:247–255. doi: 10.1046/j.1365-2265.1996.00799.x. [DOI] [PubMed] [Google Scholar]
- 2.Ferrag F, Goffin V, Buteau H, Kelly P A. Cytokines Cell Mol Ther. 1997;3:197–213. [PubMed] [Google Scholar]
- 3.Clevenger C V, Freier D O, Kline J B. J Endocrinol. 1998;157:187–197. doi: 10.1677/joe.0.1570187. [DOI] [PubMed] [Google Scholar]
- 4.Campbell G S, Argetsinger L S, Ihle J N, Kelly P A, Rillema J A, Carter-Su C. Proc Natl Acad Sci USA. 1994;91:5232–5236. doi: 10.1073/pnas.91.12.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clevenger C V, Torigoe T, Reed J C. J Biol Chem. 1994;269:5559–5565. [PubMed] [Google Scholar]
- 6.Clevenger C V, Medaglia M V. Mol Endocrinol. 1994;8:674–681. doi: 10.1210/mend.8.6.7935483. [DOI] [PubMed] [Google Scholar]
- 7.Clevenger C V, Ngo W, Luger S M, Gewirtz A M. J Biol Chem. 1995;270:13246–13253. doi: 10.1074/jbc.270.22.13246. [DOI] [PubMed] [Google Scholar]
- 8.Ihle J N. Nature (London) 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt-Ney M, Doppler W, Ball R K, Groner B. Mol Cell Biol. 1991;11:3745–3755. doi: 10.1128/mcb.11.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raught B, Liao W S, Rosen J M. Mol Endocrinol. 1995;9:1223–1232. doi: 10.1210/mend.9.9.7491114. [DOI] [PubMed] [Google Scholar]
- 11.Stoecklin E, Wissler M, Gouilleux F, Groner B. Nature (London) 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 12.Wyszomierski S L, Rosen J M. Mol Endocrinol. 2001;15:228–240. doi: 10.1210/mend.15.2.0597. [DOI] [PubMed] [Google Scholar]
- 13.Clevenger C V, Sillman A L, Prystowsky M B. Endocrinology. 1990;127:3151–3159. doi: 10.1210/endo-127-6-3151. [DOI] [PubMed] [Google Scholar]
- 14.Rao Y-P, Buckley D J, Olson M D, Buckley A R. J Cell Physiol. 1995;163:266–276. doi: 10.1002/jcp.1041630207. [DOI] [PubMed] [Google Scholar]
- 15.Hazes B, Read R J. Biochemistry. 1997;36:11051–11054. doi: 10.1021/bi971383p. [DOI] [PubMed] [Google Scholar]
- 16.Wiertz E J H J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapaport T A, Ploegh H L. Nature (London) 1997;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 17.de Virgilio M, Weninger H, Ivessar N I. J Biol Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- 18.Johannes L, Goud B. Trends Cell Biol. 1998;8:158–162. doi: 10.1016/s0962-8924(97)01209-9. [DOI] [PubMed] [Google Scholar]
- 19.Clevenger C V, Altmann S W, Prystowsky M B. Science. 1991;253:77–79. doi: 10.1126/science.2063207. [DOI] [PubMed] [Google Scholar]
- 20.Rakowicz-Szulczynska E M, Rodeck U, Herlyn M, Koprowski H. Proc Natl Acad Sci USA. 1986;83:3728–3732. doi: 10.1073/pnas.83.11.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith R M, Jarett L. Proc Natl Acad Sci USA. 1987;84:459–463. doi: 10.1073/pnas.84.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobie P E, Mertani H, Morel G, Marales-Bustos O, Norstedt G, Waters M J. J Biol Chem. 1994;269:21330–21339. [PubMed] [Google Scholar]
- 23.Jans D A, Briggs L J, Gustin S E, Jans P, Ford S, Young I G. FEBS Lett. 1997;406:316–320. doi: 10.1016/s0014-5793(97)00293-7. [DOI] [PubMed] [Google Scholar]
- 24.Rycyzyn M A, Reilly S C, O'Malley K, Clevenger C V. Mol Endocrinol. 2000;14:1175–1186. doi: 10.1210/mend.14.8.0508. [DOI] [PubMed] [Google Scholar]
- 25.Ivery M T G. Med Res Rev. 2000;20:452–484. doi: 10.1002/1098-1128(200011)20:6<452::aid-med2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 26.Price E R, Jin M, Lim D, Pati S, Walsh C T, McKeon F D. Proc Natl Acad Sci USA. 1994;91:3931–3935. doi: 10.1073/pnas.91.9.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allain F, Boutillon C, Mariller C, Spik G. J Immunol Methods. 1995;178:113–120. doi: 10.1016/0022-1759(94)00249-v. [DOI] [PubMed] [Google Scholar]
- 28.Klappa R, Freedman R B, Zimmerman R. Eur J Biochem. 1995;232:755–764. [PubMed] [Google Scholar]
- 29.Mariller C, Haendler B, Allain F, Denys A, Spik G. Biochem J. 1996;317:571–576. doi: 10.1042/bj3170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kofron J L, Kuzmic P, Kishore V, Colon-Bonilla E, Rich D H. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- 31.Kline J B, Roehrs H, Clevenger C V. J Biol Chem. 1998;274:35461–35468. doi: 10.1074/jbc.274.50.35461. [DOI] [PubMed] [Google Scholar]
- 32.Husi H, Luyten M A, Zurini M G M. J Biol Chem. 1994;269:14199–14204. [PubMed] [Google Scholar]
- 33.Lechner J, Welte T, Tomasi J K, Bruno P, Cairns C, Gustafsson J, Doppler W. J Biol Chem. 1997;272:20954–20960. doi: 10.1074/jbc.272.33.20954. [DOI] [PubMed] [Google Scholar]
- 34.Hobbs A A, Richards D A, Kessler D J, Rosen J M. J Biol Chem. 1982;257:3598–3605. [PubMed] [Google Scholar]
- 35.Chung C D, Liao J, Liu B, Rao X, Jay P, Berta J, Shuai K. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 36.Liu B, Liao J, Rao X, Kushner S A, Chung C D, Chang D D, Shuai K. Proc Natl Acad Sci USA. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosen J M, Wyszomierski S L, Hadsell D. Annu Rev Nutr. 1999;19:407–436. doi: 10.1146/annurev.nutr.19.1.407. [DOI] [PubMed] [Google Scholar]
- 38.Pfitzner E, Jahne R, Wissler M, Stoecklin E, Groner B. Mol Endocrinol. 1998;12:1582–1593. doi: 10.1210/mend.12.10.0180. [DOI] [PubMed] [Google Scholar]
- 39.Zhu M-H, John S, Berg M, Leonard W J. Cell. 1999;96:121–130. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
- 40.Hunter T. Cell. 1998;92:141–143. doi: 10.1016/s0092-8674(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 41.Yao D, Dore J J E, Leof E B. J Biol Chem. 2000;275:13149–13154. doi: 10.1074/jbc.275.17.13149. [DOI] [PubMed] [Google Scholar]
- 42.Mortillaro M J, Berezney R. J Biol Chem. 1998;273:8183–8192. doi: 10.1074/jbc.273.14.8183. [DOI] [PubMed] [Google Scholar]
- 43.Leverson J D, Ness S A. Mol Cell. 1998;1:203–211. doi: 10.1016/s1097-2765(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 44.Mamane Y, Sharma S, Petropoulos L, Lin R, Hiscott J. Immunity. 2000;12:129–140. doi: 10.1016/s1074-7613(00)80166-1. [DOI] [PubMed] [Google Scholar]
- 45.Razandi M, Peram A, Greene G L, Levin E R. Mol Endocrinol. 1999;13:307–319. doi: 10.1210/mend.13.2.0239. [DOI] [PubMed] [Google Scholar]
- 46.Clevenger C V, Plank T L. J Mammary Gland Biol Neoplasia. 1997;2:59–68. doi: 10.1023/a:1026325630359. [DOI] [PubMed] [Google Scholar]
- 47.Hankinson S E, Willett W C, Michaud D S, Manson J E, Colditz G A, Langcope C, Rosner B, Speizer F E. J Natl Cancer Inst. 1999;91:629–634. doi: 10.1093/jnci/91.7.629. [DOI] [PubMed] [Google Scholar]
- 48.Wennbo H, Kindblom J, Isaksson O G P, Tornell J. Endocrinology. 1997;138:4410–4415. doi: 10.1210/endo.138.10.5461. [DOI] [PubMed] [Google Scholar]