Abstract
A particularly well-studied evolutionary model is the vinegar fly Drosophila melanogaster, a cosmopolitan insect of ancestral southern-central African origin. Recent work suggests that it expanded out of Africa ∼9,000 years ago, and spread from the Middle East into Europe ∼1,800 years ago. During its global expansion, this human commensal adapted to novel climate zones and habitats. Despite much work on phenotypic differentiation and adaptation on several continents (especially North America and Australia), typically in the context of latitudinal clines, little is known about phenotypic divergence among European populations. Here, we sought to provide a continent-wide study of phenotypic differentiation among European populations of D. melanogaster. In a consortium-wide phenomics effort, we assayed 16 fitness-related traits on a panel of 173 isofemale lines from 9 European populations, with the majority of traits measured by several groups using semi-standardized protocols. For most fitness-related traits, we found significant differentiation among populations on a continental scale. Despite inevitable differences in assay conditions among labs, the reproducibility and hence robustness of our measurements were overall remarkably good. Several fitness components (e.g., viability, development time) exhibited significant latitudinal or longitudinal clines, and populations differed markedly in multivariate trait structure. Notably, populations experiencing higher humidity/rainfall and lower maximum temperature showed higher viability, fertility, starvation resistance, and lifespan at the expense of lower heat-shock survival, suggesting a pattern of local adaptation. Our results indicate that derived populations of this tropical fly have been shaped by pervasive spatially varying multivariate selection and adaptation to different climates on the European continent.
Keywords: phenotypic variation, fitness traits, population differentiation, adaptation, D. melanogaster, Europe
Introduction
Over the last century, the vinegar fly Drosophila melanogaster has emerged as a premier experimental model system for studying various aspects of evolution, especially the process of adaptation and its genetic and phenotypic basis (Casillas & Barbadilla, 2017; David & Capy, 1988; David et al., 2004; Flatt, 2020; Hedrick & Murray, 1983; Hoffmann & Weeks, 2007; Keller, 2007; Lachaise & Silvain, 2004; Lachaise et al., 1988; Lewontin, 1974; Parsons, 1975; Powell, 1997; Prasad & Joshi, 2003). In particular, over the past decade, many population genomics studies have improved our understanding of the demography and adaptation in natural populations of this species (e.g., Barghi et al., 2019; Bergland et al., 2014, 2016; Chen et al., 2024; Coughlan et al., 2022; Fabian et al., 2012; Garud et al., 2021; Grenier et al., 2015; Huang et al., 2014; Kapun et al., 2020, 2021; Kolaczkowski et al., 2011; Lack et al., 2015, 2016; Langley et al., 2012; Machado et al., 2021; Mackay et al., 2012; Pool et al., 2012; Turner et al., 2008), as well as using experimentally evolved populations (e.g., Burke et al., 2010; Fabian et al., 2018; Graves et al., 2017; Hoedjes et al., 2019; Kawecki et al., 2021; Orozco-terWengel et al., 2012; Schlötterer et al., 2015; Turner et al., 2011).
This cosmopolitan insect originated in the seasonally dry Miombo and Mopane woodlands of tropical southern-central Africa before expanding across and then out of the African continent, causing a bottleneck in population size (Coughlan et al., 2022; Lachaise & Silvain, 2004; Lachaise et al., 1988; Mansourian et al., 2018; Nunes et al., 2008; Pool et al., 2012; Sprengelmeyer et al., 2020). While previous work has dated the out-of-Africa split time (bottleneck) to have occurred ∼12–19k years ago (kya) (Arguello et al., 2019; Duchen et al., 2013; Li & Stephan, 2006; Thornton & Andolfatto, 2006), a new study suggests that the out-of-Africa event occurred ∼9 kya (Chen et al., 2024). This out-of-Africa split was followed by an expansion into East Asia ∼2.8–4.4 kya (Chen et al., 2024), and a spread from the Middle East into Europe ∼1.8 kya (Sprengelmeyer et al., 2020). Australia and North America were colonized even more recently, i.e., only ∼100–150 years ago (Hoffmann & Weeks, 2007; Keller, 2007; and references therein).
During this history of expansion and globalization, this ancestrally tropical insect and human commensal adapted to new climate zones and ecological niches, including equatorial tropical rainforests, savanna grasslands, deserts, temperate grasslands, and alpine areas (Fabian et al., 2015; Keller, 2007; Lachaise & Silvain, 2004; Lachaise et al., 1988; Mansourian et al., 2018; Pool et al., 2012 ; Sprengelmeyer and Pool 2021). Indeed, consistent with local adaptation (Kawecki & Ebert, 2004), a large body of work has documented spatially varying selection and clines in D. melanogaster populations along latitudinal (and sometimes also altitudinal) gradients on multiple continents (reviewed in Adrion et al., 2015; David & Capy, 1988; de Jong & Bochdanovits, 2003; Flatt, 2020; Hoffmann & Weeks, 2007; Paaby & Schmidt, 2009).
Previous studies of clinality in D. melanogaster have established multiple lines of evidence for spatially varying selection: (1) clinal differentiation among populations in fitness traits such as viability, size, pigmentation, lifespan, and reproductive diapause (e.g., Coyne & Beecham, 1987; David, 1982; David & Bocquet, 1975a, 1975b; David et al., 1977, 1985; Fabian et al., 2015; Gibert et al., 2004; Gilchrist & Partridge, 1999; Hangartner et al., 2015; Klepsatel et al., 2014; Pitchers et al., 2013; Rajpurohit & Nevded, 2013; Robinson et al., 2000; Schmidt et al., 2005a, 2005b; Schmidt & Conde, 2006; Van‘t Land et al., 1999, 2000; Zwaan et al., 2000); (2) clinal genetic variation for individual markers or polymorphisms or at the level of whole genomes (e.g., Agis & Schlötterer, 2001; Bergland et al., 2016; Betancourt et al., 2021; Bogaerts-Márquez et al., 2021; Božičević et al., 2016; Fabian et al., 2012; Kapun et al., 2016a, 2020, 2021, 2023; Kolaczkoswki et al., 2011; Mateo et al., 2018; Oakeshott et al., 1982; Reinhardt et al., 2014; Singh et al., 1982; Turner et al., 2008); and (3) functional relationships between specific clinally varying polymorphisms and fitness-related traits (e.g., Betancourt et al., 2021; Durmaz et al., 2018, 2019; Erickson et al., 2020; Glaser-Schmitt et al., 2021; Kapun et al., 2016b; Lee et al., 2013; Paaby et al., 2010, 2014; Schmidt et al., 2008; Yu & Bergland, 2022).
In addition, several studies have found pervasive genomic and phenotypic evidence for temporally (seasonally) varying selection acting in North American and European populations of D. melanogaster (e.g., Behrman et al., 2015, 2018; Bergland et al., 2014; Bitter et al., 2024; Cogni et al., 2015; Kapun et al., 2016a; Machado et al., 2021; Nunez et al., 2024; Rodrigues et al., 2021; Rudman et al., 2022). Together, these studies have greatly advanced our understanding of the mechanisms of spatially and temporally varying selection and the spatio-temporal scale of adaptation.
In contrast to North America (Schmidt et al., 2005a, 2005b), India (Rajpurohit et al., 2017), Australia (Hangartner et al., 2015; Hoffmann & Weeks, 2007), and sub-Saharan Africa (Fabian et al., 2015), however, we still have little systematic knowledge of phenotypic patterns of spatial differentiation and clinal adaptation among European populations of D. melanogaster (e.g., Ayrinhac et al., 2004; Draye & Lints, 1996; Draye et al., 1994; Imasheva et al., 1994). For instance, most work on European D. melanogaster has examined only a handful of populations and traits (reviewed in Flatt, 2020). Thus, large-scale patterns of phenotypic differentiation, such as phenotypic clines or patterns of local adaptation, remain quite poorly understood for European populations of the vinegar fly.
Here, we sought to address this major knowledge gap by leveraging the collaborative resources and workforce of the European Drosophila Population Genomics Consortium, DrosEU (https://droseu.net/) (Figure 1). In our previous work, we provided the first continent-wide analysis of patterns of genetic variation among European populations based on pool-sequencing data from 32 populations (Kapun et al., 2020). In that study, we found evidence for continent-wide selective sweeps and identified many candidate loci for local adaptation, as well as spatial frequency clines for inversion polymorphisms and transposable elements (Kapun et al., 2020; also see Kapun et al., 2021; Machado et al., 2021; Drosophila Evolution over Space and Time consortium [https://dest.bio/]). Yet, patterns of phenotypic differentiation among these European populations remained largely unknown.
Figure 1.
(A) Map of the nine locations where European populations of Drosophila melanogaster were sampled by the DrosEU consortium. PT, Portugal (Recarei = RE); ES, Spain (Gimenells = GI [Lleida]); TR, Turkey (Yesiloz = YE); DE, Germany (Munich = MU); AT, Austria (Mauternbach = MA); UA, Ukraine (Uman = UM); DK, Denmark (Karensminde = KA); FI, Finland (Akaa = AK); and RU, Russia (Valday = VA) (see Supplementary Table S1 and our GitHub website; also see Kapun et al., 2020, 2021). (B) Map showing the locations of the labs that contributed to phenotyping, an effort involving >100 researchers in 26 groups in 17 countries. Lines were maintained by É. Sucena (Instituto Gulbenkian de Ciência, Oeiras, Portugal) and shipped to recipient labs for phenotyping (Table 1; see also Supplementary Table S2).
To complement our population genomic analyses with information about fitness-relevant phenotypes, we performed a continent-wide phenotypic analysis of representative populations using an isofemale line approach (David et al., 2005; Parsons & Hosgood, 1967; see below). In summer and fall 2018, we sampled nine European populations, spanning 21° latitude and 41° longitude across the continent, and established a panel of 173 isofemale lines (∼20 lines per population; Figure 1). In early 2019, lines were shipped to participating research groups, with the bulk of the phenotyping performed in 2019 and 2020. We assayed this DrosEU Phenotyping Panel (DPP) of isofemale lines for 16 traits (this study; see Table 1), most of which represent major phenotypic components of Darwinian fitness, including traits related to growth, size, survival, stress resistance, and reproduction (Flatt, 2020).
Table 1.
Sixteen phenotypic traits assayed in our study.
| Phenotypic trait | Trait aspect | Sex | Labs (PIs) |
|---|---|---|---|
| 1. Viability | – | Mixed | PG, SG, KH, PS, MSR, BZ |
| 2. Developmental time | Egg-to-pupa | Mixed | PS |
| Egg-to-adult | M | PG, SG, KH, PS, MSR, BZ | |
| F | |||
| 3. Dry weight | – | M | HC, KH, BO |
| F | |||
| 4. Thorax length | – | M | IK, NP, MR, PS |
| F | |||
| 5. Wing area | Right | M | BO, NP, MR, MSR |
| F | |||
| Left | M | ||
| F | |||
| 6. Fertility | – | F | JCB, CF |
| 7. Lifespan | Line level | M | JP, EP |
| F | |||
| Population level | M | TF | |
| F | |||
| 8. Cold shock mortality | – | M | JG, IK, JV |
| F | |||
| 9. Chill coma recovery time | – | M | JV, JM |
| F | |||
| 10. Heat shock mortality | – | M | JP, JV |
| F | |||
| 11. Diapause | – | F | AB, TF, CS |
| 12. Locomotor activity | Activity | M | ET |
| Circadian phase | |||
| Absolute phase | |||
| Period | |||
| ND (nocturnal/diurnal ratio) | |||
| 13. Circadian eclosion timing | ZT_hours_MESA | Mixed | CW |
| ZT_hours_LSPR | |||
| Period_MESA | |||
| Period_LSPR | |||
| Rhythmicity_LSPR_amplitude | |||
| Rhythmicity_JTK_p_BH_corrected | |||
| 14. Pigmentation | Total | F | JA, PG, PS |
| Sixth tergite | |||
| Fifth tergite | |||
| Fourth tergite | |||
| 15. Starvation resistance | – | M | JG, BO, EP |
| F | |||
| 16. Parasitoid resistance | – | Mixed | JH |
Note: The following investigators and their teams contributed to the DrosEU phenotyping effort (in alphabetical order): Jessica Abbott (JA); Alan Bergland (AB); Jean-Christophe Billeter (JCB); Hervé Colinet (HC); Claudia Fricke (CF); Thomas Flatt (TF); Patricia Gibert (PG); Josefa González (JG); Sonja Grath (SG); Katja Hoedjes (KH); Jan Hrcek (JH); Iryna Kozeretska (IK); Julian Mensch (JM); Banu Onder (BO); John Parsch (JP); Elena Pasyukova (EP); Nico Posnien (NP); Michael G. Ritchie (MR); Christian Schlötterer (CS); Paul Schmidt (PS); Marina Stamenkovic-Radak (MSR); Eran Tauber (ET); Jorge Vieira (JV); Christian Wegener (CW); and Bas J. Zwaan (BZ). For more details, see Supplementary Table S2 and our GitHub website.
Measuring phenotypic traits on isofemale lines, i.e., full-sib families derived from single, inseminated females, is a convenient method for studying quantitative traits and their genetic architecture (David et al., 2005; Parsons & Hosgood, 1967). By “capturing” genotypes from a natural population, isofemale lines provide a useful “proxy” for genetic variation: Over time, as inbreeding becomes maximal, all phenotypic variation within lines eventually represents environmental variation, whereas all variation among lines eventually represents genetic variation (e.g., David et al., 2005, 2005; Falconer & Mackay, 1996; Geber, 1990; Parsons & Hosgood, 1967). A related point is that isofemale lines effectively preserve the ancestral genetic variation present in the outbred population, and the lines can be used to reconstitute the ancestral population (Nouhaud et al., 2016). Although freshly established isofemale lines often maintain segregating variation for several generations in the lab before becoming fully inbred (Endler et al., 2016), and while such variation could, in principle, contribute to lab adaptation, Nouhaud et al. (2016) were unable to detect any significant allele frequency changes between ancestral Drosophila populations and reconstituted ancestral populations even when isofemale lines were maintained for over 6 years in the lab. Further advantages of this approach include the fact that the repeatability of phenotypic measurements made on isofemale lines is typically quite high (David et al., 2005) and that panels of inbred isofemale lines can be used to perform genome-wide association studies (e.g., see Gardeux et al., 2024; Mackay et al., 2012). However, potential drawbacks of isofemale lines are that the inbreeding process can render deleterious recessive alleles homozygous and might lead to artifactual positive correlations among line means for fitness components when there was a negative correlation in the outbred population (Rose, 1984).
Our continent-wide phenotypic study of European D. melanogaster involved >100 researchers from 26 research groups in 17 countries (Figure 1). This effort resulted in >400,000 individual fly observations based on semi-standardized experimental protocols and followed by the analysis of >>100 statistical models. The high dimensionality and large size of our dataset make our study an example of “phenomics,” i.e., the acquisition of comprehensive, high-dimensional phenotype data, an important challenge and frontier in evolutionary biology (see Houle, 2010; Houle et al., 2010). As we assayed the majority of traits in multiple labs in parallel, our unprecedented phenotyping effort allowed us to examine the reproducibility of the data (see Table 1 and the Results and discussion section). The quantification of such repeatability has not been done on such a large scale before, yet it is important, particularly as phenotypic measurements of Drosophila strains are typically not replicated across labs.
Because of the massive size of our phenomic dataset, here we can only provide a summary of our most important findings. A complete description of all our methods and data, including numerous results and analyses that are not shown in the main text, can be found on our dedicated GitHub website (https://esradm.github.io/DrosEU_PhenotypingWG/).
Results and discussion
To study patterns of phenotypic variation and differentiation among the nine European populations (Figure 1), we defined 16 major traits, including several developmental, morphological, reproductive, behavioral, and stress- and survival-related traits (Table 1).
Most of these traits represent major components of fitness, including several classical life-history traits (Charlesworth, 1994; Flatt, 2020; Roff, 1992; Stearns, 1992), and might thus be phenotypic targets of selection. For several of these traits, we measured distinct “aspects” or “proxies,” as shown in the second column of Table 1.
The majority of traits were measured in multiple labs (13/16 = 81%) and both males and females. All assays were carried out at 25°C, 12 hr light:12 hr dark, and a minimum relative air humidity of 60% (unless stated otherwise; for details, see the Supplementary Materials). Because strict standardization of protocols was practically challenging to implement across all 26 participating research groups, we opted to be pragmatic and to use a “semi-standardized” study design. In practice, this meant that while we strived to use standardized protocols whenever possible, we allowed for flexibility across labs in terms of implementing experimental conditions and protocols. In part, this flexibility was also necessitated by lockdowns due to the COVID-19 pandemic.
Major components of fitness vary markedly among European populations
To quantify the extent of phenotypic differentiation among populations, we used linear models (Figure 2A). Due to inevitable differences in study design, data collection, and data structure among labs, it was often not possible to fit a single global trait-specific model that could integrate all data across labs measuring the same trait. Therefore, we fitted lab-specific models instead (see the Supplementary Materials; also see Website Section 2.1).
Figure 2.
Phenotypic differentiation among European populations of Drosophila melanogaster and reproducibility of trait measurements. (A) Percentage of phenotypic variance explained by sampling location (R2 = variance explained by the fixed-effect factor Population), for each trait (and sex, where applicable). Each dot represents the R2 value extracted from each of the 97 individual linear models. (95 dots represent marginal R2 values from linear mixed models; for two trait measurements [viability measured by the PS lab; locomotor activity ‒ absolute phase, measured by the ET lab], we used simple linear models and extracted regular R2 values.) Colored dots represent significant model p-values (⍺ = 0.05). Note that circadian eclosion timing was not analyzed using a linear modeling approach and is not shown here (see Supplementary Sections 1.6 and 2). (B) Pairwise Pearson’s correlation coefficients (r) between isofemale trait values for the same phenotype estimated by different (pairs of) labs that had measured the same trait, using line coefficients extracted from linear models. Colored dots show significant correlations (⍺ = 0.05). Traits that were only measured by a single lab are not shown. (C) Results of the meta-analyses for the effect of “Population,” showing (among-population) heterogeneity (Cochran’s Q statistic), extracted from subgroup meta-analyses based on population estimates from linear models. Colored dots represent significant differences between subgroups (= populations) after Bonferroni correction (α’ = α/n = 0.05/26 = 0.0019). As in (B), traits measured in single labs (as well as thorax length in males) did not enter these analyses. For further details, see the Supplementary Materials.
Our analyses revealed pervasive differentiation among European populations of D. melanogaster for most of the phenotypic traits measured (Figure 2A). For the great majority (∼70%) of the 97 linear models (i.e., combinations of traits, sexes, and labs), the factor Population explained a statistically significant proportion of the total phenotypic variance (Figure 2A; see Website Section 2.1 for plots of trait value estimates). The marked differences among European D. melanogaster in major fitness-related traits are consistent with adaptive differentiation among these populations. It is important to point out, however, that differences among populations in fitness-related traits measured in the lab do not necessarily imply that such differences are adaptive in nature (Lewontin, 2000a, 2000b).
While the range of marginal R2 values for the factor Population was very broad (0.009 [fertility]–0.26 [pigmentation]; mean across all estimates = 0.081; standard error [SE] = 0.006), much of the phenotypic variance in our dataset might be genetically based. This interpretation is supported by our estimates of broad-sense heritabilities (H2; estimated as “isofemale” heritabilities or intraclass correlation coefficients; David et al., 2005; Hoffmann & Parsons, 1988; Parsons, 1983) of the traits measured in individual labs (range across all individual estimates = 0.01 [locomotor activity]–0.67 [wing area]; mean = 0.36; SE = 0.018; for details, see Website Section 2.4). These estimates agree well with previous estimates in D. melanogaster (Roff & Mousseau, 1987); they are also broadly consistent with the observation of significant genetic differentiation among these nine (as well as other) European populations in our previous genomic analyses (Kapun et al., 2020, 2021; Machado et al., 2021; also see https://dest.bio/).
Trait estimates are reproducible despite differences in assay conditions
We often observed, sometimes large, differences in trait value estimates among labs (Figure 2A; also see Website Section 2.1), potentially attributable to differences in environmental conditions among labs. Such environmental variance might include uncontrolled macro-environmental differences (e.g., diet), unknown, uncontrollable micro-environmental variance (“developmental noise”), or inadvertent differences in experimental protocols and assay conditions (Ackermann et al., 2001; Crabbe et al., 1999; Falconer & Mackay, 1996; Flatt, 2005).
For example, while several traits (e.g., thorax length, pigmentation, bristle number, ovariole number) tend to be quite repeatable and rather insensitive to small, uncontrolled variability in experimental conditions, others (behavioral, physiological, or life-history traits) can be strongly sensitive to variation in conditions (Figure 2A; cf. David et al., 2005; also see Ackermann et al., 2001; Betancourt et al., 2021; Durmaz et al., 2019; Flatt et al., 2013; Leroi et al., 1994; May et al., 2019; Min et al., 2008; Rose et al., 1996). Measurements of chill-coma recovery time, for instance, are often highly variable, sometimes even under apparently identical assay conditions within the same lab (David et al., 2005; cf. Figure 2B).
The variability in trait estimates among labs is not surprising given that we used semi-standardized experimental protocols. For example, while many labs used semi-standardized diets (i.e., diets with the same composition but not using the same brands of ingredients), other groups used different, non-standardized diets. Indeed, as expected, differences in diet composition (protein [P]:carbohydrate [C] ratio) among labs had a significant effect on estimates (see Website Section 2.10). On the other hand, in the majority of cases, differences in the Wolbachia infection status of the assayed isofemale lines had no detectable impact on the studied phenotypic traits (see Website Section 2.13).
To quantify reproducibility, we calculated pairwise Pearson’s correlations between trait values estimated by labs that had measured the same trait. Despite differences in assay conditions among labs, the reproducibility of estimates was overall good, with the majority of correlations between lab estimates being significantly (albeit only weakly to moderately) positive (Figure 2B; Website Section 2.3). For example, population means for dry weight, wing area, and starvation resistance were significantly positively correlated among labs, indicating robust reproducibility. Likewise, even for some complex and/or highly environmentally sensitive traits, such as lifespan and reproductive diapause, correlations were positive and significant. Conversely, for other quantitative traits such as development time, fertility, chill-coma recovery time, and heat-shock mortality, reproducibility was low, in agreement with previous findings (e.g., see David et al., 1998, 2005). Such differences between traits in their reproducibility might reflect differences in their degree of environmental sensitivity (phenotypic plasticity versus environmental canalization) (Flatt, 2005; also see Flatt, 2020; Houle, 1992, 1998; Price & Schluter, 1991; Stearns et al., 1995). From an experimental point of view, a not mutually exclusive alternative is that some traits are more “noisy” and inherently more difficult to measure than others, especially when protocols are not strictly standardized (Ackermann et al., 2001; David et al., 1998, 2005; May et al., 2019). Thus, the fact that the significant pairwise correlations in Figure 2 tend to be only weakly to modestly positive likely reflects differences in assay conditions among labs and/or genotype-by-assay environment interactions.
To obtain estimates of phenotypic differences among populations that are unlikely to be confounded by differences in assay conditions among labs, we performed meta-analyses (Balduzzi et al., 2019) of linear models across labs (“studies”) and quantified heterogeneity among subgroups (= populations) by estimating Cochran’s Q (see the Supplementary Materials for methodological details). These analyses confirmed significant effects of population differentiation for viability, lifespan, and wing area (Figure 2C): These three complex traits show the strongest, most consistent evidence for differentiation among European populations in our data.
Generally, however, our meta-analyses seemed to be underpowered, presumably due to the relatively small number of labs and populations involved. Indeed, meta-analyses based on Cochran’s Q often tend to be underpowered (Pereira et al., 2010). Yet, despite most of our meta-analyses being nonsignificant, phenotypic differentiation among populations was significant for the majority of traits (see above: Figure 2A; ∼70% of linear models showed a significant effect of the factor Population at p < 0.05). Moreover, the clear prevalence of significantly positive correlations between labs underscores the overall high degree of reproducibility of our data across labs (see above; Figure 2B).
Our finding of high reproducibility despite variability in experimental procedures among labs is interesting given a body of work suggesting that “heterogenization” of samples or study design can improve reproducibility (Karp, 2018; Nakagawa et al., 2024; Richter et al., 2009, 2010; Usui et al., 2021; Van Der Staay et al., 2010; Voelkl & Würbel, 2016, Voelkl et al., 2018, 2020). The reason for this is the “standardization fallacy,” i.e., the notion that more stringent standardization of protocols will necessarily improve the reproducibility of experimental outcomes (Voelkl & Würbel, 2016). Contrary to this idea, many environmental factors may be difficult or impossible to rigorously standardize across labs even when this is deliberately being attempted (Crabbe et al., 1999). Individuals and measurements might therefore be less variable within a single lab than among labs; together with the fact that many environmental factors might defy strict standardization, this can lead to idiosyncratic, less reproducible results among labs (Voelkl & Würbel, 2016). Thus, paradoxically, less standardized studies that, together, cover overlapping ranges of environmental conditions, and therefore explore a broader part of the underlying reaction norm, might improve reproducibility, especially when environmental heterogeneity is introduced systematically (Nakagawa et al., 2024; Richter et al., 2009; Voelkl & Würbel, 2016). This calls for improving study designs through collaborative multi-institutional studies that perform experiments in parallel and include “heterogenization” of the design (Nakagawa et al., 2024; Richter et al., 2009; Voelkl & Würbel, 2016). Such coordinated multi-lab studies are, however, rare in evolutionary biology (e.g., Ackermann et al., 2001; for a recent small-scale example involving two research groups, see Durmaz et al., 2019; Betancourt et al., 2021).
Fitness components are genetically correlated in multivariate trait space
Many components of fitness are thought to be phenotypically, physiologically, and genetically correlated with each other: As they interact to jointly determine fitness, they should be viewed from a multivariate perspective (Charlesworth, 1993; Fabian et al., 2015; Flatt, 2020; Flatt & Heyland, 2011; Houle, 2001; Lande, 1982; Lande & Arnold, 1983; Roff, 2007; Schmidt et al., 2005b; Sinervo & Svensson, 2002; Stearns, 1992; Svensson, 2023; Svensson et al., 2021).
To study multivariate phenotypes, we derived “compound” estimates across labs for each trait and line from the linear models (Supplementary Materials). Pairwise Pearson correlations of these estimates revealed many pairs of traits that were significantly genetically correlated (Website Section 2.7). We explored the main axes of variation in the ensemble of these traits (scaled to unit variance) using principal component analyses (PCAs). Initially, we included 13 traits measured on females only plus viability (see the Supplementary Materials; Website Section 2.8); we then conducted a comparison of males and females using only the nine traits that had been measured separately in both sexes (Figure 3).
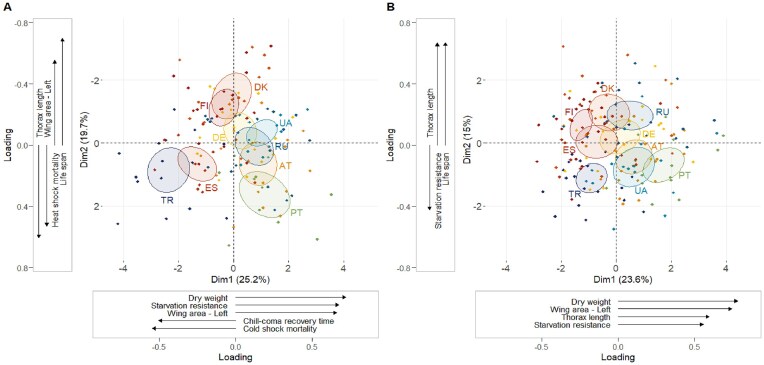
Figure 3.
Principal component analysis (PCA) plots and loadings for the first principal component (PC1) and the second principal component (PC2) in (A) males and (B) females. The same nine phenotypic traits were used in both PCAs. Confidence ellipses (95%) are drawn for each of the nine populations. Phenotypic traits with greater-than-average contributions (loadings) to a given principal component are shown in the accompanying x- and y-axis vector plots. Note that in males, the y-axis (PC2) is inverted so that the direction of phenotypic trait correlations matches across the two sexes. For further details, see the Supplementary Materials; also see Website Section 2.8.
In the 13-trait phenotype PCA, the first principal component (PC1) was defined by positive correlations of size, notably wing area (0.774), thorax length (0.592), and dry weight (0.753) as well as starvation resistance (0.502), and a negative correlation with lifespan (−0.453). PC2 revealed positive correlations between viability (0.782), fertility (0.633), starvation resistance (0.560), lifespan (0.417), and heat-shock mortality (0.435) (see plots and full loadings table in Website Section 2.8). Interestingly, the traits with the strongest loadings for these two axes, wing area (PC1: 0.774) and viability (PC2: 0.782), were also those with the largest Q values in the meta-analysis (see Figure 2C), reinforcing that these traits represent reliable markers of population differentiation in European D. melanogaster.
When comparing males and females, the overall patterns of trait correlation were similar (Figure 3). In both sexes, PC1 was defined by positive correlations between dry weight, wing area, and starvation resistance (0.783, 0.745, and 0.560 for females; 0.722, 0.664, and 0.677 for males); PC2 included a negative correlation between thorax length and lifespan (−0.424 and 0.671 for females; 0.604 and −0.699 for males). PC3 included a negative correlation between heat-shock mortality and cold-shock mortality (0.553 and −0.564 for females; −0.412 and 0.367 for males [for full loadings tables, see Website Section 2.8]). Yet, we also found several differences between the sexes. Correlations between traits were stronger in males than in females: The first four PCs explained 70.08% of the variance in males, but only 63.11% in females. Additionally, some traits showed sex-specific patterns of correlation. Interestingly, chill-coma recovery time was positively correlated with cold shock mortality in males (both PC1 and PC3), but negatively correlated with cold shock mortality in females (PC3). Furthermore, several traits (starvation resistance, wing area, and heat-shock mortality) showed sex-specific correlations along PC2 (Figure 3).
Several of the above-mentioned correlations between fitness components have been observed before, e.g., in selection experiments and/or in natural populations, such as the correlation between proxies of body size or weight and starvation resistance, or between lifespan and starvation resistance (e.g., see de Jong & Bochdanovits, 2003; Durmaz et al., 2019; Fabian et al., 2015; Flatt, 2020; Gardeux et al., 2024; Klepsatel et al., 2013; Prasad & Joshi, 2003; Stearns & Partridge, 2001). Similarly, negative correlations between size and lifespan have been found previously, yet not systematically so—this relationship is highly strain-specific and can also depend on temperature (Khazaeli et al., 2005; also see Flatt, 2020; Norry & Loeschcke, 2002).
European populations vary in multivariate trait structure
Next, we asked whether European populations of D. melanogaster might differ in their multivariate trait correlation structure; significant differences across space could indicate spatially varying selection on multivariate suites of fitness components.
Confidence ellipses for populations showed considerable separation along PC1 and PC2, particularly in males, as seen in Figure 3. To quantify the degree of multivariate differentiation among populations, we carried out population reallocation procedures and calculated Mahalanobis distances following discriminant function analysis (DFA) for both sexes separately (in females using the same set of traits as the PCA, and in males using the nine measured traits plus viability; for details, see the Supplementary Materials and Website Section 2.9).
Multivariate discrimination resulted in quite high levels of identifiability of populations (see Website Section 2.9). We found that 76.9% of male and 72.7% of female line estimates could be successfully reclassified according to their population of origin. The highest reclassification rates were for Turkey, with 100% of males and 95% of females reclassified correctly. Furthermore, among male flies, the highest intergroup Mahalanobis distances separated Turkey from Ukraine (24.05) and Finland (23.16), while low Mahalanobis values were observed separating Finland from Germany (3.25) and Austria from Russia (3.77). Among females, the highest Mahalanobis distances separated Ukraine from Spain (25.78) and Turkey from Finland (22.48), whereas low Mahalanobis values separated Russia from Germany (3.39) and Denmark (3.41) (for details, see Website Section 2.9). These results suggest that the Turkish population is the most distinct among the populations sampled, but that others, such as Ukraine, Finland, and Spain, also show strong differences in their multivariate trait structure.
Our results are thus consistent with ample scope for spatially varying, multivariate (including correlational) selection operating on European populations of D. melanogaster, similar to previous findings for North American and African populations, which differ markedly in multivariate trait structure (e.g., Fabian et al., 2015; Schmidt et al., 2005b). Notably, many of the populations that exhibit strong differences in their multivariate trait structure (i.e., Turkey, Finland, and Spain) are also among the geographically most distant populations in our dataset.
Climatic factors explain spatial patterns of trait differentiation
Many studies have documented spatially (clinally) varying selection among D. melanogaster populations on multiple continents (especially in North America and Australia), both at the genetic and phenotypic levels (see the Introduction section and references therein), but still little is known about clinal patterns on the European continent. A few studies have identified clines at the genetic level, e.g., for individual adaptive (e.g., indel) polymorphisms, neutrally evolving SNPs in short introns, as well as for transposable elements and inversion polymorphisms (Costa et al., 1992; David et al., 1986; Kapun et al., 2020, 2021; Sandrelli et al., 2007; Tauber et al., 2007), yet the evidence remains limited. Similarly, phenotypic clines in Europe remain understudied (see the Introduction section; for a recent overview, see Flatt, 2020).
We observed significant latitudinal differentiation among populations for viability, development time, wing area, thorax length, fertility, starvation resistance, heat-shock mortality, and lifespan (results depended on both lab and sex; for details, see Website Section 2.6). These results are broadly consistent with findings from other continents (see the Introductionsection). For example, the latitudinal cline for wing area is in qualitative agreement with a similar cline in wing length among populations from Eastern Europe, the Caucasus, and Central Asia (Imasheva et al., 1994). Similarly, three labs found a consistent effect of latitude of origin on viability across populations, suggesting the existence of a positive latitudinal cline for this trait in Europe (see Website Section 2.6). To the best of our knowledge, a European cline for this major fitness trait has not been reported, but a similar cline has been found in South America (Folguera et al., 2008; but see Van‘t Land et al., 1999).
Similarly, we observed longitudinal clines for particular traits such as developmental time, pigmentation, chill-coma recovery time, and female reproductive diapause (results depended on lab and sex; see Website Section 2.6). These longitudinal phenotypic clines are particularly interesting because (1) we have previously identified a pattern of major east–west genetic structure that divides the European continent into a western and an eastern cluster of populations (Kapun et al., 2020, 2021) and (2) longitudinal phenotypic clines remain practically unknown for D. melanogaster to date, with very few exceptions (see Fabian et al., 2015).
On several continents, genetic and phenotypic patterns of clinality are affected by chromosomal inversion polymorphisms (Adrion et al., 2015; de Jong & Bochdanovits, 2003; Durmaz et al., 2018; Hoffmann & Weeks, 2007; Kapun & Flatt, 2019; Kapun et al., 2016a, 2016b, 2023; Lemeunier & Aulard, 1992). For example, we have previously observed latitudinal clines for In(3L)P, In(3R)C, and In(3R)P, as well as longitudinal clines for In(2L)t and In(2R)NS, across Europe (Kapun et al., 2020). Here, we found that population-specific mean frequencies of several polymorphic inversions (In(2L)t, In(2R)NS, In(3L)P, In(3R)P, and In(3R)Mo) were significantly correlated with population-specific estimates of fitness traits (see Website Section 2.14), yet Bonferroni correction rendered these correlations nonsignificant. We also found that the presence versus the absence of inversions had significant effects on particular traits at the line level (see Website Section 2.14). Again, however, most p-values were nonsignificant after Bonferroni adjustment, except for a significant effect of In(3R)P on male heat-shock mortality. These preliminary observations suggest that inversion polymorphisms might contribute to spatial patterns of trait differentiation across the European continent.
Several major fitness components thus vary clinally across latitude and/or longitude in European populations of D. melanogaster. However, although latitude and longitude are often correlated with climatic variables such as temperature, they merely represent indirect proxies for causative climatic or other spatially varying factors. Moreover, clinality can also arise from demographic processes such as admixture or isolation by distance, not only spatially varying selection (Bergland et al., 2016; Flatt, 2016; Kapun et al., 2016a). Studying climatic variables might thus provide more direct and accurate evidence for the role of environmental factors in shaping population differentiation and adaptation.
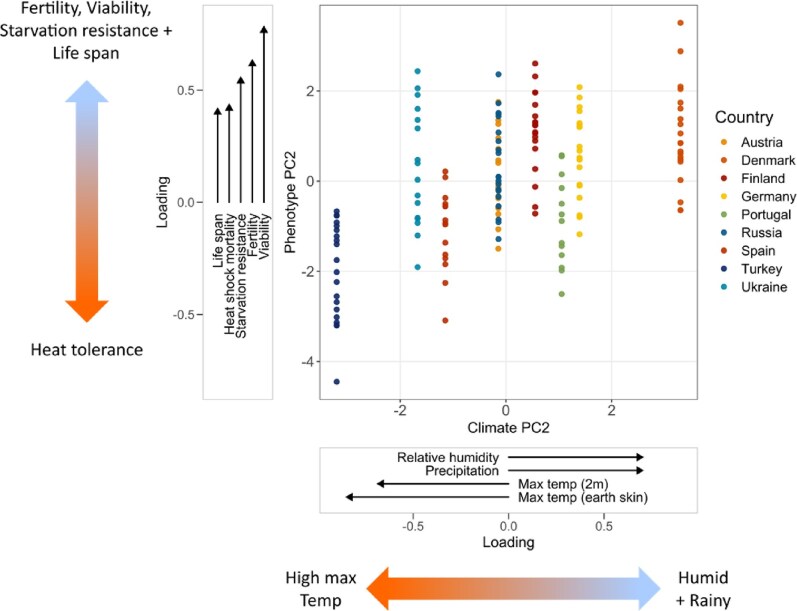
To obtain summary climate variables, data for 14 climatic measures from the last 30 days and the last 30 years were retrieved from the NASA database, and their dimensionality was reduced using separate PCAs, with the resultant PC1 and PC2 explaining 94.7% (30-day data) and 89.8% (30-year data) of the total variation. Because long-term climatic trends are more likely to shape broad patterns of phenotypic evolution, we focused on the PCs obtained using 30-year data here. PC1 was driven by a negative correlation between average temperature (earth skin temperature, temperature at 2 m, and wet bulb temperature) and the number of frost days, while PC2 was driven by a negative correlation between maximum temperature (earth skin and 2 m) and relative humidity plus precipitation (for further details, see the Supplementary Materials and Website Section 2.12). We then tested whether these two climate summary variables were useful in predicting multivariate phenotypes (PCs 1, 2, and 3 of the phenotype PCAs; Website Section 2.12) using linear models, and confirmed results with a permutation procedure.
This analysis revealed a significant association between climate PC2 and phenotype PC2 for the 13-trait PCA (F = 78.151, df = 1, p < 0.0001; permutation p = 0.009): Notably, European populations experiencing higher humidity and rainfall and lower maximum temperatures (Bogaerts-Márquez et al., 2021) exhibit higher values for major fitness-related traits, including viability, fertility, starvation resistance, and lifespan (Figure 4). However, these populations are also characterized by increased heat-shock mortality; thus, local adaptation to milder, wetter climates comes at the expense of decreased heat tolerance, suggesting that there is a trade-off between viability, fertility, starvation resistance, and lifespan on the one hand and heat-shock survival on the other hand.
Figure 4.
Correlation between phenotype principal component 2 and climate principal component 2, using all female phenotypic traits plus viability, and climatic data from the previous 30 years. Climatic and phenotypic variables with greater-than-average contributions (loadings) to a given principal component are shown in the accompanying x- and y-axis vector plots, respectively. For the corresponding results on males, which look qualitatively similar but were not significant after permutation testing, see Website Section 2.12 for details.
Thermal tolerance is mediated by the expression of heat-shock proteins (HSPs; Hoffman et al., 2013), with selection for thermal stress resistance contributing to higher constitutive levels of HSP (Sørensen et al., 2017). However, increases in HSP copy number or expression also negatively affect other phenotypic traits such as metabolic rate, fecundity, and survival (Hoekstra & Montooth, 2013; Okada et al., 2014; Roberts & Feder, 2000; Silbermann & Tatar, 2000), suggesting that pleiotropic effects of HSPs and other genes involved in thermal stress responses might be an important factor underpinning phenotypic variation across climatic gradients (Chen et al., 2018). More generally, and consistent with our findings here, previous studies of Drosophila spp. have found that increased heat resistance is often associated with reduced viability, dry weight, fecundity, fertility, cold resistance, ethanol resistance, and mating frequency (see Hoffman et al., 2003).
Our finding of a trade-off between a suite of fitness components and heat-shock survival across space (i.e., between different climates) adds to a growing number of studies in D. melanogaster that have found evidence for patterns of local adaptation driven by spatially varying selection (Anderson et al., 2003; Betancourt et al., 2021; de Jong & Bochdanovits, 2003; Durmaz et al., 2018, 2019; Fabian et al., 2015; Kapun et al., 2016b; Paaby & Schmidt, 2009; Paaby et al., 2014; Schmidt & Paaby, 2008; Schmidt et al., 2005a, 2005b ; reviewed in Flatt, 2020).
It is also noteworthy that we failed to find a negative correlation between early fertility and lifespan (see Figure 4). This is interesting, as many studies in Drosophila have reported negative genetic correlations between early fecundity and lifespan, indicative of a trade-off (e.g., reviewed in Flatt, 2011, 2020; Stearns & Partridge, 2001). Whether the positive association between fertility and lifespan observed here is artifactual, e.g., due to inbreeding (Rose, 1984) or exposure to novel lab environments (Service & Rose, 1985) or whether cooler and wetter European climates are ecologically more “benign” in terms of selectively favoring higher fertility and survival remains unclear. Whatever might be the case, negative correlations between survival and reproduction are not always found, and even when they exist, multiple confounding factors can obscure them (e.g., see discussion in Flatt, 2011, 2020; Klepsatel et al., 2013; and references therein).
The association between climate PC2 and phenotype PC2 for the female and male nine-trait phenotype PCAs was, however, not significant following permutation testing (p = 0.057 and p = 0.11, respectively), perhaps due to the absence of viability from these PCAs.
Interestingly, associations between phenotype PC2 and climate PC2 had stronger support when considering 30-day data as compared to 30-year data (significant permutations for phenotypic PCAs; see Website Section 2.12), consistent with recent observations suggesting that short-term changes in the environment on the order of a few weeks or less can drive seasonal adaptation in flies (see Bitter et al., 2024; Machado et al., 2021; Nunez et al., 2024; Rudman et al., 2022; also cf. discussion in Hoffmann & Flatt, 2022).
Summary and conclusions
Here, we have undertaken a large-scale, collaborative phenomics effort to provide the first continent-wide, systematic characterization of patterns of phenotypic differentiation and clinality among European populations of the vinegar fly D. melanogaster, a classical model system for studying fundamental questions in evolutionary biology. Our most important findings and conclusions can be summarized as follows:
European populations of D. melanogaster are significantly differentiated with respect to numerous phenotypic components of fitness, which might be subject to spatially varying (diversifying) selection.
The majority of trait estimates were significantly positively correlated between pairs of labs that measured the same trait, suggesting a high degree of reproducibility despite differences in assay conditions among labs.
PCA revealed that numerous traits were significantly correlated with each other, and DFAs showed that European populations differ markedly in their multivariate trait structure, suggesting ample scope for multivariate spatially varying selection on phenotypic components of fitness.
Consistent with spatially varying selection being driven by climatic gradients, several fitness components exhibited significant latitudinal or longitudinal clinality among populations. Most notably, egg-to-adult survival (viability) and egg-to-adult development time varied latitudinally and longitudinally, respectively.
Populations subject to higher humidity/rainfall and to lower maximum temperatures were characterized by higher values for viability, fertility, starvation resistance, and lifespan, yet exhibited lower heat-shock survival, suggesting a trade-off between these fitness components and revealing local climate adaptation. Together with previous and current genomic analyses of these populations (Bogaerts‐Márquez et al., 2021; Kapun et al., 2020, 2021; Machado et al., 2021), it will clearly be of great interest to unravel the genetic basis underlying these phenotypic patterns of climate adaptation.
Many additional analyses and results, which we could not discuss due to space limitations, can be found on our GitHub website at https://esradm.github.io/DrosEU_PhenotypingWG/; we encourage readers to explore and make use of this rich phenomic dataset and resource.
The second resource that we wish to make available to the community is our multipopulation panel of isofemale lines, the DrosEU Phenotyping Panel (DPP). The DPP might be a useful complement to other existing D. melanogaster panels, such as the Drosophila Genetic Reference Panel (DGRP; Mackay et al., 2012), the Drosophila Population Genomics Project (DPGP; Pool et al., 2012), the Drosophila Genome Nexus (DGN; Lack et al., 2015, 2016), and the Global Diversity Lines (GDL; Grenier et al., 2015). The DPP is available upon request from Élio Sucena (jesucena@ciencias.ulisboa.pt); genomic analyses of the DPP by our consortium are currently underway.
Materials and methods
A detailed description of our materials and methods is given in the Supplementary Materials associated with the manuscript and also on our dedicated GitHub website at https://esradm.github.io/DrosEU_PhenotypingWG/.
Supplementary Material
Acknowledgments
We are grateful to two anonymous reviewers and Erik Svensson for valuable comments on our manuscript. We are indebted to the members of the DrosEU community for their support and collaboration over the years, and we thank the European Society for Evolutionary Biology (ESEB) for having generously funded our activities between 2016 and 2022 with a Special Topic Networks (STN) grant. We also wish to thank Yonatan Babore, Luis Castaneda, Patrick Favre, Liam Forsythe, Anna Grandchamp, Lennart Hüper, Shahzad Khan, Ozan Kiratli, Melissa Erika Klug, Susanne Klühspies, Nadine Landgraf Koelln, Qinyang Li, Liam Miller, Margot Paris, Marisa A. Rodrigues, and Axel Wiberg for their help with our project as well as Pavlo A. Kovalenko for his assistance with collecting flies in 2018.
Contributor Information
Esra Durmaz Mitchell, Department of Biology, University of Fribourg, Fribourg, Switzerland; Functional Genomics and Metabolism Research Unit, Department of Biochemistry and Molecular Biology, University of Southern Denmark, Odense M, Denmark.
Envel Kerdaffrec, Department of Biology, University of Fribourg, Fribourg, Switzerland.
Ewan Harney, Institute of Evolutionary Biology, CSIC, Universitat Pompeu Fabra, Barcelona, Spain.
Tânia F Paulo, Instituto Gulbenkian de Ciência, Oeiras, Portugal.
Marija Savic Veselinovic, Faculty of Biology, University of Belgrade, Belgrade, Serbia.
Marija Tanaskovic, Institute for Biological Research “Siniša Stanković”, National Institute of the Republic of Serbia, University of Belgrade, Belgrade, Serbia.
Venera Tyukmaeva, Department of Evolution, Ecology and Behaviour, Institute of Infection, Veterinary, and Ecological Sciences, University of Liverpool, Liverpool, United Kingdom.
Teresa Abaurrea Fernandez de Arcaya, Centre for Biological Diversity, University of St Andrews, St. Andrews, United Kingdom.
Cansu Aksoy, Genetic Variation and Adaptation Laboratory, Department of Biology, Faculty of Science, Hacettepe University, Ankara, Turkey.
Eliza Argyridou, Division of Evolutionary Biology, Faculty of Biology, Ludwig-Maximilians-Universität München, Munich, Germany.
Tiphaine P M Bailly, Groningen Institute for Evolutionary Life Sciences, University of Groningen, Groningen, The Netherlands.
Dogus Can, Genetic Variation and Adaptation Laboratory, Department of Biology, Faculty of Science, Hacettepe University, Ankara, Turkey.
Ezgi Cobanoglu, Genetic Variation and Adaptation Laboratory, Department of Biology, Faculty of Science, Hacettepe University, Ankara, Turkey.
Nicola Cook, Centre for Biological Diversity, University of St Andrews, St. Andrews, United Kingdom.
Seda Coşkun, Genetic Variation and Adaptation Laboratory, Department of Biology, Faculty of Science, Hacettepe University, Ankara, Turkey.
Slobodan Davidovic, Institute for Biological Research “Siniša Stanković”, National Institute of the Republic of Serbia, University of Belgrade, Belgrade, Serbia.
Ekin Demir, Genetic Variation and Adaptation Laboratory, Department of Biology, Faculty of Science, Hacettepe University, Ankara, Turkey.
Tânia Dias, Instituto de Investigação e Inovação em Saúde (I3S), Universidade do Porto, Porto, Portugal; Instituto de Biologia Molecular e Celular (IBMC), Porto, Portugal.
Somayeh Rasouli-Dogaheh, Biology Centre of the Czech Academy of Sciences, Institute of Entomology, České Budějovice, Czech Republic.
Pedro Duque, Instituto de Investigação e Inovação em Saúde (I3S), Universidade do Porto, Porto, Portugal; Instituto de Biologia Molecular e Celular (IBMC), Porto, Portugal.
Katarina Eric, Institute for Biological Research “Siniša Stanković”, National Institute of the Republic of Serbia, University of Belgrade, Belgrade, Serbia.
Pavle Eric, Institute for Biological Research “Siniša Stanković”, National Institute of the Republic of Serbia, University of Belgrade, Belgrade, Serbia.
Priscilla Erickson, Department of Biology, University of Virginia, Charlottesville, United States; Department of Biology, University of Richmond, Richmond, United States.
Filip Filipovski, Faculty of Biology, University of Belgrade, Belgrade, Serbia.
Bettina Fishman, Institute of Evolution, University of Haifa, Haifa, Israel.
Amanda Glaser-Schmitt, Division of Evolutionary Biology, Faculty of Biology, Ludwig-Maximilians-Universität München, Munich, Germany.
August Goldfischer, Department of Biology, University of Pennsylvania, Philadelphia, United States.
Llewellyn Green, Institute of Evolutionary Biology, CSIC, Universitat Pompeu Fabra, Barcelona, Spain.
Sonia Janillon, Université Claude Bernard Lyon 1, CNRS, LBBE, UMR 5558, Villeurbanne, France.
Mihailo Jelic, Faculty of Biology, University of Belgrade, Belgrade, Serbia.
Hristina Kostic, Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland.
Lucas E Kreiman, IEGEBA UBA-CONICET, Universidad de Buenos Aires, Buenos Aires, Argentina.
Natacha Kremer, Université Claude Bernard Lyon 1, CNRS, LBBE, UMR 5558, Villeurbanne, France.
Manolis Lyrakis, Institut für Populationsgenetik, Vetmeduni Vienna, Vienna, Austria; Platform for Bioinformatics and Biostatistics, Department of Biological Sciences and Pathobiology, Vetmeduni Vienna, Vienna, Austria.
Oleksandr M Maistrenko, European Molecular Biology Laboratory, Structural and Computational Biology Unit, Heidelberg, Germany; Department of Marine Microbiology and Biogeochemistry, Royal Netherlands Institute for Sea Research (NIOZ), ’t Horntje (Texel), The Netherlands.
Sapho-Lou Marti, University of Rennes, CNRS, ECOBIO, UMR 6553, Rennes, France.
Megan McGunnigle, Centre for Biological Diversity, University of St Andrews, St. Andrews, United Kingdom.
Miriam Merenciano, Institute of Evolutionary Biology, CSIC, Universitat Pompeu Fabra, Barcelona, Spain.
Mário S Mira, Groningen Institute for Evolutionary Life Sciences, University of Groningen, Groningen, The Netherlands.
Vincent Montbel, Biology Centre of the Czech Academy of Sciences, Institute of Entomology, České Budějovice, Czech Republic; Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic.
Laurence Mouton, Université Claude Bernard Lyon 1, CNRS, LBBE, UMR 5558, Villeurbanne, France.
Dmitry V Mukha, Vavilov Institute of General Genetics, Russian Academy of Sciences, Moscow, Russia.
Siddharth Murali, Department of Developmental Biology, Göttingen Center for Molecular Biosciences (GZMB), University of Göttingen, Göttingen, Germany.
Aleksandra Patenkovic, Institute for Biological Research “Siniša Stanković”, National Institute of the Republic of Serbia, University of Belgrade, Belgrade, Serbia.
Oleksandra Protsenko, Taras Shevchenko National University of Kyiv, Kyiv, Ukraine; National Antarctic Scientific Center of the Ministry of Education and Science of Ukraine, Kyiv, Ukraine.
Florencia A Putero, IBBEA-UBA-CONICET, Universidad de Buenos Aires, Buenos Aires, Argentina.
Micael Reis, Université Claude Bernard Lyon 1, CNRS, LBBE, UMR 5558, Villeurbanne, France.
Natalia V Roshina, Vavilov Institute of General Genetics, Russian Academy of Sciences, Moscow, Russia; Institute of Molecular Genetics, Russian Academy of Sciences, Moscow, Russia.
Olga Y Rybina, Institute of Molecular Genetics, Russian Academy of Sciences, Moscow, Russia.
Mads F Schou, Department of Biology, Aarhus University, Aarhus, Denmark.
Thibault Schowing, Department of Biology, University of Fribourg, Fribourg, Switzerland.
Senel Selin Senkal, Genetic Variation and Adaptation Laboratory, Department of Biology, Faculty of Science, Hacettepe University, Ankara, Turkey.
Svitlana Serga, National Antarctic Scientific Center of the Ministry of Education and Science of Ukraine, Kyiv, Ukraine; CBGP, INRAE, CIRAD, IRD, SupAgro, University Montpellier, Montpellier, France.
Virginie Trieu, Department of Biology, University of Fribourg, Fribourg, Switzerland.
Alexander V Symonenko, Institute of Molecular Genetics, Russian Academy of Sciences, Moscow, Russia.
Mikhail V Trostnikov, Institute of Molecular Genetics, Russian Academy of Sciences, Moscow, Russia.
Evgenia A Tsybul'ko, Institute of Molecular Genetics, Russian Academy of Sciences, Moscow, Russia.
Joost van den Heuvel, Laboratory of Genetics, Wageningen University, Wageningen, The Netherlands.
David van Waarde, Laboratory of Genetics, Wageningen University, Wageningen, The Netherlands.
Ekaterina R Veselkina, Institute of Molecular Genetics, Russian Academy of Sciences, Moscow, Russia.
Cristina P Vieira, Instituto de Investigação e Inovação em Saúde (I3S), Universidade do Porto, Porto, Portugal; Instituto de Biologia Molecular e Celular (IBMC), Porto, Portugal.
Xiaocui Wang, Groningen Institute for Evolutionary Life Sciences, University of Groningen, Groningen, The Netherlands.
Jelle Zandveld, Department of Biology, Utrecht University, Utrecht, The Netherlands.
Jessica Abbott, Department of Biology, Lund University, Lund, Sweden.
Jean-Christophe Billeter, Groningen Institute for Evolutionary Life Sciences, University of Groningen, Groningen, The Netherlands.
Hervé Colinet, University of Rennes, CNRS, ECOBIO, UMR 6553, Rennes, France.
Mehregan Ebrahimi, College of Science and Engineering, Flinders University, Adelaide, Australia.
Patricia Gibert, Université Claude Bernard Lyon 1, CNRS, LBBE, UMR 5558, Villeurbanne, France.
Jan Hrcek, Biology Centre of the Czech Academy of Sciences, Institute of Entomology, České Budějovice, Czech Republic; Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic.
Maaria Kankare, Department of Biological and Environmental Science, University of Jyväskylä, Jyväskylä, Finland.
Iryna Kozeretska, National Antarctic Scientific Center of the Ministry of Education and Science of Ukraine, Kyiv, Ukraine.
Volker Loeschcke, Department of Biology, Aarhus University, Aarhus, Denmark.
Julián Mensch, IEGEBA UBA-CONICET, Universidad de Buenos Aires, Buenos Aires, Argentina.
Banu Sebnem Onder, Genetic Variation and Adaptation Laboratory, Department of Biology, Faculty of Science, Hacettepe University, Ankara, Turkey.
John Parsch, Division of Evolutionary Biology, Faculty of Biology, Ludwig-Maximilians-Universität München, Munich, Germany.
Elena G Pasyukova, Institute of Molecular Genetics, Russian Academy of Sciences, Moscow, Russia.
Marina Stamenkovic-Radak, Faculty of Biology, University of Belgrade, Belgrade, Serbia.
Eran Tauber, Institute of Evolution, University of Haifa, Haifa, Israel.
Cristina Vieira, Université Claude Bernard Lyon 1, CNRS, LBBE, UMR 5558, Villeurbanne, France.
Christian Wegener, Biocenter, Theodor-Boveri-Institute, Neurobiology and Genetics, University of Würzburg, Würzburg, Germany.
Katja M Hoedjes, Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland; Amsterdam Institute of Life and Environment, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
Bas J Zwaan, Laboratory of Genetics, Wageningen University, Wageningen, The Netherlands.
Andrea J Betancourt, Department of Evolution, Ecology and Behaviour, University of Liverpool, Liverpool, United Kingdom.
Claudia Fricke, Institute for Zoology, Animal Ecology, Martin-Luther University Halle-Wittenberg, Halle (Saale), Germany.
Sonja Grath, Division of Evolutionary Biology, Faculty of Biology, Ludwig-Maximilians-Universität München, Munich, Germany.
Nico Posnien, Department of Developmental Biology, Göttingen Center for Molecular Biosciences (GZMB), University of Göttingen, Göttingen, Germany.
Jorge Vieira, Instituto de Investigação e Inovação em Saúde (I3S), Universidade do Porto, Porto, Portugal; Instituto de Biologia Molecular e Celular (IBMC), Porto, Portugal.
Martin Kapun, Natural History Museum Vienna, Central Research Laboratories, Vienna, Austria; Department of Cell and Developmental Biology, Center of Anatomy and Cell Biology, Medical University of Vienna, Vienna, Austria.
Christian Schlötterer, Institut für Populationsgenetik, Vetmeduni Vienna, Vienna, Austria.
Paul Schmidt, Department of Biology, University of Pennsylvania, Philadelphia, United States.
Élio Sucena, Instituto Gulbenkian de Ciência, Oeiras, Portugal; Departamento de Biologia Animal, Faculdade de Ciências, Universidade de Lisboa, Lisbon, Portugal; Centre for Ecology, Evolution and Environmental Changes (cE3c), Faculdade de Ciências, Universidade de Lisboa, Lisbon, Portugal.
Josefa González, Institut Botànic de Barcelona, CSIC, CMCNB, Barcelona, Spain.
Alan Bergland, Department of Biology, University of Virginia, Charlottesville, United States.
Michael G Ritchie, Centre for Biological Diversity, University of St Andrews, St. Andrews, United Kingdom.
Thomas Flatt, Department of Biology, University of Fribourg, Fribourg, Switzerland.
Data and code availability
Raw data and the complete compilation of code, statistical models, analyses, and results are available on GitHub (https://esradm.github.io/DrosEU_PhenotypingWG/) and are archived on Zenodo (https://doi.org/10.5281/zenodo.15310170).
Author contributions
Contributions defined according to CRediT ontology (https://casrai.org/credit/); authors in alphabetical order: T.A.F.deA.: investigation; J.A.: investigation, methodology, writing—original draft preparation, writing—review & editing; C.A.: investigation; E.A.: investigation, methodology; T.B.: investigation; A.B.: formal analysis, investigation, methodology, software, visualization, writing—review & editing; A.B.: conceptualization, writing—review & editing; J.-C.B.: investigation; D.C.: investigation; E.C.: investigation; H.C.: investigation, methodology, data curation, supervision, writing—original draft preparation, writing—review & editing; N.C.: investigation, supervision; S.C.: investigation; S.D.: investigation; E.D.: investigation; T.D.: formal analysis, investigation, writing—review & editing; S.R.-D.: investigation; P.D.: formal analysis, investigation, writing—review & editing; E.D.M.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing—original draft preparation, writing—review & editing; M.E.: investigation; K.E.: investigation; P.E.: investigation; P.E.: investigation, methodology; F.F.: investigation; B.F.: investigation; T.F.: conceptualization, funding acquisition, investigation, methodology, project administration, resources, supervision, writing—original draft preparation, writing—review & editing; C.F.: investigation, methodology; P.G.: investigation, methodology, project administration, data curation, supervision, funding acquisition; A.G.-S.: investigation, methodology, resources, writing—review & editing; A.G.: investigation; J.G.: supervision, writing—review & editing; S.G.: investigation, methodology, writing—review & editing; L.G.: investigation; E.H.: formal analysis, visualization, writing—original draft preparation; K.M.H.: investigation, methodology, data curation, supervision, writing—original draft preparation, writing—review & editing; J.H.: methodology, funding acquisition; S.J.: investigation, resources; M.J.: investigation, methodology; M.K.: resources, writing—review & editing; M.K.: formal analysis, investigation, writing—original draft preparation, writing—review & editing; E.K.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing—original draft preparation, writing—review & editing; H.K.: investigation; I.K.: methodology, supervision, data curation, resources; L.E.K.: investigation; N.K.: investigation, methodology; V.L.: investigation; M.L.: investigation, methodology; O.M.M.: data curation, formal analysis, writing—review & editing; S.-L.M.: investigation, methodology; M.M.: investigation: J.M.: investigation, supervision, writing—review & editing; M.M.: investigation; M.S.M.: investigation; V.M.: investigation, methodology; L.M.: investigation, methodology; D.V.M.: methodology, resources; S.M.: investigation; B.S.O.: investigation, supervision, resources, methodology; J.P.: funding acquisition, resources; E.G..P.: investigation, methodology, resources, supervision, writing; A.P.: investigation, methodology, data curation; T.F.P.: investigation; N.P.: investigation, validation, writing—review & editing; O.P.: investigation; F.A.P.: investigation; M.R.: investigation, validation; M.G.R.: conceptualization, formal analysis, supervision, writing—review & editing; N.V.R.: investigation, methodology; O.Y.R.: investigation, methodology; M.S.V.: investigation, methodology; data curation; C.S.: supervision, writing—review & editing; P.S.: conceptualization, investigation; M.F.S.: investigation; T.S.: investigation; S.S.S.: investigation; S.S.: investigation, methodology, data curation, writing—original draft preparation; M.S.-R.: investigation, supervision, resources; É.S.: investigation, resources; A.V.S.: investigation, methodology, formal analysis, validation, writing; M.T.: investigation, methodology, data curation, formal analysis, writing; E.T.: investigation, formal analysis, supervision, resources, writing—review & editing; V.T.: investigation; M.V.T.: investigation, methodology; E.A.T.: investigation, methodology; V.T.: formal analysis, visualization, writing—original draft preparation; J.H.: investigation; D.W.: investigation; E.R.V.: investigation, methodology; C.P.V.: formal analysis, investigation, methodology, software, validation, writing—review & editing; C.V.: investigation, methodology; J.V.: conceptualization, formal analysis, investigation, methodology, software, validation, writing—original draft preparation, writing—review & editing; X.W.: investigation; C.W.: investigation, formal analysis, resources, writing—review & editing; J.Z.: investigation; B.Z.: investigation, funding acquisition, methodology, resources, writing—original draft preparation.
Funding
This research was supported by a Special Topics Network (STN) grant from the European Society of Evolutionary Biology (ESEB) to the DrosEU consortium as well as by individual grants and fellowships (grantees in alphabetical order): J.A.: Swedish Research Council (VR) grants 2015-04680 and 2020-05412; A.J.B.: European Research Council (ERC CoG TE_INVASION); S.D.: Ministry of Science, Technological Development and Innovation of the Republic of Serbia (NITRA) grant 451-03-66/2024-03/200007; K.E.: Ministry of Science, Technological Development and Innovation of the Republic of Serbia (NITRA) grant 451-03-66/2024-03/200007; P.E.: Ministry of Science, Technological Development and Innovation of the Republic of Serbia (NITRA) grant 451-03-66/2024-03/200007; P.E.: Jane Coffin Childs Memorial Fund for Medical Research 61-1673: T.F.: Swiss National Science Foundation (SNSF) grants 31003A-182262, 310030_219283, FZEB-0-214654; C.F.: Deutsche Forschungsgemeinschaft (DFG) grants 2973/5-1 and 2973/11; J.G.: Ministerio de Ciencia, Innovación y Universidades/Agencia Estatal de Investigación (MICIU/AEI/10.13039/501100011033/) grant PID2020-115874GB-I00; Ministerio de Ciencia, Innovación y Universidades/Agencia Estatal de Investigación (MICIU/AEI/10.13039/501100011033 and FEDER, UE) grant PID2023-148838NB-I00; Departament de Recerca i Universitats Generalitat de Catalunya grant 2021 SGR 00417; S.G.: Deutsche Forschungsgemeinschaft (DFG) grants 271330745 and 514085304; K.M.H.: Marie Skłodowska-Curie Individual Fellowship (H2020-MSCA-IF-2015) 701949; J.H.: Czech Ministry of Education, grant; European Research Council (ERC CZ LL2001); M.J.: Ministry of Science, Technological Development and Innovation of the Republic of Serbia (NITRA) grant 451-03-65/2024-03/200178; M.K.: Academy of Finland grant 322980; E.K.: European Molecular Biology Organization (EMBO) ALTF 248-2018; M.S.M.: ALW Open Programme grant 101185; J.P.: Deutsche Forschungsgemeinschaft (DFG) grants 255619725 (GR 4495/2-2) and 503272152 (GR 4495/4-1); E.G.P.: Russian state budget, assignment NRC “KI”; A.P.: Ministry of Science, Technological Development and Innovation of the Republic of Serbia (NITRA) grant 451-03-66/2024-03/200007; N.P.: Deutsche Forschungsgemeinschaft (DFG) grants PO 1648/7-1, PO 1648/3-1, PO 1648/3-2; M.S.V.: Ministry of Science, Technological Development and Innovation of the Republic of Serbia (NITRA) grant 451-03-65/2024-03/200178; C.S.: Austrian Science Fund (FWF) grants 10.55776/P32935, 10.55776/W1225, 10.55776/P33734; P.S.: National Institutes of Health (NIH) R01GM137430; M.S.-R.: Ministry of Science, Technological Development and Innovation of the Republic of Serbia (NITRA) grant 451-03-47/2023-01/200178; M.T.: Ministry of Science, Technological Development and Innovation of the Republic of Serbia (NITRA) grant 451-03-66/2024-03/200007; E.T.: Israel Science Foundation (ISF) 2121/23; C.V.: Agence Nationale de la Recherche (ANR) grant Longevity ANR-20-CE02-0015, J.V.: Fundação para a Ciência e a Tecnologia (FCT) UIDB/04293/2020; C.W.: Deutsche Forschungsgemeinschaft (DFG) grant WE 2652/7-1.
Conflict of interest
The authors declare no conflict of interest. Note: After February 24, 2022, no collaborative actions or exchanges have taken place between Ukrainian and Russian scientists within our project.
References
- Ackermann M., Bijlsma R., James A. C., Partridge L., Zwaan B. J., Stearns S. C. (2001). Effects of assay conditions in life history experiments with Drosophila melanogaster . Journal of Evolutionary Biology, 14:(2), 199–209. 10.1046/j.1420-9101.2001.00281.x [DOI] [Google Scholar]
- Adrion J. R., Hahn M. W., Cooper B. S. (2015). Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends in Genetics, 31, 434–444. 10.1016/j.tig.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agis M., Schlötterer C. (2001). Microsatellite variation in natural Drosophila melanogaster populations from New South Wales (Australia) and Tasmania. Molecular Ecology, 10, 1197–1205. 10.1046/j.1365-294X.2001.01271.x [DOI] [PubMed] [Google Scholar]
- Anderson A. R., Collinge J. E., Hoffmann A. A., Kellett M., McKechnie S. W. (2003). Thermal tolerance trade-offs associated with the right arm of chromosome 3 and marked by the hsr-omega gene in Drosophila melanogaster. Heredity, 90, 195–202. 10.1038/sj.hdy.6800220 [DOI] [PubMed] [Google Scholar]
- Arguello J. R., Laurent S., Clark A. G. (2019). Demographic history of the human commensal Drosophila melanogaster. Genome Biology and Evolution, 11, 844–854. 10.1093/gbe/evz022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrinhac A., Debat V., Gibert P., Kister A. ‐G., Legout H., Moreteau B., Vergilino R., David J. R. (2004). Cold adaptation in geographical populations of Drosophila melanogaster: Phenotypic plasticity is more important than genetic variability. Functional Ecology, 18, 700–706. 10.1111/j.0269-8463.2004.00904.x [DOI] [Google Scholar]
- Balduzzi S., Rücker G., Schwarzer G. (2019). How to perform a meta-analysis with R: A practical tutorial. Evidence Based Mental Health, 22, 153–160. 10.1136/ebmental-2019-300117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barghi N., Tobler R., Nolte V., Jakšić A. M., Mallard F., Otte K. A., Dolezal M., Taus T., Kofler R., Schlötterer C. (2019). Genetic redundancy fuels polygenic adaptation in Drosophila. PLoS Biology, 17, e3000128. 10.1371/journal.pbio.3000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman E. L., Howick V. M., Kapun M., Staubach F., Bergland A. O., Petrov D. A., Lazzaro B. P., Schmidt P. S. (2018). Rapid seasonal evolution in innate immunity of wild Drosophila melanogaster. Proceedings of the Royal Society of London B, 285, 20172599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman E. L., Watson S. S., O'Brien K. R., Heschel M. S., Schmidt P. S. (2015). Seasonal variation in life history traits in two Drosophila species. Journal of Evolutionary Biology, 28, 1691–1704. 10.1111/jeb.12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Behrman E. L., O'Brien K. R., Schmidt P. S., Petrov D. A. (2014). Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genetics, 10, e1004775. 10.1371/journal.pgen.1004775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Tobler R., González J., Schmidt P., Petrov D. (2016). Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Molecular Ecology, 25, 1157–1174. 10.1111/mec.13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt N. J., Rajpurohit S., Durmaz E., Fabian D. K., Kapun M., Flatt T., Schmidt P. (2021). Allelic polymorphism at foxo contributes to local adaptation in Drosophila melanogaster. Molecular Ecology, 30, 2817–2830. 10.1111/mec.15939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter M. C., Berardi S., Oken H., Huynh A., Lappo E., Schmidt P., Petrov D. A. (2024). Continuously fluctuating selection reveals fine granularity of adaptation. Nature, 634, 389–396., 10.1038/s41586-024-07834-x [DOI] [PubMed] [Google Scholar]
- Bogaerts‐Márquez M., Guirao‐Rico S., Gautier M., González J. (2021). Temperature, rainfall and wind variables underlie environmental adaptation in natural populations of Drosophila melanogaster. Molecular Ecology, 30, 938–954. 10.1111/mec.15783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Božičević V., Hutter S., Stephan W., Wollstein A. (2016). Population genetic evidence for cold adaptation in European Drosophila melanogaster populations. Molecular Ecology, 25, 1175–1191. 10.1111/mec.13464 [DOI] [PubMed] [Google Scholar]
- Burke M. K., Dunham J. P., Shahrestani P., Thornton K. R., Rose M. R., Long A. D. (2010). Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature, 467, 587–590. 10.1038/nature09352 [DOI] [PubMed] [Google Scholar]
- Casillas S., Barbadilla A. (2017). Molecular population genetics. Genetics, 205, 1003–1035. 10.1534/genetics.116.196493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. (1993). Natural selection on multivariate traits in age-structured populations. Proceedings of the Royal Society B, 251, 47–52. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. (1994). Evolution in age-structured populations(2nd ed.). Cambridge University Press. [Google Scholar]
- Chen B., Feder M. E., Kang L. (2018). Evolution of heat-shock protein expression underlying adaptive responses to environmental stress. Molecular Ecology, 27, 3040–3054. 10.1111/mec.14769 [DOI] [PubMed] [Google Scholar]
- Chen J., Liu C., Li W., Zhang W., Wang Y., Clark A. G., Lu J. (2024). From sub-Saharan Africa to China: Evolutionary history and adaptation of Drosophila melanogaster revealed by population genomics. Science Advances, 10, eadh3425. 10.1126/sciadv.adh3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogni R., Kuczynski K., Lavington E., Koury S., Behrman E. L., O'brien K. R., Schmidt P. S., Eanes W. F. (2015). Variation in Drosophila melanogaster central metabolic genes appears driven by natural selection both within and between populations. Proceedings of the Royal Society B: Biological Sciences, 282, 20142688. 10.1098/rspb.2014.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R., Peixoto A. A., Barbujani G., Kyriacou C. P. (1992). A latitudinal cline in a Drosophila clock gene. Proceedings of the Royal Society B, 250, 43–49. [DOI] [PubMed] [Google Scholar]
- Coughlan J. M., Dagilis A. J., Serrato-Capuchina A., Elias H., Peede D., Isbell K., Castillo D. M., Cooper B. S., Matute D. R. (2022). Patterns of population structure and introgression among recently differentiated Drosophila melanogaster populations. Molecular Biology and Evolution, 39, msac223. 10.1093/molbev/msac223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Beecham E. (1987). Heritability of two morphological characters within and among natural populations of Drosophila melanogaster. Genetics, 117, 727–737. 10.1093/genetics/117.4.727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe J. C., Wahlsten D., Dudek B. C. (1999). Genetics of mouse behavior: Interactions with laboratory environment. Science, 284, 1670–1672. 10.1126/science.284.5420.1670 [DOI] [PubMed] [Google Scholar]
- David J. R. (1982). Latitudinal variability of Drosophila melanogaster: Allozyme frequencies divergence between European and Afrotropical populations. Biochemical Genetics, 20, 747–761. 10.1007/BF00483971 [DOI] [PubMed] [Google Scholar]
- David J. R., Bocquet C. (1975a). Similarities and differences in latitudinal adaptation of two Drosophila sibling species. Nature, 257, 588–590. 10.1038/257588a0 [DOI] [PubMed] [Google Scholar]
- David J. R., Bocquet C. (1975b). Evolution in a cosmopolitan species: Genetic latitudinal clines in Drosophila melanogaster wild populations. Experientia, 31, 164–166. 10.1007/BF01990682 [DOI] [PubMed] [Google Scholar]
- David J. R., Capy P. (1988). Genetic variation of Drosophila melanogaster natural populations. Trends in Genetics, 4, 106–111. 10.1016/0168-9525(88)90098-4 [DOI] [PubMed] [Google Scholar]
- David J. R., Allemand R., Capy P., Chakir M., Gibert P., Pétavy G., Moreteau B. (2004). Comparative life histories and ecophysiology of Drosophila melanogaster and D. simulans. Genetica, 120, 151–163. 10.1023/B:GENE.0000017638.02813.5a [DOI] [PubMed] [Google Scholar]
- David J. R., Capy P., Payant V., Tsakas S. (1985). Thoracic trident pigmentation in Drosophila melanogaster: Differentiation of geographical populations. Genetics Selection Evolution, 17, 211–224. 10.1186/1297-9686-17-2-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J. R., Gibert P., Legout H., Pétavy G., Capy P., Moreteau B. (2005). Isofemale lines in Drosophila: An empirical approach to quantitative trait analysis in natural populations. Heredity, 94, 3–12. 10.1038/sj.hdy.6800562 [DOI] [PubMed] [Google Scholar]
- David J. R., Gibert P., Pla E., Petavy G., Karan D., Moreteau B. (1998). Cold stress tolerance in Drosophila: Analysis of chill coma recovery in D. melanogaster. Journal of Thermal Biology, 23(5), 291–299. 10.1016/S0306-4565(98)00020-5 [DOI] [Google Scholar]
- David J., Bocquet C., de Scheemaeker-Louis M. (1977). Genetic latitudinal adaptation of Drosophila melanogaster: New discriminative biometrical traits between European and equatorial African populations. Genetical Research, 30, 247–255. 10.1017/S0016672300017651 [DOI] [Google Scholar]
- David J., Merçot H., Capy P., McEvey S., Van Herrewege J. (1986). Alcohol tolerance and Adh gene frequencies in European and African populations of Drosophila melanogaster. Genetics Selection Evolution, 18, 405–416. 10.1186/1297-9686-18-4-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong G., Bochdanovits Z. (2003). Latitudinal clines in Drosophila melanogaster: Body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. Journal of Genetics, 82, 207–223. 10.1007/BF02715819 [DOI] [PubMed] [Google Scholar]
- Draye X., Lints F. (1996). Geographic variations of life history strategies in Drosophila melanogaster. III. New data. Experimental Gerontology, 31, 717–733. 10.1016/S0531-5565(96)00073-3 [DOI] [PubMed] [Google Scholar]
- Draye X., Bullens P., Lints F. A. (1994). Geographic variations of life history strategies in Drosophila melanogaster. I. Analysis of wild-caught populations. Experimental Gerontology, 29, 205–222. 10.1016/0531-5565(94)90052-3 [DOI] [PubMed] [Google Scholar]
- Duchen P., Živković D., Hutter S., Stephan W., Laurent S. (2013). Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population. Genetics, 193, 291–301. 10.1534/genetics.112.145912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmaz E., Benson C., Kapun M., Schmidt P., Flatt T. (2018). An inversion supergene in Drosophila underpins latitudinal clines in survival traits. Journal of Evolutionary Biology, 31, 1354–1364. 10.1111/jeb.13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmaz E., Rajpurohit S., Betancourt N., Fabian D. K., Kapun M., Schmidt P., Flatt T. (2019). A clinal polymorphism in the insulin signaling transcription factor foxo contributes to life-history adaptation in Drosophila. Evolution, 73, 1774–1792. 10.1111/evo.13759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler L., Betancourt A. J., Nolte V., Schlötterer C. (2016). Reconciling differences in pool-GWAS between populations: A case study of female abdominal pigmentation in Drosophila melanogaster. Genetics, 202, 843–855. 10.1534/genetics.115.183376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P. A., Weller C. A., Song D. Y., Bangerter A. S., Schmidt P., Bergland A. O. (2020). Unique genetic signatures of local adaptation over space and time for diapause, an ecologically relevant complex trait, in Drosophila melanogaster. PLoS Genetics, 16, e1009110. 10.1371/journal.pgen.1009110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian D. K., Garschall K., Klepsatel P., Santos-Matos G., Sucena É., Kapun M., Lemaitre B., Schlötterer C., Arking R., Flatt T. (2018). Evolution of longevity improves immunity in Drosophila. Evolution Letters, 2, 567–579. 10.1002/evl3.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian D. K., Kapun M., Nolte V., Kofler R., Schmidt P. S., Schlötterer C., Flatt T. (2012). Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Molecular Ecology, 21, 4748–4769. 10.1111/j.1365-294X.2012.05731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian D. K., Lack J. B., Mathur V., Schlötterer C., Schmidt P. S., Pool J. E., Flatt T. (2015). Spatially varying selection shapes life history clines among populations of Drosophila melanogaster from sub-Saharan Africa. Journal of Evolutionary Biology, 28, 826–840. 10.1111/jeb.12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C. (1996). Introduction to quantitative genetics(4th ed.). Longman. [Google Scholar]
- Flatt T., Heyland A. (Eds). (2011). Mechanisms of life history evolution—The genetics and physiology of life history traits and trade-offs. Oxford University Press. [Google Scholar]
- Flatt T. (2005). The evolutionary genetics of canalization. The Quarterly Review of Biology, 80, 287–316. 10.1086/432265 [DOI] [PubMed] [Google Scholar]
- Flatt T. (2011). Survival costs of reproduction in Drosophila. Experimental Gerontology, 46, 369–375. 10.1016/j.exger.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Flatt T. (2016). Genomics of clinal variation in Drosophila: Disentangling the interactions of selection and demography. Molecular Ecology, 25, 1023–1026. 10.1111/mec.13534 [DOI] [PubMed] [Google Scholar]
- Flatt T. (2020). Life-history evolution and the genetics of fitness components in Drosophila melanogaster. Genetics, 214, 3–48. 10.1534/genetics.119.300160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T., Amdam G. V., Kirkwood T. B. L., Omholt S. W. (2013). Life-history evolution and the polyphenic regulation of somatic maintenance and survival. The Quarterly Review of Biology, 88, 185–218. 10.1086/671484 [DOI] [PubMed] [Google Scholar]
- Folguera G., Ceballos S., Spezzi L., Fanara J. J., Hasson E. (2008). Clinal variation in developmental time and viability, and the response to thermal treatments in two species of Drosophila. Biological Journal of the Linnean Society, 95, 233–245. 10.1111/j.1095-8312.2008.01053.x [DOI] [Google Scholar]
- Gardeux V., Bevers R. P. J., David F. P. A., Rosschaert E., Rochepeau R., Deplancke B. (2024). DGRPool, a web tool leveraging harmonized Drosophila Genetic Reference Panel phenotyping data for the study of complex traits. eLife, 12, RP88981. 10.7554/eLife.88981.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud N. R., Messer P. W., Petrov D. A. (2021). Detection of hard and soft selective sweeps from Drosophila melanogaster population genomic data. PLoS Genetics, 17, e1009373. 10.1371/journal.pgen.1009373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geber M. A. (1990). The cost of meristem limitation in Polygonum arenastrum: Negative genetic correlations between fecundity and growth. Evolution, 44, 799–819. [DOI] [PubMed] [Google Scholar]
- Gibert P., Capy P., Imasheva A., Moreteau B., Morin J. P., Pétavy G., David J. R. (2004). Comparative analysis of morphological traits among Drosophila melanogaster and D. simulans: Genetic variability, clines and phenotypic plasticity. Genetica, 120, 165–179. 10.1023/B:GENE.0000017639.62427.8b. [DOI] [PubMed] [Google Scholar]
- Gilchrist A. S., Partridge L. (1999). A comparison of the genetic basis of wing size divergence in three parallel body size clines of Drosophila melanogaster. Genetics, 153, 1775–1787. 10.1093/genetics/153.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser-Schmitt A., Wittmann M. J., Ramnarine T. J. S., Parsch J. (2021). Sexual antagonism, temporally fluctuating selection, and variable dominance affect a regulatory polymorphism in Drosophila melanogaster. Molecular Biology and Evolution, 38, 4891–4907. 10.1093/molbev/msab215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. L. Jr., Hertweck K. L., Phillips M. A., Han M. V., Cabral L. G., Barter T. T., Greer L. F., Burke M. K., Mueller L. D., Rose M. R.. (2017). Genomics of parallel experimental evolution in Drosophila. Molecular Biology and Evolution, 34, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier J. K., Arguello J. R., Moreira M. C., Gottipati S., Mohammed J., Hackett S. R., Boughton R., Greenberg A. J., Clark A. G. (2015). Global Diversity Lines—A five-continent reference panel of sequenced Drosophila melanogaster strains. G3 Genes|Genomes|Genetics, 5, 593–603. 10.1534/g3.114.015883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangartner S. B., Hoffmann A. A., Smith A., Griffin P. C. (2015). A collection of Australian Drosophila datasets on climate adaptation and species distributions. Scientific Data, 2, 150067. 10.1038/sdata.2015.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., Murray E. (1983). Selection and measures of fitness. In Ashburner M., Carson H. L., Thompson J. N. (Eds.), The genetics and biology of Drosophila(vol. 3d) (pp. 61–104.). Jr. Academic Press. [Google Scholar]
- Hoedjes K. M., van den Heuvel J., Kapun M., Keller L., Flatt T., Zwaan B. J. (2019). Distinct genomic signals of lifespan and life history evolution in response to postponed reproduction and larval diet in Drosophila. Evolution Letters, 3, 598–609. 10.1002/evl3.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra L. A., Montooth K. L. (2013). Inducing extra copies of the hsp70 gene in Drosophila melanogaster increases energetic demand. BMC Evolutionary Biology, 13, 68. 10.1186/1471-2148-13-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Flatt T. (2022). The rapid tempo of adaptation. Science, 375(6586), 1226–1227. 10.1126/science.abo1817 [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Parsons P. A. (1988). The analysis of quantitative variation in natural populations with isofemale strains. Genetics Selection Evolution, 20, 87–98. 10.1186/1297-9686-20-1-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Weeks A. R. (2007). Climatic selection on genes and traits after a 100-year-old invasion: A critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica, 129, 133–147. 10.1007/s10709-006-9010-z [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Chown S. L., Clusella‐Trullas S. (2013). Upper thermal limits in terrestrial ectotherms: How constrained are they?. Functional Ecology, 27, 934–949. 10.1111/j.1365-2435.2012.02036.x [DOI] [Google Scholar]
- Hoffmann A. A., Sørensen J. G., Loeschcke V. (2003). Adaptation of Drosophila to temperature extremes: Bringing together quantitative and molecular approaches. Journal of Thermal Biology, 28, 175–216. 10.1016/S0306-4565(02)00057-8 [DOI] [Google Scholar]
- Houle D. (1992). Comparing evolvability and variability of quantitative traits. Genetics, 130, 195–204. 10.1093/genetics/130.1.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D. (1998). How should we explain variation in the genetic variance of traits?. Genetica, 102–103, 241–253. 10.1023/A:1017034925212 [DOI] [PubMed] [Google Scholar]
- Houle D. (2001). Characters as the units of evolutionary change. In Wagner G. P. (Ed.), The character concept in evolutionary biology(pp. 109–140.). Academic Press. 10.1016/B978-012730055-9/50015-X [DOI] [Google Scholar]
- Houle D. (2010). Numbering the hairs on our heads: The shared challenge and promise of phenomics. Proceedings of the National Academy of Sciences, 107, 1793–1799. 10.1073/pnas.0906195106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D., Govindaraju D. R., Omholt S. (2010). Phenomics: The next challenge. Nature Reviews Genetics, 11, 855–866. 10.1038/nrg2897 [DOI] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ràmia M., Tarone A. M., Turlapati L., Zichner T., Zhu D., Lyman R. F., Magwire M. M., Blankenburg K., Carbone M. A., Chang K., Ellis L. L., Fernandez S., Han Yi, Highnam G., Hjelmen C. E., … Mackay T. F. C. (2014). Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Research, 24, 1193–1208. 10.1101/gr.171546.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imasheva A. G., Bubli O. A., Lazebny O. E. (1994). Variation in wing length in Eurasian natural populations of Drosophila melanogaster. Heredity, 72, 508–514. 10.1038/hdy.1994.68 [DOI] [PubMed] [Google Scholar]
- Kapun M., Flatt T. (2019). The adaptive significance of chromosomal inversion polymorphisms in Drosophila melanogaster. Molecular Ecology, 28, 1263–1282. 10.1111/mec.14871 [DOI] [PubMed] [Google Scholar]
- Kapun M., Barrón M. G., Staubach F., Obbard D. J., Wiberg R. A. W., Vieira J., Goubert C., Rota-Stabelli O., Kankare M., Bogaerts-Márquez M., Haudry A., Waidele L., Kozeretska I., Pasyukova E. G., Loeschcke V., Pascual M., Vieira C. P., Serga S., Montchamp-Moreau C., … González J. (2020). Genomic analysis of European Drosophila melanogaster populations reveals longitudinal structure, continent-wide selection, and previously unknown DNA viruses. Molecular Biology and Evolution, 37, 2661–2678. 10.1093/molbev/msaa120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., Fabian D. K., Goudet J., Flatt T. (2016a). Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Molecular Biology and Evolution, 33, 1317–1336. 10.1093/molbev/msw016 [DOI] [PubMed] [Google Scholar]
- Kapun M., Mitchell E. D., Kawecki T. J., Schmidt P., Flatt T. (2023). An ancestral balanced inversion polymorphism confers global adaptation. Molecular Biology and Evolution, 40, msad118. 10.1093/molbev/msad118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., Nunez J. C. B., Bogaerts-Márquez M., Murga-Moreno J., Paris M., Outten J., Coronado-Zamora M., Tern C., Rota-Stabelli O., Guerreiro M. P. G., Casillas S., Orengo D. J., Puerma E., Kankare M., Ometto L., Loeschcke V., Onder B. S., Abbott J. K., Schaeffer S. W., … Bergland A. O. (2021). Drosophila Evolution over Space and Time (DEST): A new population genomics resource. Molecular Biology and Evolution, 38, 5782–5805. 10.1093/molbev/msab259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., Schmidt C., Durmaz E., Schmidt P. S., Flatt T. (2016b). Parallel effects of the inversion In(3R)Payne on body size across the North American and Australian clines in Drosophila melanogaste r. Journal of Evolutionary Biology, 29, 1059–1072. 10.1111/jeb.12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp N. A. (2018). Reproducible preclinical research—Is embracing variability the answer?. PLoS Biology, 16, e2005413. 10.1371/journal.pbio.2005413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki T. J., Ebert D. (2004). Conceptual issues in local adaptation. Ecology Letters, 7, 1225–1241. 10.1111/j.1461-0248.2004.00684.x [DOI] [Google Scholar]
- Kawecki T. J., Erkosar B., Dupuis C., Hollis B., Stillwell R. C., Kapun M. (2021). The genomic architecture of adaptation to larval malnutrition points to a trade-off with adult starvation resistance in Drosophila. Molecular Biology and Evolution, 38, 2732–2749. 10.1093/molbev/msab061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A. (2007). Drosophila melanogaster’s history as a human commensal. Current Biology, 17, R77–R81. 10.1016/j.cub.2006.12.031 [DOI] [PubMed] [Google Scholar]
- Khazaeli A., Vanvoorhies W., Curtsinger J. (2005). The relationship between life span and adult body size is highly strain-specific in Drosophila melanogaster. Experimental Gerontology, 40, 377–385. 10.1016/j.exger.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Klepsatel P., Gáliková M., De Maio N., Ricci S., Schlötterer C., Flatt T. (2013). Reproductive and post-reproductive life history of wild-caught Drosophila melanogaster under laboratory conditions. Journal of Evolutionary Biology, 26, 1508–1520. 10.1111/jeb.12155 [DOI] [PubMed] [Google Scholar]
- Klepsatel P., Gáliková M., Huber C. D., Flatt T. (2014). Similarities and differences in altitudinal versus latitudinal variation for morphological traits in Drosophila melanogaster. Evolution, 68, 1385–1398. 10.1111/evo.12351 [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J. (2011). Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics, 187, 245–260. 10.1534/genetics.110.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D., Silvain J. F. (2004). How two afrotropical endemics made two cosmopolitan commensals: The Drosophila melanogaster-D. simulans paleogeographic riddle. Genetica, 120, 17–39. 10.1023/B:GENE.0000017627.27537.ef [DOI] [PubMed] [Google Scholar]
- Lachaise D., Cariou M. L., David J. R., Lemeunier F., Tsacas L., Ashburner M. (1988). Historical biogeography of the Drosophila melanogaster species subgroup. Evolutionary Biology, 22, 159–225. 10.1007/978-1-4613-0931-4_4 [DOI] [Google Scholar]
- Lack J. B., Cardeno C. M., Crepeau M. W., Taylor W., Corbett-Detig R. B., Stevens K. A., Langley C. H., Pool J. E. (2015). The Drosophila genome nexus: A population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics, 199, 1229–1241. 10.1534/genetics.115.174664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack J. B., Lange J. D., Tang A. D., Corbett-Detig R. B., Pool J. E. (2016). A thousand fly genomes: An expanded Drosophila genome nexus. Molecular Biology and Evolution, 33, 3308–3313. 10.1093/molbev/msw195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. (1982). A quantitative genetic theory of life history evolution. Ecology, 63, 607–615. 10.2307/1936778 [DOI] [Google Scholar]
- Lande R., Arnold S. J. (1983). The measurement of selection on correlated characters. Evolution, 37, 1210–1226. 10.2307/2408842 [DOI] [PubMed] [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C. G., Schrider D. R., Pool J. E., Langley S. A., Suarez C., Corbett-Detig R. B., Kolaczkowski B., Fang S., Nista P. M., Holloway A. K., Kern A. D., Dewey C. N., Song Y. S., Hahn M. W., Begun D. J. (2012). Genomic variation in natural populations of Drosophila melanogaster. Genetics, 192, 533–598. 10.1534/genetics.112.142018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F., Eyre‐Walker Y. C., Rane R. V., Reuter C., Vinti G., Rako L., Partridge L., Hoffmann A. A. (2013). Polymorphism in the neurofibromin gene, Nf1, is associated with antagonistic selection on wing size and development time in Drosophila melanogaster. Molecular Ecology, 22, 2716–2725. 10.1111/mec.12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeunier F., Aulard S. (1992). Inversion polymorphism in Drosophila melanogaster. In Krimbas C. B., Powell J. R. (Eds.), Drosophila inversion polymorphism(pp. 339–405.). CRC Press. [Google Scholar]
- Leroi A. M., Chippindale A. K., Rose M. R. (1994). Long-term laboratory evolution of a genetic life-history trade-off in Drosophila melanogaster. 1. The role of genotype-by-environment interaction. Evolution, 48, 1244–1257. [DOI] [PubMed] [Google Scholar]
- Lewontin R. C. (1974). The genetic basis of evolutionary change. Columbia University Press. [Google Scholar]
- Lewontin R. C. (2000a). The problems of population genetics. In Singh R. S., Krimbas C. B. (Eds.), Evolutionary genetics—Fom molecules to morphology(pp. 5–23.). Cambridge University Press. [Google Scholar]
- Lewontin R. C. (2000b). What do population geneticists know and how do they know it?In Creath R., Maienschein J. (Eds.), Biology and epistemology(pp. 191–214.). Cambridge University Press. [Google Scholar]
- Li H., Stephan W. (2006). Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genetics, 2, e166. 10.1371/journal.pgen.0020166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado H. E., Bergland A. O., Taylor R., Tilk S., Behrman E., Dyer K., Fabian D. K., Flatt T., González J., Karasov T. L., Kim B., Kozeretska I., Lazzaro B. P., Merritt T. Js, Pool J.E., O'brien K., Rajpurohit S., Roy P. R., Schaeffer S. W., … Petrov D. A. (2021). Broad geographic sampling reveals the shared basis and environmental correlates of seasonal adaptation in Drosophila. eLife, 10, e67577. 10.7554/eLife.67577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., Zhu D., Casillas S., Han Yi, Magwire M. M., Cridland J. M., Richardson M. F., Anholt R. R. H., Barrón M., Bess C., Blankenburg K. P., Carbone M. A., Castellano D., Chaboub L., Duncan L., … Gibbs R. A. (2012). The Drosophila melanogaster Genetic Reference Panel. Nature, 482, 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansourian S., Enjin A., Jirle E. V., Ramesh V., Rehermann G., Becher P. G., Pool J. E., Stensmyr M. C. (2018). Wild African Drosophila melanogaster are seasonal specialists on Marula fruit. Current Biology, 28, 3960–3968.e3963. 10.1016/j.cub.2018.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo L., Rech G. E., González J. (2018). Genome-wide patterns of local adaptation in Western European Drosophila melanogaster natural populations. Scientific Reports, 8, 16143. 10.1038/s41598-018-34267-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May C. M., van den Heuvel J., Doroszuk A., Hoedjes K. M., Flatt T., Zwaan B. J. (2019). Adaptation to developmental diet influences the response to selection on age at reproduction in the fruit fly. Journal of Evolutionary Biology, 32, 425–437. 10.1111/jeb.13425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K. J., Yamamoto R., Buch S., Pankratz M., Tatar M. (2008). Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell, 7, 199–206. 10.1111/j.1474-9726.2008.00373.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Lagisz M., Yang Y., Drobniak S. M. (2024). Finding the right power balance: Better study design and collaboration can reduce dependence on statistical power. PLoS Biology, 22, e3002423. 10.1371/journal.pbio.3002423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norry F. M., Loeschcke V. (2002). Temperature-induced shifts in associations of longevity with body size in Drosophila melanogaster. Evolution, 56, 299–306. [DOI] [PubMed] [Google Scholar]
- Nouhaud P., Tobler R., Nolte V., Schlötterer C. (2016). Ancestral population reconstitution from isofemale lines as a tool for experimental evolution. Ecology and Evolution, 6, 7169–7175. 10.1002/ece3.2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes M. D. S., Neumeier H., Schlötterer C. (2008). Contrasting patterns of natural variation in global Drosophila melanogaster populations. Molecular Ecology, 17, 4470–4479. 10.1111/j.1365-294X.2008.03944.x [DOI] [PubMed] [Google Scholar]
- Nunez J. C. B., Lenhart B. A., Bangerter A., Murray C. S., Mazzeo G. R., Yu Y., Nystrom T. L., Tern C., Erickson P. A., Bergland A. O. (2024). A cosmopolitan inversion facilitates seasonal adaptation in overwintering Drosophila. Genetics, 226, iyad207. 10.1093/genetics/iyad207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeshott J. G., Gibson J. B., Anderson P. R., Knibb W. R., Anderson D. G., Chambers G. K. (1982). Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Ddrosophila melanogaster on different continents. Evolution, 36, 86–96. 10.2307/2407970 [DOI] [PubMed] [Google Scholar]
- Okada Y., Teramura K., Takahashi K. H. (2014). Heat shock proteins mediate trade-offs between early-life reproduction and late survival in Drosophila melanogaster. Physiological Entomology, 39, 304–312. 10.1111/phen.12076 [DOI] [Google Scholar]
- Orozco‐Terwengel P., Kapun M., Nolte V., Kofler R., Flatt T., Schlötterer C. (2012). Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles. Molecular Ecology, 21, 4931–4941. 10.1111/j.1365-294X.2012.05673.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., Schmidt P. S. (2009). Dissecting the genetics of longevity in Drosophila melanogaster. Fly, 3, 29–38. 10.4161/fly.3.1.7771 [DOI] [PubMed] [Google Scholar]
- Paaby A. B., Bergland A. O., Behrman E. L., Schmidt P. S. (2014). A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution, 68, 3395–3409. 10.1111/evo.12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., Blacket M. J., Hoffmann A. A., Schmidt P. S. (2010). Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Molecular Ecology, 19, 760–774. 10.1111/j.1365-294X.2009.04508.x [DOI] [PubMed] [Google Scholar]
- Parsons P. A. (1975). The comparative evolutionary biology of the sibling species, Drosophila melanogaster and D. simulans. The Quarterly Review of Biology, 50, 151–169. 10.1086/408437 [DOI] [PubMed] [Google Scholar]
- Parsons P. A. (1983). The evolutionary biology of colonizing species. Cambridge University Press. [Google Scholar]
- Parsons P. A., Hosgood S. M. W. (1967). Genetic heterogeneity among the founders of laboratory populations of Drosophila. Genetica, 38, 328–339. 10.1007/BF01507465 [DOI] [PubMed] [Google Scholar]
- Pereira T. V., Patsopoulos N. A., Salanti G., Ioannidis J. P. A. (2010). Critical interpretation of Cochran’s Q test depends on power and prior assumptions about heterogeneity. Research Synthesis Methods, 1, 149–161. 10.1002/jrsm.13 [DOI] [PubMed] [Google Scholar]
- Pitchers W., Pool J. E., Dworkin I. (2013). Altitudinal clinal variation in wing size and shape in African Drosophila melanogaste r: One cline or many?. Evolution, 67, 438–452. 10.1111/j.1558-5646.2012.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., Crepeau M. W., Duchen P., Emerson J. J., Saelao P., Begun D. J., Langley C. H. (2012). Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genetics, 8, e1003080. 10.1371/journal.pgen.1003080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R. (1997). Progress and prospects in evolutionary biology—The Drosophila model. Oxford University Press. [Google Scholar]
- Prasad N. G., Joshi A. (2003). What have two decades of laboratory life-history evolution studies on Drosophila melanogaster taught us?. Journal of Genetics, 82, 45–76. 10.1007/BF02715881 [DOI] [PubMed] [Google Scholar]
- Price T., Schulter D. (1991). On the low heritability of life-history traits. Evolution, 45, 853–861. 10.2307/2409693 [DOI] [PubMed] [Google Scholar]
- Rajpurohit S., Nedved O. (2013). Clinal variation in fitness related traits in tropical drosophilids of the Indian subcontinent. Journal of Thermal Biology, 38, 345–354. 10.1016/j.jtherbio.2013.04.004 [DOI] [Google Scholar]
- Rajpurohit S., Zhao X., Schmidt P. S. (2017). A resource on latitudinal and altitudinal clines of ecologically relevant phenotypes of the Indian Drosophila. Scientific Data, 4, 170066. 10.1038/sdata.2017.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt J. A., Kolaczkowski B., Jones C. D., Begun D. J., Kern A. D. (2014). Parallel geographic variation in Drosophila melanogaster. Genetics, 197, 361–373. 10.1534/genetics.114.161463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S. H., Garner J. P., Würbel H. (2009). Environmental standardization: Cure or cause of poor reproducibility in animal experiments?. Nature Methods, 6, 257–261. 10.1038/nmeth.1312 [DOI] [PubMed] [Google Scholar]
- Richter S. H., Garner J. P., Auer C., Kunert J., Würbel H. (2010). Systematic variation improves reproducibility of animal experiments. Nature Methods, 7, 167–168. 10.1038/nmeth0310-167 [DOI] [PubMed] [Google Scholar]
- Roberts S. P., Feder M. E. (2000). Changing fitness consequences of hsp70 copy number in transgenic Drosophila larvae undergoing natural thermal stress. Functional Ecology, 14, 353–357. 10.1046/j.1365-2435.2000.00429.x [DOI] [Google Scholar]
- Robinson S. J., Zwaan B., Partridge L. (2000). Starvation resistance and adult body composition in a latitudinal cline of Drosophila melanogaster. Evolution, 54, 1819–1824. [DOI] [PubMed] [Google Scholar]
- Rodrigues M. F., Vibranovski M. D., Cogni R. (2021). Clinal and seasonal changes are correlated in Drosophila melanogaster natural populations. Evolution, 75, 2042–2054. 10.1111/evo.14300 [DOI] [PubMed] [Google Scholar]
- Roff D. A. (1992). The evolution of life histories. Theory and analysis. Chapman and Hall. [Google Scholar]
- Roff D. A. (2007). Contributions of genomics to life-history theory. Nature Reviews Genetics, 8, 116–125. 10.1038/nrg2040 [DOI] [PubMed] [Google Scholar]
- Roff D. A., Mousseau T. A. (1987). Quantitative genetics and fitness: Lessons from Drosophila. Heredity, 58, 103–118. 10.1038/hdy.1987.15 [DOI] [PubMed] [Google Scholar]
- Rose M. R. (1984). Genetic covariation in Drosophila life history: Untangling the data. The American Naturalist, 123, 565–569. 10.1086/284222 [DOI] [Google Scholar]
- Rose M. R., Nusbaum T. J., Chippindale A. K. (1996). Laboratory evolution: The experimental wonderland and the Cheshire cat syndrome. Chapter 7. In Rose M. R., Lauder G. V. (Eds.), Adaptation(pp. 221–-241.). Academic Press. [Google Scholar]
- Rudman S. M., Greenblum S. I., Rajpurohit S., Betancourt N. J., Hanna J., Tilk S., Yokoyama T., Petrov D. A., Schmidt P. (2022). Direct observation of adaptive tracking on ecological time scales in Drosophila. Science, 375(6586), eabj7484. 10.1126/science.abj7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrelli F., Tauber E., Pegoraro M., Mazzotta G., Cisotto P., Landskron J., Stanewsky R., Piccin A., Rosato E., Zordan M., Costa R., Kyriacou C. P., (2007). A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science, 316, 1898–1900. 10.1126/science.1138426 [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Kofler R., Versace E., Tobler R., Franssen S. U. (2015). Combining experimental evolution with next-generation sequencing: A powerful tool to study adaptation from standing genetic variation. Heredity, 114, 431–440. 10.1038/hdy.2014.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. S., Conde D. R. (2006). Environmental heterogeneity and the maintenance of genetic variation for reproductive diapause in Drosophila melanogaster. Evolution, 60, 1602–1611. [PubMed] [Google Scholar]
- Schmidt P. S., Paaby A. B. (2008). Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution, 62, 1204–1215. 10.1111/j.1558-5646.2008.00351.x [DOI] [PubMed] [Google Scholar]
- Schmidt P., Matzkin L., Ippolito M., Eanes W. (2005a). Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution, 59, 1721–1732. [PubMed] [Google Scholar]
- Schmidt P., Paaby A., Heschel M. (2005b). Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evolution, 59, 2616–2625. [PubMed] [Google Scholar]
- Schmidt P., Zhu C., Das J., Batavia M., Yang L., Eanes W. F. (2008). An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proceedings of the National Academy of Sciences, 105, 16207–16211. 10.1073/pnas.0805485105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service P. M., Rose M. R. (1985). Genetic covariation among life-history components: The effects of novel environments. Evolution, 39, 943–945. 10.2307/2408694 [DOI] [PubMed] [Google Scholar]
- Silbermann R., Tatar M. (2000). Reproductive costs of heat shock protein in transgenic Drosophila melanogaster. Evolution, 54, 2038–2045. [DOI] [PubMed] [Google Scholar]
- Sinervo B., Svensson E. (2002). Correlational selection and the evolution of genetic architecture. Heredity, 89, 329–338. 10.1038/sj.hdy.6800148 [DOI] [PubMed] [Google Scholar]
- Singh R. S., Hickey D. A., David J. (1982). Genetic differentiation between geographically distant populations of Drosophila melanogaster. Genetics, 101, 235–256. 10.1093/genetics/101.2.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen J. G., Schou M. F., Loeschcke V. (2017). Evolutionary adaptation to environmental stressors: A common response at the proteomic level. Evolution, 71, 1627–1642. 10.1111/evo.13243 [DOI] [PubMed] [Google Scholar]
- Sprengelmeyer Q. D., Pool J. E. (2021). Ethanol resistance in Drosophila melanogaster has increased in parallel cold-adapted populations and shows a variable genetic architecture within and between populations. Ecology and Evolution, 11, 15364–15376. 10.1002/ece3.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengelmeyer Q. D., Mansourian S., Lange J. D., Matute D. R., Cooper B. S., Jirle E. V., Stensmyr M. C., Pool J. E. (2020). Recurrent collection of Drosophila melanogaster from wild African environments and genomic insights into species history. Molecular Biology and Evolution, 37, 627–638. 10.1093/molbev/msz271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns S. C. (1992). The evolution of life histories. Oxford University Press. [Google Scholar]
- Stearns S. C., Partridge L. (2001). The genetics of aging in Drosophila. In Masoro E., Austad S. (Eds.), Handbook of the biology of aging(pp. 353–368.). Academic Press. [Google Scholar]
- Stearns S. C., Kaiser M., Kawecki T. J. (1995). The differential genetic and environmental canalization of fitness components in Drosophila melanogaster. Journal of Evolutionary Biology, 8, 539–557. 10.1046/j.1420-9101.1995.8050539.x [DOI] [Google Scholar]
- Svensson E. I. (2023). Phenotypic selection in natural populations: What have we learned in 40 years?. Evolution, 77, 1493–1504. 10.1093/evolut/qpad077 [DOI] [PubMed] [Google Scholar]
- Svensson E. I., Arnold S. J., Bürger R., Csilléry K., Draghi J., Henshaw J. M., Jones A. G., De Lisle S., Marques D. A., Mcguigan K., Simon M. N., Runemark A. (2021). Correlational selection in the age of genomics. Nature Ecology & Evolution, 5, 562–573. 10.1038/s41559-021-01413-3 [DOI] [PubMed] [Google Scholar]
- Tauber E., Zordan M., Sandrelli F., Pegoraro M., Osterwalder Nicolò, Breda C., Daga A., Selmin A., Monger K., Benna C., Rosato E., Kyriacou C. P., Costa R. (2007) Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science, 316, 1895–1898. 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- Thornton K., Andolfatto P. (2006). Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics, 172, 1607–1619. 10.1534/genetics.105.048223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Levine M. T., Eckert M. L., Begun D. J. (2008). Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics, 179, 455–473. 10.1534/genetics.107.083659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Stewart A. D., Fields A. T., Rice W. R., Tarone A. M. (2011). Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genetics, 7, e1001336. 10.1371/journal.pgen.1001336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T., Macleod M. R., McCann S. K., Senior A. M., Nakagawa S. (2021). Meta-analysis of variation suggests that embracing variability improves both replicability and generalizability in preclinical research. PLoS Biology, 19, e3001009. 10.1371/journal.pbio.3001009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Staay F. J., Arndt S. S., Nordquist R. E. (2010). The standardization-generalization dilemma: A way out. Genes, Brain and Behavior, 9, 849–855. 10.1111/j.1601-183X.2010.00628.x [DOI] [PubMed] [Google Scholar]
- Van‘t Land J., Van Putten P., Zwaan Kamping, Van Delden W. (1999). Latitudinal variation in wild populations of Drosophila melanogaster: Heritabilities and reaction norms. Journal of Evolutionary Biology, 12, 222–232. 10.1046/j.1420-9101.1999.00029.x [DOI] [Google Scholar]
- Van't Land J., Van Putten W. F., Villarroel H., Kamping A., van Delden W. (2000). Latitudinal variation for two enzyme loci and an inversion polymorphism in Drosophila melanogaster from Central and South America. Evolution, 54, 201–209. [PubMed] [Google Scholar]
- Voelkl B., Würbel H. (2016). Reproducibility crisis: Are we ignoring reaction norms?. Trends in Pharmacological Sciences, 37, 509–510. 10.1016/j.tips.2016.05.003 [DOI] [PubMed] [Google Scholar]
- Voelkl B., Altman N. S., Forsman A., Forstmeier W., Gurevitch J., Jaric I., Karp N. A., Kas M. J., Schielzeth H., Van De Casteele T., Würbel H. (2020). Reproducibility of animal research in light of biological variation. Nature Reviews Neuroscience, 21, 384–393. 10.1038/s41583-020-0313-3 [DOI] [PubMed] [Google Scholar]
- Voelkl B., Vogt L., Sena E. S., Würbel H. (2018). Reproducibility of preclinical animal research improves with heterogeneity of study samples. PLoS Biology, 16, e2003693. 10.1371/journal.pbio.2003693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Bergland A. O. (2022). Distinct signals of clinal and seasonal allele frequency change at eQTLs in Drosophila melanogaster. Evolution, 76, 2758–2768. 10.1111/evo.14617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaan B. J., Azevedo R. B. R., James A. C., Van 'T Land J., Partridge L. (2000). Cellular basis of wing size variation in Drosophila melanogaster: A comparison of latitudinal clines on two continents. Heredity, 84, 338–347. 10.1046/j.1365-2540.2000.00677.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data and the complete compilation of code, statistical models, analyses, and results are available on GitHub (https://esradm.github.io/DrosEU_PhenotypingWG/) and are archived on Zenodo (https://doi.org/10.5281/zenodo.15310170).