Abstract
Glutamate receptor-interacting protein 1 (GRIP1) is an adaptor protein composed of seven PDZ (postsynaptic density-95/Discs large/zona occludens-1) domains, capable of mediating diverse protein–protein interactions. GRIP1 has been implicated in the regulation of neuronal synaptic function, but its physiologic roles have not been defined in vivo. We find that elimination of murine GRIP1 results in embryonic lethality. GRIP1−/− embryos develop abnormalities of the dermo-epidermal junction, resulting in extensive skin blistering around day 12 of embryonic life. Ultra-structural characterization of the blisters (or bullae) revealed cleavage of the dermo-epidermal junction below the lamina densa, an alteration reminiscent of the dystrophic form of human epidermolysis bullosa. Blisters were also observed in the lateral ventricle of the brain and in the meninges covering the cerebral cortex. These genetic data suggest that the GRIP1 scaffolding protein is required for the formation and integrity of the dermo-epidermal junction and reveal the importance of PDZ domains in the organization of supramolecular structures essential for mammalian embryonic development.
Protein interaction domains are a common feature of polypeptides involved in signal transduction (1, 2). Intracellular proteins containing multiple interaction domains can serve as scaffolds to recruit the components of a signaling network into larger molecular complexes. The resulting supramolecular structures localize cell surface receptors and their cytoplasmic targets to defined subcellular sites, in a fashion that is essential for rapid and specific intracellular signaling (3).
Postsynaptic density-95/Discs large/zona occludens-1 (PDZ) domains are typical interaction domains; they have a modular structure of 80–100 aa and are found in many distinct cytoplasmic proteins, usually in conjunction with other interaction domains (3). They typically bind peptide motifs located at the C termini of interacting proteins, notably transmembrane receptors, adhesion molecules, and ion channels (4–6). In this mode, PDZ domains recognize a C-terminal hydrophobic residue and at least three preceding amino acids, with different PDZ domains preferentially binding to distinct C-terminal sequences (6–8). PDZ domains can also interact with internal β-turn motif and can mediate homomultimerization or heteromultimerization (9, 10). Interestingly, PDZ domains are scarce in yeast, but abundant in metazoan organisms, suggesting that they organize more complex properties of cells in multicellular animals.
INAD (inactivation-no-afterpotential D) and glutamate receptor-interacting protein 1 (GRIP1) are the Drosophila and mammalian prototypes, respectively, for a family of proteins composed exclusively of PDZ domains. The five PDZ domains of Drosophila INAD bind proteins involved in visual phototransduction, including the TRP calcium channel, phospholipase Cβ, eye-specific protein kinase C, and the NINAC myosin III, in a fashion that is essential for the efficient response of photoreceptors to quanta of light. Thus INAD colocalizes regulatory proteins in a way that is critical for phototransduction (11–13).
Mammalian GRIP1 has seven PDZ domains and interacts through PDZ4 and PDZ5 with the C-terminal -SVKI motif of the GluR2 and GluR3 subunits of the postsynaptic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type glutamate receptors (14, 15). GRIP1 participates in the localization and assembly of AMPA receptors, and interfering with the GRIP PDZ-GluR2/3 prevents AMPA receptor recruitment to the synapse (16). GRIP1 is present both at synapses and in intracellular compartments, including putative transport vesicles of neurons (14, 15). Murine GRIP1 expression can be detected by embryonic day (E) 10 and precedes that of AMPA receptors; in contrast, the expression of a closely related gene product, GRIP2/ABP (15, 17), parallels that of AMPA receptors and increases postnatally (18). These data suggest that GRIPs regulate the trafficking and postsynaptic clustering of AMPA receptors. However, GRIP1 and GRIP2 may play a larger role in synaptic signaling, because they can bind through PDZ6 to EphB2/EphA7 receptor tyrosine kinases and the ephrin-B1 ligand (19–21). Furthermore, GRIP1 PDZ7 binds a Ras guanine nucleotide exchange factor, GRASP1 (22). GRIP1 may therefore play a multifaceted role in assembling and localizing postsynaptic signaling complexes, although its in vivo functions have not been determined.
To explore the physiological role of this mammalian PDZ domain-only protein, we developed a GRIP1-deficient mouse strain. Surprisingly, morphologic, immuno-histochemical, and ultra-structural observations demonstrate that loss of GRIP1 function causes a skin blistering disorder during embryogenesis, reminiscent of human dystrophic epidermolysis bullosa (EB). These data indicate an essential role for the GRIP1 PDZ domain protein in the structural and functional integrity of the dermo-epidermal junction.
Materials and Methods
Generation of GRIP1−/− Mouse Mutants.
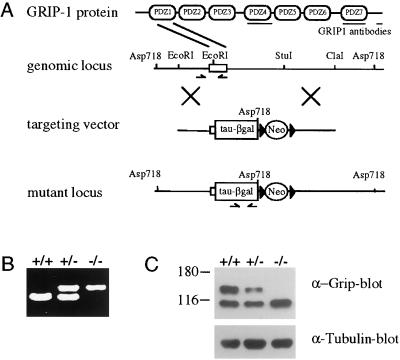
A genomic library from 129/Sv mice was probed with a cDNA fragment coding for PDZ domains 5 and 6 of the GRIP1 gene. To construct the targeting vector a 15-kb clone from a 129/Sv murine genomic library, containing the exon coding for the 5′ end of the first PDZ domain, was used. An EcoRI restriction site within this exon was used to fuse a tau-LacZ-loxP-Neo-CMV-loxP cassette (23) in-frame to the coding GRIP1 sequence. A 2.1-kb EcoRI and a 4.5-kb StuI–ClaI fragment were used as the short and long arms, respectively. The tau-LacZ-loxP-Neo-CMV-loxP cassette replaced a 3.6-kb genomic sequence, including the 3′ fragment of the exon encoding the first PDZ domain (see above). Upon electroporation with the linearized vector R1 embryonic stem (ES) cells were selected with G418.
PCR Screening of the Mice.
Template DNA for genomic PCR was isolated from ear tissue as described (24). Four primers were used to screen for wild-type (wt), heterozygous (+/−), and null (−/−) mice: primer 1, tacctagacagcctttctcagcacc; primer 2, ctccttcttcatcaactccaccactg; primer 3, ctgattgaagcagaagcctgcgatg; and primer 4, tattggcttcatccaccacatacagg. Primers 1 and 2 amplified a 220-bp region of the GRIP1 wt locus, and primers 3 and 4 amplified a 280-bp fragment of the β-galactosidase gene.
Western Blotting.
E12.5 embryos were homogenized in phospho lipase C lysis buffer by using a Teflon homogenizer as described (24). Western blot analysis was performed as described (24).
β-Galactosidase Staining of Whole Embryos and Tissue Sections.
β-Galactosidase activity was detected as described (24).
Antibodies.
Antibodies specifically recognizing the following molecules were used: GRIP1 (Upstate Biotechnology, used in Western blotting), platelet-endothelial cell adhesion molecule 1 (PECAM) (PharMingen), α-actin smooth muscle cell (Sigma), pan-cadherin (Sigma), β4-integrin (PharMingen), laminin (Sigma), and collagen types IV (abcam, Cambridge, U.K.) and VII (abcam). Antibodies specifically recognizing murine GRIP1 were obtained from rabbits immunized with two different glutatione S-transferase fusion proteins, containing residues 387–483 of GRIP1 PDZ4 (GenBank accession no. AB051560) and residues 902–992 of PDZ7.
Immunostaining and Electron Microscopy.
Immunostaining and electron microscopy analysis was performed as described (24).
Results
Targeted Disruption of the GRIP1 Gene Results in Embryonic Lethality.
Homologous recombination in murine ES cells was performed by using a targeting vector designed to replace part of the exon encoding the first PDZ domain of GRIP1 with an in-frame tau-LacZ coding sequence and a neomycin expression cassette (Fig. 1A). Five of 600 G418-resistant ES cell lines were identified by Southern blot analysis (data not shown). Two of the targeted ES cell lines were injected into blastocysts and independently gave rise to two male chimaeras, which transmitted the mutant allele through the germ line upon mating to ICR females. The resulting (129/J × ICR)F1 GRIP1+/− mice were intercrossed in an effort to generate GRIP1−/− animals. Genotype analysis by PCR (Fig. 1B) failed to detect any GRIP1−/− mutants after birth (P30) or at E7.5. However, GRIP1−/− mutants were present with approximately Mendelian frequencies at day 3.5 postcoitum. Further backcrossing with ICR mice resulted in a significant difference in the time of embryonic lethality. Intercrossing of (129/J × ICR)F3 heterozygous GRIP1 mutants yielded GRIP1−/− embryos in numbers close to the expected Mendelian frequencies at E12. Furthermore, low, but significant, numbers of viable GRIP1−/− embryos could be identified up to E16 (Table 1). Therefore, depending on the genetic background, in vivo ablation of GRIP1 function results in two distinct phenotypes, characterized by lethality at different stages of embryonic development. In this study, we have focused our analysis on the later embryonic phenotype observed in the progeny of intercrosses between (129/J × ICR)F3 GRIP1 heterozygous mice.
Figure 1.
Generation of a GRIP1-deficient mouse strain. (A) Schematic representation of GRIP1 gene-targeting strategy. Restriction map of the wt GRIP1 locus, targeting vector, and targeted locus. The targeting vector contains a tau–LacZ reporter gene fused in-frame with the exon encoding the first PDZ domain. Antibodies used for Western blot analysis are indicated. (B) Genotypic analysis of E12 embryos by PCR. To identify wt and targeted loci, primers were designed in the deleted wt locus and the β-galactosidase region, respectively. (C) Expression of GRIP1 was assessed by Western blot analysis of protein extracts from E12 embryos. The antibody used to detect GRIP1 expression is specific for the C-terminal portion of GRIP1 and does not cross-react with GRIP2 (18). The 100-kDa cross-reactive band bound by the C-terminal peptide-specific antibody was not recognized by antisera raised to the PDZ4 or the PDZ7 domains of GRIP1 (data not shown) and is therefore not GRIP1 specific.
Table 1.
Influence of the genetic background on the time of GRIP1−/− embryonic lethality
| Age | (129/J × ICR) F1
|
Age | (129/J × ICR) F3
|
||||
|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | +/+ | +/− | −/− | ||
| E 3.5 | 13 | 24 | 11 | E 12 | 66 | 121 | 35 |
| E 7.5 | 23 | 51 | 0 | E 14 | 71 | 153 | 31 |
| P 30 | 108 | 203 | 0 | E 16 | 28 | 67 | 4 |
| P 30 | 64 | 110 | 0 | ||||
The frequencies of +/+, +/− and −/− GRIP1 mutants were compared in the progenies of (129/J × ICR)F1 and F3 intercrosses. Approximate Mendelian frequencies of GRIP1 mutants were obtained at E3.5 and E12 in the progeny of F1 and F3 intercrosses, respectively. The viability of GRIP1−/− embryos was significantly prolonged by backcrossing into the ICR background.
The GRIP1 protein was identified in wt and heterozygous embryos, but was undetectable in GRIP1−/− embryos, when using antibodies specific for the C-terminal peptide (Fig. 1C) and the GRIP1 PDZ4 domain (data not shown), which recognize all known splice variants of GRIP1 (25). Therefore, the targeted mutation entirely ablates production of the GRIP1 polypeptide.
GRIP1 Is Expressed in the Developing Nervous System and Epidermis.
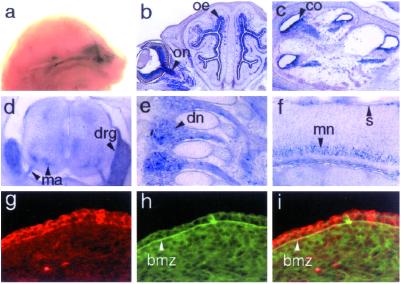
The pattern of GRIP1 expression was previously investigated by Western blotting and radioactive in situ hybridization in rat embryos (20). To clarify which cell types express GRIP1 during embryogenesis, we used tau–LacZ in the GRIP1 targeting vector. The resulting GRIP1tau-LacZ fusion protein, expressed under the control of the endogenous GRIP1 promoter, appears to faithfully reflect expression of wt GRIP1, as indicated by the overlapping staining patterns of β-galactosidase and GRIP1-specific antibodies (data not shown). Analysis of GRIP1tau-LacZ expression by whole-mount staining for β-galactosidase activity revealed expression in the developing nervous system of GRIP1+/− embryos. In particular, GRIP1tau-LacZ was strongly expressed in the forming olfactory tract (Fig. 2a), olfactory epithelium, eye (Fig. 2b), and cochlea (Fig. 2c), and in motor neurons and parasympathetic ganglia (Fig. 2 d–f). Strong GRIP1 expression was also detected outside the nervous system, particularly in skeletal muscles (data not shown) and at the dermo-epidermal junction (Fig. 2 f and g). Further immunohistochemical analysis of skin sections using antibodies specific for GRIP1 or cadherin revealed colocalization of GRIP1 with cadherin (Fig. 2 g–i), indicating that GRIP1 is a component of the basal cell layer of the developing epidermis. In GRIP1−/− mutants, the expression of GRIP1 in the epidermis is abrogated (data not shown). This site of GRIP1 expression is of particular significance in light of the phenotype observed in GRIP1−/− embryos, discussed below.
Figure 2.
GRIP1 is expressed in the developing nervous system and epidermis. Whole-mount staining of GRIP1+/− E16 embryos shows GRIP1tau-LacZ expression in the olfactory nerve (a). GRIP1 is also expressed in the optic nerve (b, on), olfactory epithelium (b, oe), cochlea (c, co), motor axons (d, ma), dorsal root ganglia (d, drg), nerve of the dorsal root ganglia (e, dn), and motor neurons (f, mn), as assessed by β-galactosidase staining on transverse (b–d) and sagittal (e and f) sections. Notably, β-galactosidase activity was also present in the skin (f, s). Immunostaining with antibodies specific for GRIP 1 (g) and pan-cadherin (h) of skin sections from wt embryos revealed GRIP1 expression in the basal epithelial cells (g). The expression of cadherin in the BMZ (h) colocalizes with that of GRIP1 in the BMZ (arrowheads), as shown by overlapping staining of the GRIP1 and pan-cadherin antibodies (i). Sections were obtained from GRIP1+/− E16 (b and c), E14 (d–f), and wt E12 (g–i) embryos. (Magnifications: a, ×3; b, ×7; c and d, ×15; e and f, ×20; and g–i, ×60.)
GRIP1−/− Embryos Are Affected by a Severe Bullous Disorder.
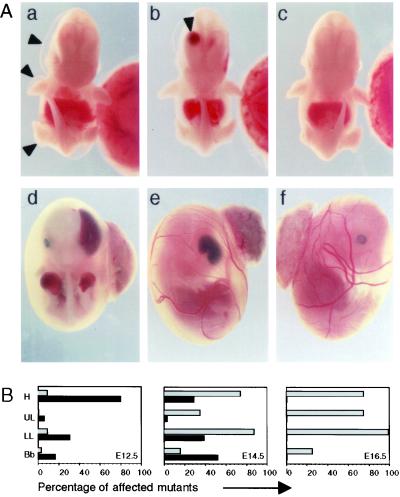
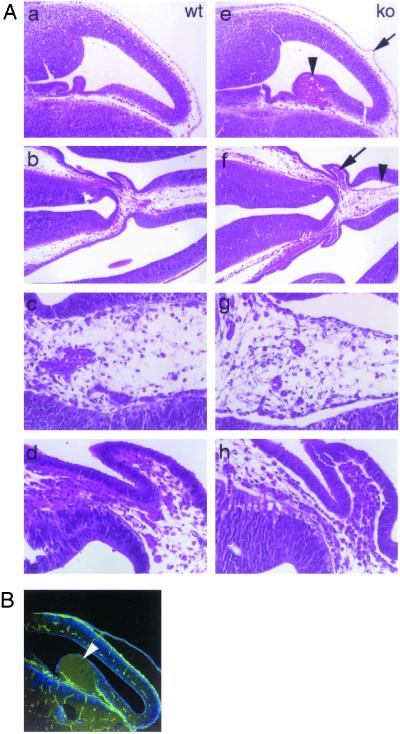
The most striking morphological feature observed in GRIP1−/− embryos was the presence of multiple fluid-filled raised areas clearly visible on the body surface, which can be defined as blisters or bullae. These bullae could be classified on the basis of their location, time of appearance, and type of fluid content (Fig. 3B). Typically, the blisters formed on the head (including the developing palate, eye, and vomero-nasal cavity), forelimbs, hind limbs, and spinal cord (Fig. 3A a and b). They first became visible around E12, when most blisters visible on the body surface were filled with fluid devoid of cellular content and therefore defined as serous blisters. However, intracranial haemorrhagic bullae could be observed through the frontal area of the developing skull (Fig. 3Ab). At later time points, blisters were haemorrhagic (Fig. 3A d and e) and contained many nucleated erythrocytes (not shown). The heart did not appear edematous at any time point. Taken together, these findings indicate that GRIP1−/− embryos develop a severe bullous disorder, which, by rapid evolution of the serous into haemorrhagic bullae, results in massive blood loss, hypovolemia, and death in utero around gestational day 14.5.
Figure 3.
Development of a severe bullous disorder in GRIP1−/− embryos. (A) Gross morphology of GRIP1−/− E12 embryos, showing large serous bullae on the surface of the head (a and b, arrowheads), upper and lower limbs (a) and with a large haemorrhagic bulla in the right lateral ventricle of the brain (b, arrowhead). In GRIP1−/− E14 embryos (shown inside the yolk sac), haemorrhagic bullae were found in similar locations: head (d and e), upper limbs (d). wt littermate controls are shown for comparison (c and f). (Magnifications: a–c, ×4; d–f, ×25.) (B) Frequency of serous (black bars) and haemorrhagic (gray bars) bullae in the head (H), upper limbs (UL), lower limbs (LL), and back bone (Bb) of GRIP1−/− embryos at different stages of embryonic development. The data derive from the cumulative observation of 35, 31, and four GRIP1−/− embryos at E12, E14, and E16, respectively.
Cleavage of the Epithelial-Mesenchymal Junction Results in the Formation of Bullae in GRIP1−/− Embryos.
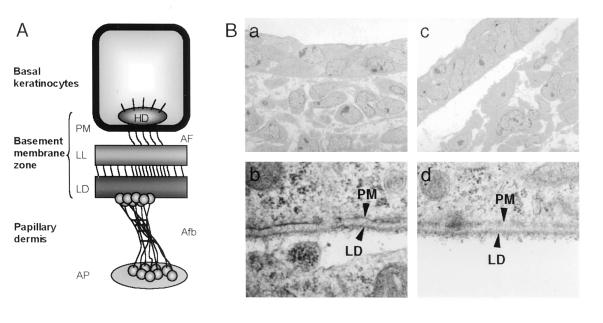
Inherited bullous disorders in humans, collectively defined as EB, are characterized by great heterogeneity in their clinical manifestations and morphologic properties, and are classified according to the level at which detachment of the epithelial cell layer occurs (26). Histopathological analysis of the skin blisters observed in GRIP1−/− embryos revealed an extensive detachment of the epithelial monolayer from the underlying mesenchymal tissue (Fig. 4A c and d). Interestingly, separation between the dermis and epithelial cell layer could also be observed in skin areas without macroscopically detectable blisters (Fig. 4A c and d). As attachment to the basement membrane is essential for survival of epithelial cells at later stages of development (E16) (27), we investigated whether epithelial cells lining the blisters of E12 GRIP1−/− embryos underwent apoptosis, but could not find any evidence of increased cell death (data not shown). We next investigated the site at which the epidermis detaches from the dermis in GRIP1−/− embryos (see the schematic representation of the dermo-epidermal junction in Fig. 6A). Immunostaining with antibodies specific for β4-integrin, laminin, or collagen types IV and VII was used to identify the basal epidermal cell layer, lamina lucida, lamina densa (LD), and sublamina densa compartment, respectively. We found that β4-integrin, type IV collagen (data not shown), and laminin (Fig. 4Bd) all segregated on the roof of the bullae, whereas type VII collagen was present on both the roof and the floor of the bullae (Fig. 4Be). These findings indicate that tissue separation in the serous bullae occurs below the LD (also see Fig. 6Bd).
Figure 4.
Cleavage at the dermal side of the cutaneous BMZ is responsible for skin blistering in GRIP1−/− mutants. (A) Hematoxylin-eosin-stained sections of lower limbs of wt (a and b) and GRIP1−/− (c and d) E12 embryos. Serous blisters are clearly visible on both sides of the limb (c). At higher magnification, the epidermis appears diffusely detached from the underlying dermis in blistered (arrowhead) and nonblistered areas (arrow). (B) Immunostaining of limb sections of wt (a–c) and GRIP1−/− (d–f) E12 embryos with antibodies recognizing laminin (a and d), collagen type VII (b and e), and PECAM (c and f; counterstaining with Hoechst 33258). Laminin staining segregates on the roof of the bulla (d, arrowheads), whereas collagen type VII is present on both sides of the tissue cleft (e, arrowheads). (Magnifications: A a and c, ×20, b and d, ×60; B, a–f, ×60.)
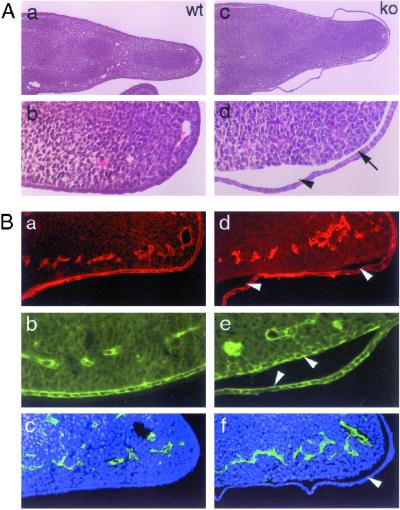
Figure 6.
GRIP1−/− embryos develop dystrophic EB. (A) Schematic representation of the ultrastructure of the dermo-epidermal junction (as described in the text). (B) Ultrastructural analysis of skin from wt (a and b) and GRIP1−/− mutants (c and d). In GRIP1−/− mutants, nonblistered areas exhibit a large gap between epidermis and dermis (c), a feature absent in the skin of littermate controls. Analysis of the roof of the tissue cleft at higher magnification (d) reveals the presence of the LD. This type of lesion is typical of the dystrophic form of EB. (Magnifications: a and b, ×5,500; c, ×44,500; d, ×45,000.)
As both serous and haemorrhagic bullae shared the same locations and appeared sequentially in GRIP1−/− embryos, we investigated whether they are related. Immunostaining with antibodies to PECAM and α-actin smooth muscle cell, which are specific markers for endothelial and mesenchymal cells in the vascular system, was used to explore whether the blisters have a vascular origin. However, neither serous (Fig. 4Bf) nor haemorrhagic (data not shown) bullae showed direct evidence of vascular cell types. Furthermore, as with the serous bullae, β4-integrin was found on the roof of the haemorrhagic lesions (data not shown), supporting the hypothesis that serous and haemorrhagic bullae share a common origin and represent increasingly advanced stages of the same pathological defect. The evolution of serous blister into hemorrhagic bullae likely occurs upon rupture of neighboring vessels into the blisters.
We also pursued the origin of the intracranial bullae, seen in E12 mutant embryos. Tissue sectioning localized these lesions to the lateral ventricle of the brain, in close proximity to the developing choroid plexus (Fig. 5Ae). The surfaces of the bullae were PECAM negative (Fig. 5B), indicating that the bullae were not of endothelial origin. The possibility that presumptive PECAM-positive cells lining the bullae had undergone apoptosis cannot be ruled out, but is unlikely, as further histopathological examination of the brains of GRIP1−/− embryos revealed the presence of numerous serous blisters. Furthermore, we observed diffuse detachment of neuroepithelial cells, which in wt mice express GRIP1 (data not shown), from the underlying mesenchymal tissue (Fig. 5A f–h). These findings are compatible with the hypothesis that blisters in both skin and lateral ventricles of the brain have a common pathogenesis and result from loss of GRIP1-dependent cohesiveness of epithelial cells to the underlying mesenchyma.
Figure 5.
Development of bullae in the lateral ventricle of the brain. (A) Hematoxylin-eosin staining of brain sections from wt (a–d) and GRIP1−/− (e–h) E12 embryos. Large haemorrhagic bulla (e, arrowhead) and blister on the surface of the meninges (e, arrow). A large serous blister (f, arrowhead; g, higher magnification) and extensive detachment of the neural epithelium from the underlying mesenchymal tissue (f, arrow; h, higher magnification) can be observed in the lateral ventricle, in close proximity to the developing choroid plexus. (B) PECAM immunostaining of a large bulla (arrowhead) protruding into the cerebral ventricle. Lack of PECAM staining on the surface of the bulla rules out its vascular origin. (Magnifications: A a, b, e, and f, ×20; c, d, g, and h, ×50; B, ×20.)
Skin Lesions in GRIP1−/− Mutants Resemble the Dystrophic Form of EB.
To define the perturbations observed in GRIP1 mutant embryos more precisely, we performed an electron microscopic analysis of skin blisters and peri-lesional skin. Ultrastructural analysis of the skin (Fig. 6B) confirmed the presence of the LD on the roof of the blister and clearly showed cleavage in the sublamina densa area at the dermal side of the cutaneous basement membrane zone (BMZ; Fig. 6Bd). Interestingly, electron microscopy of skin areas without macroscopically detectable blisters revealed diffuse detachment of the developing epidermis from the mesenchyma (Fig. 6Bc). The bullous disorder observed in GRIP1−/− murine embryos therefore has pathological features similar to those of human patients affected by dystrophic EB.
Discussion
PDZ domain proteins appear to play a pivotal role in the assembly, targeting, and spatial arrangement of signaling complexes at the cell membrane (3). To test the biological functions of a mammalian multi-PDZ adapter, we have generated mouse strains carrying a mutation in the GRIP1 locus that coordinately ablates GRIP1 coding potential and marks the pattern of GRIP1 expression by insertion of a tau–LacZ cassette expressed under the control of the endogenous GRIP1 promoter. GRIP1 was highly expressed in the developing nervous system. However, this does not exclude the possibility that GRIP1 may have a role in other cell types, and indeed, GRIP1 was also expressed in non-neuronal tissues, notably at the dermo-epidermal junction.
Consistent with this diverse expression GRIP1 apparently plays multiple roles during embryonic development. GRIP1 mutants on an (129/J × ICR)F1 background died shortly after implantation, whereas further backcrossing onto the ICR background to obtain (129/J × ICR)F3 resulted in an increase in embryonic survival up to E16. These phenotypic differences are likely to result from modifier gene(s) (28) inherited with the 129/J and ICR backgrounds; both phenotypic variants were independently obtained from two targeted ES cell lines. The development of severe skin blistering in the GRIP1−/− embryos on the ICR background indicates that GRIP1 plays an essential role in the stability of the dermo-epidermal junction. These data underscore the biological importance of scaffolding proteins composed of PDZ interaction domains.
During embryogenesis, the epidermis consists of basal epithelial cells and periderm, a layer of thin, endothelium-like cells that develops between E9 and E11 (29). To resist mechanical shearing forces, the epidermis is anchored to the dermis at the dermo-epidermal junction (see Fig. 6A). Basal keratinocytes are strongly anchored to the papillary dermis through a complex system of attachment units organized inside and around the BMZ, comprising plasma membrane, lamina lucida, and LD. The hemidesmosomes are electron-dense cytoplasmic plaques connected intracellularly with keratin intermediate filaments and extracellularly through anchoring filaments to the anchoring fibrils (Afb) in the LD. Afb extend from the LD to the anchoring plaque (AP) in the dermis. A number of large oligomeric proteins, including α6β4 integrin, laminin 5, and collagens VII and XVII form these anchoring complexes and have been implicated in the pathogenesis of clinical variants of EB (26).
Dystrophic EB (DEB), the type of disorder apparently observed in GRIP1−/− embryos, is characterized by tissue cleavage below the LD. In humans, DEB comprises two dominantly and four recessively inherited disorders, associated with mutations of the collagen VII gene, COL7A1 (26). Targeted deletion of collagen VII in the mouse also results in DEB-type skin lesions and death within 2 weeks of birth (30). In Col7A1−/− newborn skin adhesion of the epidermis and dermis occurs, but is very fragile and exhibits low resistance to external shearing forces. In contrast, in the absence of GRIP1, the cohesion between dermis and epidermis seems to be entirely abrogated, and cleavage of the two layers occurs in an environment, which is highly protected from mechanical stress by the amniotic fluid. There is presently no evidence for GRIP1 mutations in inherited human blistering disorders, but the role of human GRIP1 in skin formation warrants study.
In addition to the epidermal blistering, defects in the brain, characterized by the formation of bullae protruding into the lateral ventricle and at the level of the meninges covering the cerebral cortex, were observed. Interestingly loss of basement membrane components has been previously associated with brain defects. Absence of Hsp47 (31), a stress-inducible glycoprotein, which transiently binds to newly synthesized procollagen, results in abnormal orientation of the neuroepithelium with invasion of the underlying mesenchyme. This defect is probably caused by altered collagen I and IV synthesis and loss of the basement membrane. Deletion of β1-integrin in neurons and glia causes abnormal remodeling of the meningeal basement membrane (32). The structural similarity between the epidermal-dermal junction and the basement membrane of meninges and the neuroepithelium of the lateral ventricle supports the hypothesis that skin blistering and intracranial bullae in GRIP1 mutants may have a common pathogenetic mechanism. Despite the expression of GRIP1 in the olfactory epithelium and retina, these tissues did not exhibit obvious abnormalities. A compensatory role of GRIP2 and/or background-dependent factors may have a protective effect on these epithelial structures.
Our findings raise a number of questions as to the in vivo function of GRIP1. It is intriguing that the lack of an intracellular protein results in detachment of the dermal-epidermal junction below the LD. Furthermore, our preliminary observations that GRIP1−/− embryos have altered collagen type VII immunostaining and reduced plasma membrane electron density suggest a profound structural disorganization of the BMZ. GRIP1 could participate in multiple ways in the organization of the BMZ. First, GRIP1 may be involved in the scaffolding or localization of adhesion complexes and anchoring molecules responsible for epidermal-dermal cohesion. In addition, GRIP1 is implicated in vesicular trafficking (3) and could potentially regulate secretion and/or processing of extracellular matrix proteins. In any event, the phenotype of GRIP1−/− embryos strongly suggests that the architecture of the epidermal-dermal junction requires PDZ domain-mediated interactions, likely to organize junctional components into a functional structure.
The importance of GRIP1 to the integrity of the dermo-epidermal junction is not at odds with a role in the nervous system. As our findings indicate an essential role of GRIP1 in organizing the basal domain of epithelial cells and previous reports have shown its involvement in synaptic activity (16), it is tempting to speculate that GRIP1 may have functional similarities to Caenorhabditis elegans LIN-10 (33). This PDZ domain-containing protein is required for both basolateral localization of the LET-23 receptor tyrosine kinase in vulval precursor epithelial cells and postsynaptic localization of the glutamate receptor GLR-1 (34, 35). It will be of interest to explore the extent to which the basal domain of polarized epithelial cells is functionally analogous to the dendritic compartment of neurons.
Acknowledgments
We thank Lee Adamson and Yuqing Zhou for sharing their knowledge and expertise in embryonic angiogenesis, Douglas Holmyard for electron microscopy, Ken Harpal for technical assistance, and Arnoud Sonnenberg for critically reading the manuscript. This work was supported by a Terry Fox Program Project grant from the National Institute of Canada and by the Canadian Institutes of Health Research. F.B. was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft. T.P. is a Distinguished Scientist of the Canadian Institutes of Health Research.
Abbreviations
- GRIP1
glutamate receptor-interacting protein 1
- PDZ
postsynaptic density-95/Discs large/zona occludens-1
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- En
embryonic day n
- ES
embryonic stem
- wt
wild type
- PECAM
platelet-endothelial cell adhesion molecule 1
- LD
lamina densa
- BMZ
basement membrane zone
- EB
epidermolysis bullosa
References
- 1.Pawson T, Scott J D. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T, Nash P. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- 3.Sheng M, Sala C. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Kim E, Niethammer M, Rothschild A, Jan Y N, Sheng M. Nature (London) 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 5.Kornau H C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 6.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 7.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 8.Stricker N L, Christopherson K S, Yi B A, Schatz P J, Raab R W, Dawes G, Bassett D E, Jr, Bredt D S, Li M. Nat Biotechnol. 1997;15:336–342. doi: 10.1038/nbt0497-336. [DOI] [PubMed] [Google Scholar]
- 9.Brenman J E, Chao D S, Gee S H, McGee A W, Craven S E, Santillano D R, Wu Z, Huang F, Xia H, Peters M F, et al. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 10.Hillier B J, Christopherson K S, Prehoda K E, Bredt D S, Lim W A. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 11.Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker C S. Nature (London) 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 12.Chevesich J, Kreuz A J, Montell C. Neuron. 1997;18:95–105. doi: 10.1016/s0896-6273(01)80049-0. [DOI] [PubMed] [Google Scholar]
- 13.Scott K, Zuker C S. Nature (London) 1998;395:805–808. doi: 10.1038/27448. [DOI] [PubMed] [Google Scholar]
- 14.Dong H, O'Brien R J, Fung E T, Lanahan A A, Worley P F, Huganir R L. Nature (London) 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 15.Wyszynski M, Valtschanoff J G, Naisbitt S, Dunah A W, Kim E, Standaert D G, Weinberg R, Sheng M. J Neurosci. 1999;19:6528–6537. doi: 10.1523/JNEUROSCI.19-15-06528.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, Kerchner G A, Sala C, Wei F, Huettner J E, Sheng M, Zhuo M. Nat Neurosci. 1999;2:972–977. doi: 10.1038/14771. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava S, Ziff E B. Ann NY Acad Sci. 1999;868:561–564. doi: 10.1111/j.1749-6632.1999.tb11329.x. [DOI] [PubMed] [Google Scholar]
- 18.Dong H, Zhang P, Song I, Petralia R S, Liao D, Huganir R L. J Neurosci. 1999;19:6930–6941. doi: 10.1523/JNEUROSCI.19-16-06930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin D, Gish G D, Songyang Z, Pawson T. J Biol Chem. 1999;274:3726–3733. doi: 10.1074/jbc.274.6.3726. [DOI] [PubMed] [Google Scholar]
- 20.Bruckner K, Pablo Labrador J, Scheiffele P, Herb A, Seeburg P H, Klein R. Neuron. 1999;22:511–524. doi: 10.1016/s0896-6273(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 21.Torres R, Firestein B L, Dong H, Staudinger J, Olson E N, Huganir R L, Bredt D S, Gale N W, Yancopoulos G D. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- 22.Ye B, Liao D, Zhang X, Zhang P, Dong H, Huganir R L. Neuron. 2000;26:603–617. doi: 10.1016/s0896-6273(00)81198-8. [DOI] [PubMed] [Google Scholar]
- 23.Mombaerts P, Wang F, Dulac C, Chao S K, Nemes A, Mendelsohn M, Edmondson J, Axel R, Dong H, Zhang P, et al. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 24.Lai K M, Pawson T. Genes Dev. 2000;14:1132–1145. [PMC free article] [PubMed] [Google Scholar]
- 25.Yamazaki M, Fukaya M, Abe M, Ikeno K, Kakizaki T, Watanabe M, Sakimura K. Neurosci Lett. 2001;304:81–84. doi: 10.1016/s0304-3940(01)01766-9. [DOI] [PubMed] [Google Scholar]
- 26.Pulkkinen L, Uitto J. Matrix Biol. 1999;18:29–42. doi: 10.1016/s0945-053x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- 27.DiPersio C M, van der Neut R, Georges-Labouesse E, Kreidberg J A, Sonnenberg A, Hynes R O. J Cell Sci. 2000;113:3051–3062. doi: 10.1242/jcs.113.17.3051. [DOI] [PubMed] [Google Scholar]
- 28.Nadeau J H. Nat Rev Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- 29.M'Boneko V, Merker H J. Acta Anat (Basel) 1988;133:325–336. doi: 10.1159/000146662. [DOI] [PubMed] [Google Scholar]
- 30.Heinonen S, Mannikko M, Klement J F, Whitaker-Menezes D, Murphy G F, Uitto J. J Cell Sci. 1999;112:3641–3648. doi: 10.1242/jcs.112.21.3641. [DOI] [PubMed] [Google Scholar]
- 31.Nagai N, Hosokawa M, Itohara S, Adachi E, Matsushita T, Hosokawa N, Nagata K. J Cell Biol. 2000;150:1499–1506. doi: 10.1083/jcb.150.6.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graus-Porta D, Blaess S, Senften M, Littlewood-Evans A, Damsky C, Huang Z, Orban P, Klein R, Schittny J C, Muller U. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 33.Kim S K, Horvitz H R. Genes Dev. 1990;4:357–371. doi: 10.1101/gad.4.3.357. [DOI] [PubMed] [Google Scholar]
- 34.Rongo C, Whitfield C W, Rodal A, Kim S K, Kaplan J M. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- 35.Whitfield C W, Benard C, Barnes T, Hekimi S, Kim S K. Mol Biol Cell. 1999;10:2087–2100. doi: 10.1091/mbc.10.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]