Abstract
Explanations for biogeographic disjunctions involving South America and Africa typically invoke vicariance of western Gondwanan biotas or long distance dispersal. These hypotheses are problematical because many groups originated and diversified well after the last known connection between Africa and South America (≈105 million years ago), and it is unlikely that “sweepstakes” dispersal accounts for many of these disjunctions. Phylogenetic analyses of the angiosperm clade Malpighiaceae, combined with fossil evidence and molecular divergence-time estimates, suggest an alternative hypothesis to account for such distributions. We propose that Malpighiaceae originated in northern South America, and that members of several clades repeatedly migrated into North America and subsequently moved via North Atlantic land connections into the Old World during episodes starting in the Eocene, when climates supported tropical forests. This Laurasian migration route may explain many other extant lineages that exhibit western Gondwanan distributions.
Malpighiaceae contains trees, shrubs, and vines that are distributed widely in tropical and subtropical forests and savannas of the Old and New Worlds (ref. 1; Fig. 1). Approximately 85% of the species occur in the New World where they are pollinated by specialized oil-collecting bees that are absent from the Old World. This distribution has invited the development of alternative theories for the origin and diversification of Malpighiaceae. Vogel (2) proposed the “Gondwanian aborigine” hypothesis, in which the current distribution resulted from the break-up of western Gondwana (the supercontinent comprising Africa and South America). This hypothesis implies that the Malpighiaceae originated before western Gondwana divided [i.e., ≈105 mya (3)], as did several included lineages with disjunct distributions between the New and Old Worlds. In contrast, Anderson (1) and others (4) have favored an “American colonist” hypothesis, according to which Malpighiaceae originated in northern South America, in isolation from Africa, and dispersed eastward across the Atlantic at least twice. This hypothesis predicts that the ages of Malpighiaceae and of divergences between New World and Old World lineages are younger than the last known land connection between South America and Africa.
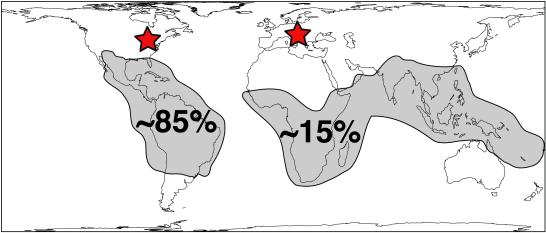
Figure 1.
Current geographic distribution of Malpighiaceae estimated from Arénes (22). Malpighiaceae, with ≈1,250 species (1), are most diverse in northern South America (22). The ≈180 Old World species, belonging to six lineages (see Fig. 2) are variously represented in Africa (47 species), Australasia (17 species), India (43 species), and Madagascar (80 species) [from Arénes (22)]. Red stars indicate fossil localities from Hungary and Slovenia (17), and Tennessee (21).
To elucidate the biogeographic history of Malpighiaceae, we used maximum likelihood to estimate the phylogeny for the group by using DNA nucleotide data from chloroplast ndhF and nuclear phytochrome C (PHYC) sequences (see Materials and Methods). This data set included sequences from one outgroup and 70 species of Malpighiaceae. To test these biogeographic hypotheses, it is necessary to attach a temporal dimension to the phylogeny to infer the timing (i) of the origin of Malpighiaceae and (ii) of the disjunctions between New and Old World lineages. Although molecular divergence estimates must be viewed with caution, these approaches may identify a window of time for branching events that will help select among competing hypotheses.
Materials and Methods
Our data set included 71 ndhF sequences and 71 PHYC sequences, which were readily aligned by eye. This data set included sequences from one outgroup and 70 species of Malpighiaceae representing the majority of genera, including multiple accessions from morphologically diverse or putatively non-monophyletic genera. Most ndhF sequences were previously obtained by using the methodology published in Davis, Anderson, and Donoghue (5) and in Davis (6). Two additional ndhF sequences were also generated for this study (GenBank accession nos. AF500495 and AF500496). The PHYC sequences are newly generated (GenBank accession nos. AF500522– AF500582), except for AF436794–AF436804 from Davis (6), and were obtained by using previously detailed (7) PCR, cloning, and sequencing procedures. We screened up to five clones from several species representing most of the major lineages within Malpighiaceae (GenBank accession nos. AF500497–AF500521) and found no evidence of duplication events within PHYC, consistent with previous findings by Mathews and Donoghue (7).
To assess the level of congruence between the ndhF and PHYC data sets, we used the incongruence length difference test (8) implemented in paup* v. 4.0b8 (9) for UNIX as the partition-homogeneity test. We used simple taxon addition (saving 10 trees per replicate), tree-bisection-reconnection branch-swapping, and heuristic searches with 999 repartitions of the data. The results (P = 0.55) indicated that ndhF and PHYC were congruent and so they were combined for further analysis. The combined data set included 1,833 aligned sites. Data matrices analyzed in this study are available from the first author and from TreeBASE (http://www.treebase.org).
Phylogenetic analyses by using maximum likelihood were conducted on the combined data set with paup*. Tree searches were conducted with 300 random sequence addition replicates and tree-bisection-reconnection branch swapping. We performed 300 bootstrap replicates to assess clade support. To choose a model of sequence evolution we performed likelihood ratio tests (10) with likelihood trees generated by using a series of models with increasing complexity. The GTR+I+Γ model had a higher likelihood than other models and was used to evaluate molecular rate constancy.
To infer the location of disjunctions between New and Old World lineages, ancestral areas were reconstructed by using dispersal-vicariance analysis (DIVA; ref. 11). DIVA reconstructs ancestral areas by minimizing the number of dispersal and extinction events needed to explain the distribution pattern. Vicariance is the default mode of speciation in DIVA, and such events are not counted as steps in identifying optimal solutions; inferred dispersal and extinction events are counted as one step each. Our data matrix used to assess ancestral areas was constructed by scoring terminals for presence in either the New World or the Old World, and the analysis was carried out by using diva v. 1.1 [ref. 12; available by anonymous FTP from Uppsala University (ftp.uu.se or ftp.systbot.uu.se)]. The outgroup for Malpighiaceae was scored as New World based on the close phylogenetic relationship of the neotropical endemic Whittonia guianensis Sandw (Peridiscaceae) (13).
Branch lengths and an associated likelihood score were calculated for the respective models of sequence evolution in the absence of a molecular clock. We used trees resulting from maximum likelihood searches to test for rate constancy among lineages under the assumption of a molecular clock by using 2(−lnL1–lnL0) as a test statistic. This test statistic was compared with a χ2 distribution (with n–2 degrees of freedom; n = number of taxa) to assess significance. Finding that a clock was rejected (P < 0.05), we used a nonparametric rate-smoothing algorithm (14) to estimate divergence times, focusing specifically on New World-Old World disjunctions inferred from our DIVA reconstructions.
To estimate standard errors associated with divergence dates, we used a parametric bootstrapping strategy similar to the three-step procedure of Baldwin and Sanderson (15). Their method was a nonparametric procedure to estimate the stochastic error associated with sampling a finite number of nucleotide characters. We adopted a parametric bootstrapping approach, as follows: (i) 100 data sets were simulated on the maximum likelihood tree with the computer software seq-gen v. 1.2.3 (16); (ii) the resulting simulated data sets were imported into paup*, and divergence times were estimated on the original tree topology with the sequence model and parameters estimated from the original data; and (iii) the resulting branch length estimates from the simulated data sets were used to calculate the variance in divergence time estimates. These error estimates reflect the stochastic error associated with the molecular evolution process.
A fossil species of Tetrapterys recovered from the early Oligocene (33 mya; ref. 17) from Hungary and Slovenia provides a reliable divergence estimate for the Tetrapterys clade and was used to calibrate our tree. Several other excellent Malpighiaceae fossils are known from the Eocene and provide important information on its distribution in the past (see Results and Discussion). However, because these fossils are not readily assignable to any extant genus, they are difficult to include as calibration points.
Results and Discussion
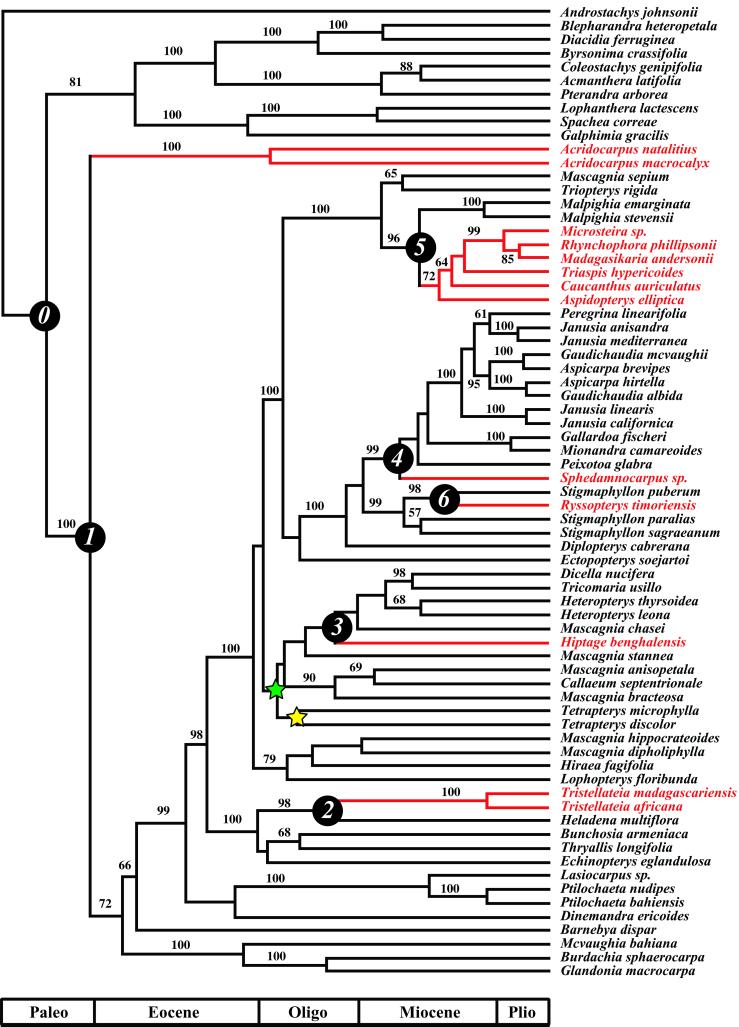
Our phylogeny (Fig. 2) is robust and congruent with previous parsimony analyses based on four chloroplast genes (5, 18). The basal split in Malpighiaceae separates the wholly New World subfamily Byrsonimoideae from a major clade containing both New and Old World species. Within this clade, the Old World Malpighiaceae belong to six separate lineages (Fig. 2), all of which contain species with winged-fruits. A fossil species of Tetrapterys recovered from the early Oligocene (33 mya; ref. 17) suggests that the divergence between Tetrapterys and its sister lineage had occurred by this time (Fig. 2; green star). Based on this assumption, the minimum age of Malpighiaceae is estimated to be 63.6 ± 5.8 mya, and the six New World-Old World divergences would have occurred between the early Eocene (55.1 ± 6.0 mya) and the middle Miocene (12.9 ± 0.85 mya). If, instead, we assume that the age of the fossil corresponds to the basal split within Tetrapterys (Fig. 2; yellow star), we obtain maximum age estimates that are ≈4 Myr older than the estimated minimum ages. Similar results are obtained by placing the fossil at less likely positions deeper in the tree.
Figure 2.
Maximum likelihood tree topology (−ln L = 17,333.97) for combined ndhF and PHYC data with bootstrap values (>50%) indicated on branches. Old World lineages are shaded red. Divergence times were calculated on the rate-smoothed topology by calibrating the two nodes indicated by the colored stars with a fossil of Tetrapterys (33 mya; ref. 17). The origin of Malpighiaceae (node 0) is reconstructed as New World and is estimated at 63.6 ± 5.8 mya. The nodes labeled 1–6 correspond to the New/Old World disjunctions estimated from the DIVA reconstruction. Age estimates for nodes 1 through 6 (calibrated with the fossil placed at the green star) are, respectively: 55.1 ± 6.0 mya, 30.4 ± 2.6, 29.4 ± 2.1, 19.1 ± 1.5, 15.1 ± 1.2, and 12.9 ± 0.85. The scale bar indicates major Tertiary epochs (Paleo = Paleocene; Oligo = Oligocene; Plio = Pliocene).
Our divergence estimates indicate that Malpighiaceae originated well after the last known connection between Africa and South America (≈105 mya). These estimates parallel those reported by Magallón and Sanderson (19), which suggest that many major angiosperm lineages have radiated during more recent times in the Tertiary, and do not extend back to the Cretaceous. In fact, our fossil calibration points would have to be approximately 20 Myr older to make the ancestral node within Malpighiacecae consistent with a Gondwanan origin (i.e., 105 mya). Divergence estimates for the New World-Old World disjunctions are even younger, rendering the “Gondwanian aborigine” explanation untenable (Fig. 2; nodes 1–6).
In contrast, our divergence estimates are potentially consistent with a series of episodic long-distance dispersal events as suggested by Anderson (1) and others (4). However, our results indicate that the “American colonist” scenario would require at least six dispersal events across the Atlantic. The plausibility of this scenario is further diminished by excellent Eocene and Oligocene fossils of Malpighiaceae from several localities throughout the Northern Hemisphere and Africa: Perisyncolporites from northern South America and Nigeria (20), Eoglandulosa from Tennessee (21), and Tetrapterys from Hungary and Slovenia (17). Our phylogenetic results, together with the fossil evidence, suggest migration through Laurasia as a new explanation for the present distribution of Malpighiaceae.
These data imply that the Malpighiaceae originated and began to diversify in northern South America in the early Paleocene (≈64 mya). Anderson (1) also postulated a South American origin, which is consistent with the high levels of diversity and endemism in Malpighiaceae of Guyana (22). From South America, various wing-fruited species may have migrated into North America via scattered continental and/or volcanic islands that connected North and South America at various times through the Tertiary, starting in the Eocene (23). This route apparently played a central role in facilitating the distribution of Antillean land-mammals (24), and would most likely have been available to Malpighiaceae during this timeframe. From North America, these lineages could migrate eastward across Laurasia into the Old World via a series of connections across the North Atlantic (the “North Atlantic Land Bridge”; ref. 25), subsequently diversifying in Africa (and later in Madagascar) and in some cases in Asia (see Fig. 3; red arrow). Populations in eastern North America, Europe, and other northern regions were presumably eliminated as tropical/subtropical climates disappeared from these areas during the Miocene and later epochs.
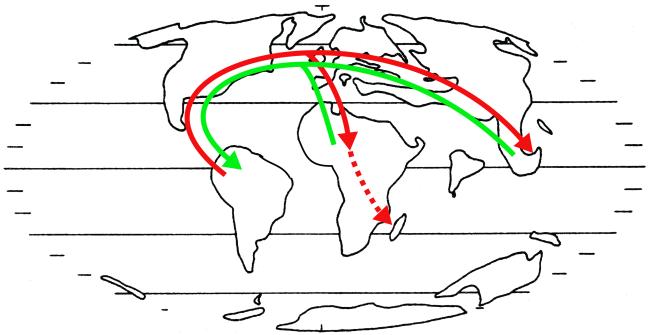
Figure 3.
Eocene paleogeographic map [after Parrish (41); modified from Doyle and Le Thomas (35)] summarizing hypothesized Laurasian migration of western Gondwana taxa. The distribution of Malpighiaceae is proposed to have resulted by migration from South America to the Old World (red arrow); green arrow shows a possible pattern of dispersion from the Old World to the New World taken by other tropical angiosperm clades.
In addition to the “North Atlantic Land Bridge,” Beringia facilitated the spread of the northern hemisphere biota throughout much of the Tertiary, and may represent an alternative migration pathway for Malpighiaceae. It apparently played an important role in determining the distribution of mammals of Asian origin that migrated to North America during periods of climatic warming in the late Palaeocene-early Eocene (26). Similarly, there may have been repeated dispersion of temperate and boreal plants through Beringia from the Eocene to the present (27). Although most of our estimates of divergence times indicate that Malpighiaceae could have migrated via Beringia, its high latitude (between 69° and 75° N; ref. 28) almost certainly prohibited its use by thermophilic lineages for most of this time, and fossil localities do not support this route for the dispersion of Malpighiaceae. Moreover, Old World Malpighiaceae are almost entirely restricted to Africa and Madagascar with a few, relatively minor, excursions into Asia. Together, paleoclimatic, phylogenetic, and fossil data are most consistent with an easterly migration across the Atlantic rather than a westerly movement via Beringia.
The North Atlantic pathway apparently served as a conduit for migration of several lineages of Malpighiaceae at different times. Dispersion episodes leading to nodes 1, 2, and 3 (Fig. 2) broadly coincide with Eocene/Oligocene thermal maxima and the dominance of a “boreotropical flora” across the North Atlantic (29). Significant cooling during the Oligocene (30) resulted in southward retreats and the extirpation of some lineages comprising this flora (31–33). Warming during the Miocene resulted in the return of several thermophilic lineages to North America and Europe (28), probably including lineages of Malpighiaceae labeled 4, 5, and 6 (Fig. 2).
These findings have general implications for the origin of disjunctions between South America and Africa (and elsewhere in the Old World tropics). Vicariance resulting from the break-up of western Gondwana cannot explain those disjunct angiosperm lineages (of which there may be many) that originated and diversified after the last direct continental connection. Dispersal directly across the Atlantic Ocean, although perhaps feasible in some cases, seems unlikely to be a general explanation because vagility varies greatly among lineages. Additionally, dispersal through Antarctica into southern Africa seems unlikely in view of the ancient split between Africa and Australasia (162–165 mya; ref. 3). Moreover, none of these theories explain the existence of undisputed Laurasian fossils for some Gondwanan disjuncts. Dispersion through Laurasia at times during the Tertiary when climatic conditions supported tropical vegetation may provide the best explanation for many organisms that show the classic western Gondwanan disjunction pattern. Malpighiaceae are here interpreted as a case of iterative spread from South America to the Old World through Laurasia. Recent phylogenetic analyses of several other tropical angiosperm clades appear to provide examples of spread either in the same direction (Melastomeae within Melastomataceae; ref. 34) or in the opposite direction, from the Old to the New World [Annonaceae (35); Lauraceae (36); Fig. 3, green arrow]. Other groups may have originated within subtropical North America and migrated into the Old World, and perhaps much later into South America [dichrostachioid Leguminosae (37); dalbergioid Leguminosae (38)].
The existence of a “boreotropical” connection across the North Atlantic during the Eocene has long been viewed as a key to understanding patterns of disjunction around the Northern Hemisphere [e.g., close relationships between Eastern Asian and Eastern North American plants (25, 27, 28, 39)]. Our analysis indicates that this pathway may also have played an important role in explaining the global distribution of tropical groups. Were it not for such Laurasian dispersion, many familiar pantropical clades might now be restricted to one hemisphere or the other. Conversely, the restriction of some major plant lineages to one hemisphere [e.g., Cactaceae in the New World (40)] may reflect their origin and radiation during a time when Laurasia was no longer a viable conduit for migration.
Acknowledgments
We thank W. Anderson, D. Baum, J. Cracraft, B. Moore, S. Renner, and C. Webb for helpful discussions, and M. Chase for providing critical material. A. Knoll and three anonymous reviewers provided helpful comments on an earlier version of this manuscript. H. Jonsson and P. Virketis provided technical assistance. This work was supported by National Science Foundation Grant DEB-0073299.
Abbreviation
- DIVA
dispersal-vicariance analysis
Footnotes
References
- 1.Anderson W R. Mem N Y Bot Gard. 1990;64:210–224. [Google Scholar]
- 2.Vogel S. Mem N Y Bot Gard. 1990;55:130–142. [Google Scholar]
- 3.McLoughlin S. Aust J Bot. 2001;49:271–300. [Google Scholar]
- 4.Raven P H, Axelrod D I. Ann Missouri Bot Gard. 1974;61:539–673. [Google Scholar]
- 5.Davis C C, Anderson W R, Donoghue M J. Am J Bot. 2001;88:1830–1846. [PubMed] [Google Scholar]
- 6.Davis C C. Am J Bot. 2002;89:723–730. doi: 10.3732/ajb.89.4.699. [DOI] [PubMed] [Google Scholar]
- 7.Mathews S, Donoghue M J. Science. 1999;286:947–950. doi: 10.1126/science.286.5441.947. [DOI] [PubMed] [Google Scholar]
- 8.Farris J S, Källersjö M, Kluge A G, Bult C. Cladistics. 1994;10:315–319. [Google Scholar]
- 9.Swofford D L. paup* Sunderland, MA: Sinauer; 1999. , Version 4.0b8. [Google Scholar]
- 10.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 11.Ronquist F. Syst Biol. 1997;46:195–203. [Google Scholar]
- 12.Ronquist F. diva (Uppsala Univ., Uppsala), Version 1.1. 1996. [Google Scholar]
- 13.Savolainen V, Fay M F, Albach D C, Backlund A, van der Bank M, Cameron K M, Johnson S A, Lledó M D, Pintaud J C, Powell M, et al. Kew Bull. 2000;55:257–309. [Google Scholar]
- 14.Sanderson M J. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- 15.Baldwin B G, Sanderson M J. Proc Natl Acad Sci USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rambaut A, Grassly N C. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- 17.Hably L, Manchester S R. Rev Palaeobot Palynol. 2000;111:93–101. doi: 10.1016/s0034-6667(00)00019-1. [DOI] [PubMed] [Google Scholar]
- 18.Cameron K M, Chase M W, Anderson W R, Hills H G. Am J Bot. 2001;88:1847–1862. [PubMed] [Google Scholar]
- 19.Magallón S, Sanderson M J. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 20.Germeraad J H, Hopping C A, Muller J. Rev Palaeobot Palynol. 1968;6:189–348. [Google Scholar]
- 21.Taylor D W, Crepet W. Am J Bot. 1987;74:274–286. [Google Scholar]
- 22.Arènes J. Compt Rend Sommaire Séances Soc Biogéogr. 1957;290:81–108. [Google Scholar]
- 23.Iturralde-Vinent M A, MacPhee R D E. Bull Am Mus Nat Hist. 1999;238:1–95. [Google Scholar]
- 24.MacPhee R D E, Iturralde-Vinent M A. Am Mus Novit. 1995;3141:1–31. [Google Scholar]
- 25.Tiffney B H. J Arnold Arbor. 1985;66:243–273. [Google Scholar]
- 26.Beard K C. Bull Carnegie Mus Nat Hist. 1998;34:5–39. [Google Scholar]
- 27.Donoghue M J, Bell C D, Li J. Int J Plant Sci. 2001;162:S41–S52. [Google Scholar]
- 28.Tiffney B H. J Arnold Arbor. 1985;66:73–94. [Google Scholar]
- 29.Wolfe J A. Ann Missouri Bot Gard. 1975;62:264–279. [Google Scholar]
- 30.Berggren W A, Prothero D R. In: Eocene–Oligocene Climatic and Biotic Evolution. Prothero D R, Berggren W A, editors. Princeton: Princeton Univ. Press; 1992. pp. 1–28. [Google Scholar]
- 31.Collinson M E. In: Eocene–Oligocene Climatic and Biotic Evolution. Prothero D R, Berggren W A, editors. Princeton: Princeton Univ. Press; 1992. pp. 437–450. [Google Scholar]
- 32.Wolfe J A. In: Eocene–Oligocene Climatic and Biotic Evolution. Prothero D R, Berggren W A, editors. Princeton: Princeton Univ. Press; 1992. pp. 421–436. [Google Scholar]
- 33.Manchester S R. Ann Missouri Bot Gard. 1999;86:472–522. doi: 10.3417/2014033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renner S S, Clausing G, Meyer K. Am J Bot. 2001;88:1290–1300. [PubMed] [Google Scholar]
- 35.Doyle J A, Le Thomas A. Géogr Phys Quatern. 1997;51:353–361. [Google Scholar]
- 36.Chanderbali A S, van der Werff H, Renner S S. Ann Missouri Bot Gard. 2001;88:104–134. [Google Scholar]
- 37.Lavin M, Luckow M. Am J Bot. 1993;80:1–14. [Google Scholar]
- 38.Lavin M, Thulin M, Labat J-N, Pennington R T. Syst Bot. 2000;25:449–467. [Google Scholar]
- 39.Wen J. Ann Rev Ecol Syst. 1999;30:421–455. [Google Scholar]
- 40.Nyffeler R. Am J Bot. 2002;89:312–326. doi: 10.3732/ajb.89.2.312. [DOI] [PubMed] [Google Scholar]
- 41.Parish J T. In: The Origins of Angiosperms and Their Biological Consequences. Friis E M, Chaloner W G, Crane P R, editors. Cambridge, U.K.: Cambridge Univ. Press; 1987. pp. 51–73. [Google Scholar]