Abstract
Segregation Distorter (SD) is a meiotic drive system in Drosophila that causes preferential transmission of the SD chromosome from SD/SD+ males owing to the induced dysfunction of SD+ spermatids. The key distorter locus, Sd, is a dominant neomorphic allele encoding a truncated, but enzymatically active, RanGAP (RanGTPase-activating protein) whose nuclear mislocalization underlies distortion by disrupting the Ran signaling pathway. Here, we show that even wild-type RanGAP can cause segregation distortion when it is overexpressed in the male germ line or when the gene dosage of a particular modifier locus is increased. Both manipulations result in substantial nuclear accumulation of RanGAP. Distortion can be suppressed by overexpression of Ran or Ran guanine nucleotide exchange factor (RanGEF) in the male germ line, indicating that the primary consequence of nuclear mislocalization of RanGAP is reduction of intranuclear RanGTP levels. These results prove that segregation distortion does not depend on any unique properties of the mutant RanGAP encoded by Sd and provide a unifying explanation for the occurrence of distortion in a variety of experimental situations.
A fundamental principle of Mendelian genetics is the equal transmission of both homologues or alleles from a heterozygous pair. Nonetheless, meiotic drive systems, in which this principle is regularly violated by the preferential transmission of a particular chromosome or allele at the expense of its partner, exist in nature (1). Segregation distorter (SD) is a naturally occurring meiotic drive system located on the second chromosome of Drosophila melanogaster (2–4). SD/SD+ males transmit the SD chromosome to almost 100% of the progeny. Distortion at full strength requires not only the primary locus, Sd, but also several upward modifiers including Enhancer of SD [E(SD)], Modifier of SD [M(SD)], and Stabilizer of SD [St(SD)] (2, 3, 5–7). The target of Sd and the upward modifiers is the Responder (Rsp) locus: chromosomes that carry a sensitive (Rsps) or supersensitive (Rspss) allele of Responder are sensitive to the action of SD, whereas those carrying an insensitive allele (Rspi) are resistant (6, 8–10). The basic mechanism of distortion is sperm dysfunction, which involves a failure of chromatin condensation in SD+-bearing spermatids, leading to subsequent defects in spermatid elongation and maturation (11, 12).
Sd, a dominant neomorphic mutation, encodes a truncated RanGTPase-activating protein (RanGAP) lacking 234 aa at the C terminus (Sd-RanGAP; ref. 13). Ran is a small GTPase located predominantly in the nucleus. Along with its cofactors, RanGAP and RanGEF (Ran guanine nucleotide exchange factor), Ran is essential for nuclear transport (14) as well as for other nuclear functions, including cell cycle regulation, mitotic spindle formation, and postmitotic nuclear envelope assembly (15–17). The cytoplasmic localization of RanGAP and the nuclear localization of RanGEF establish a concentration gradient of RanGTP across the nuclear envelope that is critical for proper function of the Ran signaling pathway. We previously showed that Sd-RanGAP retains essentially normal enzymatic activity but is mislocalized to nuclei. Both enzymatic activity and nuclear localization of Sd-RanGAP are required for distortion (18). Overexpression of either Ran or RanGEF in the male germ line suppresses distortion, suggesting that Sd-RanGAP causes distortion by diminishing the concentration of nuclear RanGTP, thereby disrupting Ran-dependent functions. In particular, nuclear transport may be impaired (18).
By analogy with results of recent studies on yeast RanGAP (19), nuclear mislocalization of Sd-RanGAP can be explained if the usual cytoplasmic distribution of RanGAP is the outcome of a dynamic mechanism in which RanGAP shuttles in and out of nuclei. Normally, this process favors nuclear export over nuclear localization. Shuttling of yeast RanGAP is mediated by a nuclear localization signal (NLS) and two nuclear export signals (NESs) that are located in evolutionarily conserved regions of the protein. Amino acid alignment between yeast and Drosophila RanGAP suggests that these NLSs and NESs are conserved in Drosophila RanGAP. The C-terminal deletion in Sd-RanGAP removes one of the putative NESs, which could shift the equilibrium in favor of a predominant nuclear localization (18). If this idea is correct, then there are no intrinsic functional differences between Sd-RanGAP and wild-type RanGAP other than their contrasting subcellular distributions. Thus, even wild-type RanGAP might be capable of causing segregation distortion if its subcellular distribution were perturbed such that there was significant accumulation inside nuclei. Alternatively, distortion could depend on entirely novel attributes of the truncated Sd-RanGAP that could not be mimicked in any simple way by the wild-type protein.
Here, we demonstrate that wild-type RanGAP is capable of causing segregation distortion when it is overexpressed in the male germ line, a manipulation that results in aberrant nuclear accumulation of this enzyme. Moreover, presence of an extra dose of E(SD) causes segregation distortion even when Sd is absent. In this case, too, we find that there is substantial accumulation of wild-type RanGAP inside nuclei. Distortion in these cases can be suppressed by overexpression of Ran or RanGEF in the male germ line, arguing that, as in the case of SD, a reduction in the level of intranuclear RanGTP is the primary cause of distortion. These results prove that nuclear localization of RanGAP activity is sufficient to cause distortion and provide a unifying explanation for the occurrence of distortion in a variety of experimental situations.‡
Materials and Methods
Fly Genetics and Germ-Line Transformation.
Flies were maintained on standard medium, and all crosses were carried out at 25°C. SD-5R7 and SD-5R16 are γ-ray-induced revertant chromosomes of SD-5 (6). SD-5R7 has Sd+ at the position of Sd; thus, this chromosome carries Sd+ E(SD) Rspi M(SD) St(SD) (20). SD-5R16 contains a strong dominant suppressor of SD; thus, this chromosome carries Su(SD) Sd E(SD) Rspi M(SD) St(SD). The ReR-5 cn chromosome was isolated as a recombinant chromosome that carries Sd+ E(SD) Rsps M(SD)+ St(SD)+ (21). SD+ chromosomes have the genotype Sd+ E(SD)+ M(SD)+ St(SD)+. The specific Rsp allele present depends on the particular chromosome. Rspss cn is the standard Rspss chromosome. Rspi cn bw carries a Rspi allele (6). cn bw is the standard tester second chromosome that carries Rsps and two recessive eye color markers that produce a white-eyed phenotype when homozygous. TM3 is a multiply inverted third chromosome balancer marked with Sb. All of the transgenic lines used in these experiments except for the β2-Ran and β2-RanGEF lines were generated on this chromosome. Descriptions of all standard chromosomes and markers are described in Lindsley and Zimm (22).
Segregation ratios were measured as in Kusano et al. (18). Briefly, k values for each transgenic line were calculated as the proportion of Rspi cn+-bearing progeny among the total progeny. For k tests in Table 1, males from each independent transgenic line were individually crossed to cn bw tester females. Each transgenic line was tested in a SD-5R7/Rspss cn, SD-5R7/Rspi cn bw, or SD-5R16/Rspss cn genetic background by scoring the progeny for appropriate eye color markers. All transgenic lines carried insertions on the TM3 chromosome. To eliminate any extrinsic viability effects on the measured segregation ratios, k values were corrected for viability differences as in Kusano et al. (18). For k tests in Table 2, males carrying β2-Ran or β2-RanGEF in appropriate genetic background were individually crossed to cn bw tester females. To eliminate viability effects with the β2-Ran or β2-RanGEF transgenes, which are located on the X chromosome, only male progeny (which did not receive this chromosome) were counted for k tests. All k values are presented as the mean ± SD.
Table 1.
Overexpression of wild-type RanGAP in the male germ line causes segregation distortion
| Transgene | SD-5R7/Rspss cn [E(SD) Rspi M(SD) St(SD)/Rspss] | SD-5R7/Rspi cn bw [E(SD) Rspi M(SD) St(SD)/Rspi] | SD-5R16/Rspss cn [Su(SD) Sd E(SD) Rspi M(SD) St(SD)/Rspss] |
|---|---|---|---|
| — | 0.524 ± 0.035 (n = 1483) | 0.474 ± 0.084 (n = 1192) | 0.475 ± 0.038 (n = 960) |
| Sd-RanGAP | 1.0 ± 0.000 (n = 938) | 0.474 ± 0.044 (n = 1398) | 0.559 ± 0.072 (n = 826) |
| β2-RanGAP | 0.895 ± 0.056 (n = 1484) | 0.474 ± 0.027 (n = 1288) | 0.518 ± 0.045 (n = 1123) |
| β2-RanGAP | 0.933 ± 0.031 (n = 1637) | 0.479 ± 0.045 (n = 891) | 0.504 ± 0.040 (n = 1587) |
| β2-RanGAP R87A | 0.510 ± 0.041 (n = 1426) | ||
| β2-RanGAP R87A | 0.472 ± 0.030 (n = 1678) |
Segregation ratios were determined in males carrying enzymatically active or inactive (R87A) RanGAP transgenes driven from the β2-tubulin promoter. Each row represents an independent insertion line. The k values (mean ± SD) were calculated from the proportion of the total offspring (n) that received the Rspi cn+ chromosome. The SD components present in each of the tested genotypes are shown in brackets. See Materials and Methods for details of fly crosses and genetic markers.
Table 2.
Germ-line overexpression of Ran or RanGEF suppresses segregation distortion caused by wild-type RanGAP
| Transgene | SD-5R7/Rspss cn; Sd-RanGAP [E(SD) Rspi M(SD) St(SD)/Rspss; Sd-RanGAP] | SD-5R7/Rspss cn; β2-RanGAP [E(SD) Rspi M(SD) St(SD)/Rspss; β2-RanGAP] | SD-5R7/ReR-5 cn [E(SD) Rspi M(SD) St(SD)/E(SD) Rsps] |
|---|---|---|---|
| — | 1.0 ± 0.00 (n = 1322) | 0.933 ± 0.031 (n = 1637) | 0.740 ± 0.014 (n = 1947) |
| β2-Ran | 0.472 ± 0.066 (n = 768) | 0.444 ± 0.059 (n = 860) | 0.573 ± 0.032 (n = 1816) |
| β2-RanGEF | 0.518 ± 0.055 (n = 764) | 0.412 ± 0.045 (n = 773) | 0.562 ± 0.037 (n = 1231) |
Segregation ratios were determined in males expressing Ran or RanGEF driven from the β2-tubulin promoter (18). The k values (mean ± SD) are calculated from the proportion of the total offspring (n) that received the Rspi cn+ chromosome. The SD components present in each of the tested genotypes are shown in brackets. See Materials and Methods for details of fly crosses and chromosome markers.
P element-mediated transformation techniques were performed as described in Spradling (23) with modification. Briefly, dechorinated embryos were injected with each plasmid construct at 0.8 mg/ml in 5 mM KCl, 0.1 mM PO4 (pH 7.8), with 3% Durkee green food coloring.
Plasmid Construction.
To overexpress wild-type RanGAP in the male germ line, we used the β2-tubulin promoter, which specifically drives high levels of expression in the postmitotic male germ line (24, 25). The ORF of wild-type RanGAP flanked with EcoRI sites was cloned into the EcoRI site of the testis vector, which contains the β2-tubulin promoter inserted into the pCasPeR4 transformation vector. In Schizosaccharomyces pombe RanGAP, residue R74, corresponding to R87 of the Drosophila protein, is essential for activity (26). We mutated R87 in Drosophila RanGAP to A and assayed enzyme activity in vitro (18).
Immunostaining of Testes Squashes.
Immunostaining of testes squashes was performed as described in Kusano et al. (18). Testes were incubated in rabbit anti-RanGAP antibody or mouse monoclonal HA.11 antibody (Covance, Princeton, NJ) at appropriate concentrations and subsequently in the secondary antibody (Alexa 488-conjugated goat anti-rabbit IgG or Alexa 488-conjugated goat anti-mouse IgG; Molecular Probes). Chromatin was stained with propidium iodide. Confocal images were collected on a MRC1024 confocal microscope (Bio-Rad).
Western Blot Analysis.
Western blots were performed as described in Kusano et al. (18). To assess the degree of overexpression of wild-type RanGAP, serial dilutions of protein samples were compared with the endogenous expression levels of RanGAP.
Results
Overexpression of Wild-Type RanGAP in Male Germ Line Causes Segregation Distortion.
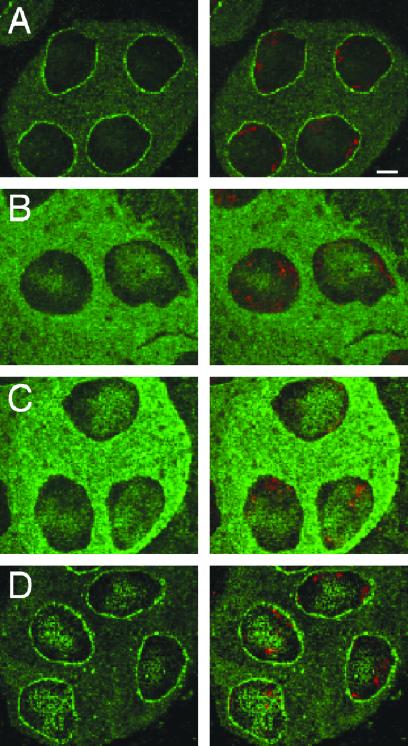
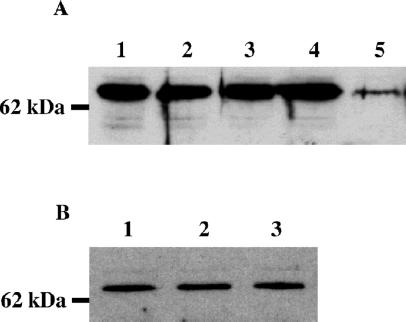
Sd-RanGAP differs from its wild-type counterpart by a C-terminal deletion of 234 aa (13), which confers this protein with dominant neomorphic properties. Although Sd-RanGAP appears to retain essentially normal enzymatic activity, its subcellular distribution is markedly altered, and substantial amounts of the mutant protein inappropriately localize to nuclei (ref. 18; Fig. 1 A and B). This ectopically localized protein requires RanGAP enzymatic activity to cause distortion (18), but it remains unclear whether distortion also involves some novel and undefined activities associated with the truncated protein. To investigate whether even wild-type RanGAP is capable of causing segregation distortion if it could somehow be made to accumulate inside nuclei, we examined the effect on chromosome transmission of overexpressing wild-type RanGAP in the male germ line. For this purpose, we drove expression of wild-type RanGAP from the β2-tubulin promoter, which is expressed specifically in the male germ line (24, 25). Expression of RanGAP in the testes of transgenic males was increased about 20-fold compared with controls (Fig. 2A).
Figure 1.
Immunolocalization of RanGAP in primary spermatocytes. Confocal images show staining with anti-RanGAP antibody (A, C, and D) or anti-HA (B) antibody on the Left and merged images with propidium iodide staining of chromatin on the Right. (A) Wild-type males. (B) Males expressing Sd-RanGAP tagged with an HA (influenza hemagglutinin) epitope (see ref. 18). (C) Males expressing β2-RanGAP. (D) Males carrying two doses of E(SD). Scale bar = 5 μm.
Figure 2.
Western blot analysis of RanGAP expression in testes. Each lane contains the total protein from two pairs of testes boiled in SDS sample buffer, separated electrophoretically, transferred onto poly(vinylidene difluoride)-plus membrane, and probed with anti-RanGAP antibody. (A) Expression of RanGAP from the β2-tubulin promoter results in an approximate 20-fold increase in RanGAP expression. Lanes 1 and 2, Two independent transgenic lines expressing β2-RanGAP; lanes 3 and 4, two independent transgenic lines expressing β2-RanGAP R87A; lane 5, Cantons-S (control). (B) Expression of RanGAP is not increased in males carrying extra doses of E(SD). Lane 1, Males carrying two doses of E(SD); lane 2, males carrying one dose of E(SD); lane 3, Canton-S (control) males lacking E(SD).
As shown in Table 1, expression of the β2-RanGAP transgene in a background containing all of the upward modifiers of distortion and heterozygous for Rspi vs. Rspss results in very strong distortion against the Rspss chromosome, yielding k values of 0.895 and 0.933 in two independent transgenic lines. Thus, germ line overexpression of wild-type RanGAP causes segregation distortion.
Distortion Caused by Overexpressed Wild-Type RanGAP Has the Same Properties as That Caused by SD.
To determine whether segregation distortion caused by germ-line overexpression of wild-type RanGAP is equivalent to that caused by a native SD chromosome, we carried out additional experiments to examine the functional properties of this distortion. Distortion caused by overexpressed RanGAP specifically involves operation of the SD system because no distortion was observed in comparable crosses in which the target chromosomes carried a Rspi allele rather than a Rsps or Rspss allele (Table 1, column 3). Moreover, in the presence of SD-5R16, which carries a strong suppressor of SD (6), no distortion against a Rspss-bearing target chromosome was caused by germ-line overexpression of wild-type RanGAP (Table 1, column 4).
We have previously shown that distortion caused by Sd-RanGAP depends on RanGAP enzymatic activity because the R87A mutation, which eliminates this activity, ablates the ability to cause distortion (18, 26). Introduction of the R87A mutation into wild-type RanGAP also abolishes its ability to cause distortion when it is overexpressed in the male germ line (Fig. 2A and Table 1, rows 5 and 6). Thus, distortion caused by β2-RanGAP appears in every respect to be fully equivalent to that caused by transgenic Sd-RanGAP and by native SD chromosomes.
Nuclear Mislocalization of Overexpressed Wild-Type RanGAP.
Distortion by Sd-RanGAP depends on its ectopic nuclear localization (ref. 18; Fig. 1B). Immunolocalization of RanGAP in primary spermatocytes from flies expressing the β2-RanGAP transgene revealed a similar alteration (Fig. 1C). In contrast with the usual localization of RanGAP at the cytoplasmic periphery of the nuclear envelope (Fig. 1A), in flies expressing β2-RanGAP the protein shows prominent nuclear localization in addition to abundant distribution throughout the cytoplasm, a pattern very reminiscent of that observed for Sd-RanGAP. Thus, overexpression of wild-type RanGAP in the male germ line results in elevated levels of nuclear RanGAP. The likely consequence of this elevation would be to stimulate RanGTPase activity in the nucleus with the resultant depletion of nuclear RanGTP. If this is the ultimate cause of distortion, as we have hypothesized for Sd-RanGAP, overexpression of Ran or RanGEF in the male germ line, which is expected to have an opposing effect on nuclear RanGTP levels, should suppress distortion caused by β2-RanGAP. As shown in Table 2 (columns 2 and 3), expression of β2-Ran or β2-RanGEF transgenes fully suppresses distortion caused by β2-RanGAP in exactly the same manner as these transgenes suppress distortion caused by a native SD chromosome or by an Sd-RanGAP transgene (18). Thus, the underlying mechanism of distortion appears to be the same in these cases.
Increased Dosage of E(SD) Causes Segregation Distortion Associated with Nuclear Accumulation of RanGAP.
The discovery that even wild-type RanGAP can cause distortion under appropriate conditions offered a possible explanation for another situation in which distortion has been observed. Previous studies have shown that, in the appropriate background, two doses of E(SD) can cause distortion even though only Sd+ alleles are present (ref. 21; Table 2, row 1, column 4). Although the basis of this distortion has remained mysterious, it could now be explained if increased dosage of E(SD) enhanced expression of wild-type RanGAP. However, Western blot analysis to measure RanGAP expression levels in males bearing extra doses of E(SD) did not detect any increased expression of RanGAP compared with control males (Fig. 2B). Although expression of RanGAP was not increased, immunolocalization in primary spermatocytes revealed that, in the presence of two doses of E(SD), there was a pronounced increase in the levels of nuclear RanGAP (Fig. 1D). Moreover, distortion caused by increased dosage of E(SD) is again completely suppressed by β2-Ran or β2-RanGEF transgenes (Table 2). These results indicate that E(SD) may encode a factor that enhances active nuclear import or localization of RanGAP and that increased dosage of E(SD) causes distortion by significantly elevating the level of nuclear RanGAP.
Discussion
We have shown that overexpression of wild-type RanGAP in the male germ line causes strong segregation distortion. This distortion exhibits all of the properties characteristic of that associated with native SD chromosomes: it requires RanGAP enzymatic activity, it depends on the presence of upward modifiers and a sensitive Rsp target, and it is eliminated by known suppressors of segregation distortion as well as by overexpression of Ran or RanGEF in the male germ line. Most strikingly, overexpression of wild-type RanGAP resulted in substantial mislocalization of the protein to nuclei in primary spermatocytes, similar to what has been observed for normal levels of expression of Sd-RanGAP (18). This result suggests that wild-type RanGAP is capable of entering nuclei and most likely does so to some degree even at endogenous levels of expression. Although this conclusion is contrary to the prevalent view that RanGAP is localized exclusively in the cytoplasm, it is in agreement with studies in yeast indicating that the normal cytoplasmic localization of RanGAP in the outcome of a dynamic equilibrium in which RanGAP shuttles in and out of the nucleus (19). This process is thought to be mediated by the activities of an NLS and two NESs that are located in evolutionarily conserved portions of the protein (19). Amino acid alignments with Drosophila RanGAP suggest that these elements are also present in the Drosophila protein. We have previously argued that the truncated RanGAP encoded by Sd deletes one of the NESs, thereby biasing the subcellular distribution from cytoplasm to nucleus. In the present case, 10- to 20-fold overexpression of wild-type RanGAP is apparently sufficient to alter the usual equilibrium such that an excessive amount of RanGAP accumulates inside nuclei.
The notion that nuclear mislocalization of enzymatically active RanGAP is responsible for distortion can also account for the distortion caused by two doses of E(SD) (21). Increased dosage of the modifier element E(SD) results in a marked accumulation of wild-type RanGAP inside spermatocyte nuclei even though the protein is expressed at endogenous levels. On the basis of this observation, it seems likely that E(SD) encodes a factor that facilitates nuclear import of RanGAP, inhibits its export, or in some other way enhances the nuclear localization of RanGAP. Because distortion associated with wild-type RanGAP is elicited by two doses of E(SD) but not one, we imagine that the physiological effect of this protein is dose dependent and that a certain threshold amount is required to accumulate a sufficient level of wild-type RanGAP in germ-line nuclei to produce distortion. A single dose of E(SD) substantially increases the strength of distortion caused by Sd-RanGAP (6, 13). Our results suggest that E(SD) may do so by further increasing the propensity of Sd-RanGAP to localize to nuclei. Because E(SD) behaves as a dominant neomorphic mutation (6, 21), what function is normally subserved by the wild-type protein remains an intriguing question. Unfortunately, the location of E(SD) within the heterochromatin of the second chromosome (3, 6) will greatly complicate molecular identification and characterization of this gene.
Our results argue that, aside from its altered subcellular distribution, there is nothing intrinsically unusual about the truncated Sd-RanGAP compared with the wild-type enzyme. Although the physical basis for nuclear mislocalization of RanGAP differs for Sd-RanGAP, β2-RanGAP, and increased dosage of E(SD), the underlying mechanism of distortion is apparently the same in all of these cases. In the appropriate genetic background, all that is required to cause segregation distortion in Drosophila is an excess of enzymatically active RanGAP in nuclei of the male germ line. The information gained from this study provides a unifying explanation for the occurrence of distortion and will be essential in elucidating the remaining mechanistic details of this phenomenon.
Acknowledgments
We thank Rayla Temin, Rob Reenan, and Michael Palladino for helpful comments on the manuscript, Sean Carroll for use of the confocal microscope, Young Ho Koh for assistance with the confocal microscopy, and Bob Kreber for technical assistance. This work was supported by National Science Foundation Grant DMB-9905974.
Abbreviations
- RanGAP
RanGTPase-activating protein
- RanGEF
RanGTPase guanine exchange factor
- SD
segregation distorter
- E(SD)
enhancer of SD
- M(SD)
modifier of SD
- St(SD)
stabilizer of SD
- NES
nuclear export signal
- NLS
nuclear localization signal
Footnotes
This is paper no. 3587 from the Laboratory of Genetics, University of Wisconsin-Madison.
References
- 1.Lyttle T W. Annu Rev Genet. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- 2.Hartl D L, Hiraizumi Y. In: The Genetics and Biology of Drosophila. Ashburner M, Novitski E, editors. 1b. New York: Academic; 1976. pp. 615–666. [Google Scholar]
- 3.Temin R G, Ganetzky B, Powers P A, Lyttle T W, Pimpinelli S, Wu C-I, Hiraizumi Y. Am Nat. 1990;137:287–331. [Google Scholar]
- 4.Lyttle T W. Trends Genet. 1993;9:205–210. doi: 10.1016/0168-9525(93)90120-7. [DOI] [PubMed] [Google Scholar]
- 5.Sandler L, Hiraizumi Y. Genetics. 1960;45:1671–1689. doi: 10.1093/genetics/45.12.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganetzky B. Genetics. 1977;86:321–355. [PMC free article] [PubMed] [Google Scholar]
- 7.Hiraizumi Y, Martin D W, Eckstrand I A. Genetics. 1980;95:693–706. doi: 10.1093/genetics/95.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C-I, Lyttle T W, Wu M-L, Lin G-F. Cell. 1988;54:179–189. doi: 10.1016/0092-8674(88)90550-8. [DOI] [PubMed] [Google Scholar]
- 9.Lyttle T W. Genetics. 1989;121:751–763. doi: 10.1093/genetics/121.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pimpinelli S, Dimitri P. Genetics. 1989;121:765–772. doi: 10.1093/genetics/121.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl D L, Hiraizumi Y, Crow J. Proc Natl Acad Sci USA. 1967;58:2240–2245. doi: 10.1073/pnas.58.6.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokuyasu K T, Peacock W J, Hardy R W. J Ultrastruct Res. 1977;58:96–101. doi: 10.1016/s0022-5320(77)80011-7. [DOI] [PubMed] [Google Scholar]
- 13.Merrill C, Bayraktaroglu L, Kusano A, Ganetzky B. Science. 1999;283:1742–1745. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- 14.Görlich D, Kutay U. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 15.Sazer S, Dasso M. J Cell Sci. 2000;113:1111–1118. doi: 10.1242/jcs.113.7.1111. [DOI] [PubMed] [Google Scholar]
- 16.Moore J D. BioEssays. 2001;23:77–85. doi: 10.1002/1521-1878(200101)23:1<77::AID-BIES1010>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 17.Dasso M. Cell. 2001;104:321–324. doi: 10.1016/s0092-8674(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 18.Kusano A, Staber C, Ganetzky B. Dev Cell. 2001;1:351–361. doi: 10.1016/s1534-5807(01)00042-9. [DOI] [PubMed] [Google Scholar]
- 19.Feng W, Benko A L, Lee J-H, Stanford D R, Hopper A K. J Cell Sci. 1999;112:330–347. doi: 10.1242/jcs.112.3.339. [DOI] [PubMed] [Google Scholar]
- 20.Powers P A, Ganetzky B. Genetics. 1991;129:133–144. doi: 10.1093/genetics/129.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Temin R G. Genetics. 1991;128:339–356. doi: 10.1093/genetics/128.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 23.Spradling A C. In: Drosophila: A Practical Approach. Roberts D B, editor. Oxford: IRL; 1986. pp. 175–196. [Google Scholar]
- 24.Hoyle H D, Raff E. J Cell Biol. 1990;111:1009–1026. doi: 10.1083/jcb.111.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoyle H D, Hutchens J A, Turner F R, Raff E C. Dev Genet. 1995;16:148–170. doi: 10.1002/dvg.1020160208. [DOI] [PubMed] [Google Scholar]
- 26.Hillig R C, Renault L, Vetter I R, Drell T, 4th, Wittinghofer A, Becker J. Mol Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]