Abstract
Precancerous cells that enter S phase without appropriate growth and viability factors undergo programmed cell death, suggesting that apoptosis may help guarantee organismic integrity [Evan, G. & Littlewood, T. (1998) Science 281, 1317–1322]. However, the connection between proliferation and cell death has remained unclear. Here, we show that the positive cell cycle regulator cyclin D3 [Matsushime H., Roussel M. F., Ashmun, R. A. & Sherr, C. J. (1991) Cell 65, 701–713] interacts with the death enzyme Caspase 2 [Wang, L., Miura, M., Bergeron, L., Zhu, H. & Yuan, J. (1994) Cell 78, 739–750]. Directed expression of cyclin D3 and Caspase 2 in human cells potentiated apoptosis compared with expression of Caspase 2 alone. Cyclin D3 expression increased the amount of cleaved (active) Caspase 2. We describe a PCR mutagenesis/ligation/two-hybrid/green fluorescent protein approach that facilitates the isolation of missense mutant proteins defective in interaction with particular partners absent other phenotypes or knowledge of the system. We used this approach to isolate Caspase 2 mutants that did not bind cyclin D3 (noninteractors). Noninteractors were sensitive to apoptosis-dependent proteolysis, but did not potentiate apoptosis. Noninteractors did not block apoptosis caused by wild-type Caspase 2. Our results are consistent with the idea that an interaction with cyclin D3 may stabilize Caspase 2, and suggest that a physical interaction between cyclin D3 and Caspase 2 connects the genetic networks that govern cell-cycle progression with those that govern cell death.
Interaction mating two-hybrid methods and coprecipitation mass-spectrometric approaches (1, 2) are now used to find protein–protein interactions on a large scale. The development of these high-throughput methods has made it important to devise ways to rapidly characterize the functional significance of the detected interactions. One way to assess the importance of a particular interaction is to break it with dominant agents such as peptide aptamers (3, 4). Another is to break the interaction by a missense mutation in one of the interacting partners (5). Here, we describe a simple method that combines PCR mutagenesis, PCR ligation, yeast recombination, and two-hybrid analysis to facilitate the isolation of noninteracting missense mutants.
We used this method to study possible connections between two processes, cell proliferation and apoptotic cell death. A number of lines of evidence suggested that cell proliferation and cell death might be coupled. In particular, overexpression of c-Myc, E2F, mutants of human retinoblastoma protein (Rb), and deprivation of growth factors by exposure to low serum (refs. 6–12, and see for example ref. 13) can induce apoptosis in many cell types, whereas wild-type (wt) Rb and inhibitors of cyclin-dependent kinases can block apoptosis (14, 15). Both processes are largely regulated by protein–protein interactions, which prompted us to look for protein interactions that might connect them.
Progress through the cell cycle requires cyclins, positive regulatory proteins that activate cyclin-dependent kinases. During G1, the phase in which the cell may commit to cell division, a number of cyclins participate, including cyclin D3 (16). Cyclin D3 is a member of the cyclin D family, which regulates the initial G1 to S transition by inhibiting Rb and activating E2F proteins (16). Apoptosis requires caspases, a family of cysteine proteases (17). Caspases have been placed into three groups based on their substrate specificity (17). Group I caspases (1, 4, 5, and 13) participate in cytokine maturation and are thought to not directly participate in apoptosis (17). Group II (“executioner”) caspases (2, 3, and 7) cleave substrates directly responsible for causing apoptosis (17). Group III caspases or “activator” caspases (6, 8, 9, and 10) activate both group II and group III caspases (17). Here, we identify an interaction between cyclin D3 and a group II caspase, Caspase 2, generate loss-of-interaction missense mutants of Caspase 2, and we use the mutants to test the significance of the interaction.
Materials and Methods
Interaction Mating.
We introduced, into strain EGY42, which carried the pSH18–34 lexAop-lacZ reporter, bait plasmids expressing human caspases and D-type cyclins, and pEG202, the vector control. We then mated these transformants to EGY48, which carried human caspase preys, D-type cyclin preys, and the pJG4–5 vector control as described (1, 18). We amplified fragments containing Caspase 1, 2, and 3 from a human cDNA library (17) by using Vent polymerase and primer pairs (5′-CCGGAATTCATGGCCGACAAGGTCCTGAAGGAGAAGAGAAAGC-3′, 5′-CCGGTCGACGGATCCTTAATGTCCTGGGAAGAGGTAGAAACATCTTGTC-3′), (5′-CCCCAATTGATGGCCGCTGACAGGGGACGCAGGATATTGG GAG-3′,5′-CCCCTCGAGTCATGTGGGAGGGTGTCCTGGGAACAGGTAGAGG-3′), and (5′-GCGGGGAATTCATGGAGAACACTGAAAACTCAGTG-3′,5′-GCGCCCCTCGAGTTAGTGATAAAAATAGAGTTCTTTTGTG-3′), respectively. We cut the PCR fragments with EcoRI/XhoI, MfeI/XhoI, and EcoRI/XhoI, respectively, and ligated them into prey vector pJG4–5 and bait vector pEG202 cut with EcoRI/XhoI.
Expression of Cyclin D3 in Mammalian Cells and Apoptosis Assay.
We amplified human cyclin D3 by PCR using PfuTurbo polymerase (Stratagene) and primers 5′-ATTGGAATTCGCCACCATGGAGCTGCTGTGTTGCGAAGGC-3′ and 5′-CGCCGCTCGAGCTACAGGTGTATGGCTGTGACATC-3′ from pA7R containing human cyclin D3 (a gift from Andrew Arnold, ref. 19). We cut the PCR fragment with EcoRI/XhoI and introduced it into mammalian expression vector pACL-4 (A. Colman-Lerner, unpublished data, see http://www.molsci.org/∼amendelsohn/acl4.html for construction details) cut with MfeI/XhoI. In the resulting plasmid, the cytomegalovirus (CMV) promoter drives the expression of cyclin D3 and the phosphoglycerate kinase promoter drives the expression of green fluorescent protein (GFP). We used the same Caspase 2–Myc construct and vector control (pcDNA3.1) as in Fig. 2. We then transfected human embryonic kidney (HEK) 293 cells overnight with calcium phosphate by using with 0.5 μg of DNA of each construct for a total of 1 μg of DNA.
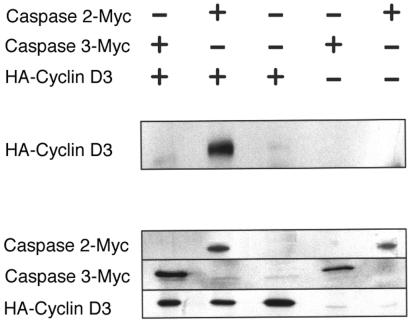
Figure 2.
Caspase 2 interacts with cyclin D3 in mammalian fibroblasts by transfection and immunoprecipitation (IP) Western analysis. IP Western showing that cyclin D3 interacts with Caspase 2 in HEK 293 cells and not with Caspase 2-Myc. (Top) HEK 293 cells transfected with the indicated plasmids. + indicates the plasmid labeled at the right; − indicates the corresponding vector control. (Middle) Cell lysates immunoprecipitated with anti-Myc and immunoblotted with anti-HA. HA-cyclin D3 is detected at 33 kDa. (Bottom) Expression of the corresponding proteins. For Caspase 2-Myc, we immunoprecipitated cell lysates with anti-Myc antibody and probed a blot of the resulting gel with anti-Myc antibody. For Caspase 3-Myc and HA-cyclin D3, we probed the blot with anti-Myc and anti-HA antibody, respectively. Panel shows that cyclin D3 coprecipitates with Caspase 2-Myc, but not with the Myc-tagged vector control, and not with Caspase 3-Myc.
Cyclin D3 and Caspase 2 Expression in Mammalian Cells and Immunoprecipitation Western Assay.
For the transfection, we amplified Caspases 2 and 3, and cyclin D3 with PfuTurbo (Stratagene) PCR using primer pairs (5′-CTCTCAATTGGCCACCATG- GCCGCTGACAGGGGACGCAG-3′,5′-GGCCTCGAGTGTGGGAGGGTGTCCTGGG-3′), (5′-GCGAATTCCCAC- CGATGGAGAACACTGAAAACTCAG-3′,5′-GGCCTCGAGGTGATAAAAATAGAGTTCTTTTGTGAG-3′), and (5′-ATTGGAATTCGCCACCATGGAGCTGCTGTGTTGCGAAGGC-3′ and 5′-CGCCGCTCGAGCTACAGGTGTATGGCTGTGACATC-3′), respectively. We digested the Caspase 2 and 3 fragments with MfeI/XhoI and EcoRI/XhoI respectively and introduced them into the EcoRI/XhoI-cut Myc-tag expression vector pcDNA3.1/Myc-his (Invitrogen) to create (pcDNA3.1-caspase-2). In this vector, the CMV promoter drives the expression of proteins with a Myc tag fused to their C termini. We cut the cyclin D3 fragment with EcoRI/XhoI and introduced it into EcoRI/XhoI-digested pBC147 (20), a vector in which the CMV promoter directs the synthesis of fusion proteins containing an N-terminal fusion of the influenza virus hemagglutinin (HA) tag, to make pBC147-cyclinD3. We purified DNA by two successive CsCl equilibrium gradients and transfected HEK 293 cells overnight with 10 μg of each plasmid, or vector control for a total of 20 μg per transfection, by using the Clontech CalPhos version of the standard calcium phosphate method.
For the immunoprecipitation Westerns, we lysed cells in RIPA buffer (21) and immunoprecipitated them with a 1:100 dilution of anti-Myc 9E10 monoclonal antibody (Santa Cruz Biotechnology) on prewashed protein G Sepharose beads (Sigma). We boiled the beads in SDS sample buffer for 5 min, ran the supernatant on 10% SDS gels, and electrotransferred the proteins on those gels to nitrocellulose membranes. We probed the membranes with a 1:1000 dilution of 12CA5 anti-HA antibody (21) and detected the signal with goat anti-mouse IgG conjugated to horseradish peroxidase as provided by the emission coupled luminescence kit (ECL, Amersham Pharmacia). We performed control Westerns as above and probing them with either a 1:1000 dilution of 9E10 anti-Myc antibody or 1:1000 dilution of the anti-HA antibody.
Western Analysis of Caspase 2 Expression in Cyclin D3 Cotransfected HEK 293 Cells.
We transfected HEK 293 cells with DNA for 24 h with calcium phosphate as described above (for amounts, see Table 1). We lysed cells in RIPA buffer, and then boiled the lysate for 10 min. Western analysis was performed as described above.
Table 1.
Amounts of DNA used for HEK transfection
| Plasmids | DNA, μg
|
||||
|---|---|---|---|---|---|
| Exp. A | Exp. B | Exp. C | Exp. D | Exp. E | |
| pBC147–cyclin D3 | 0 | 18.75 | 0 | 6.25 | 18.75 |
| pcDNA3.1–caspase-2 | 0 | 0 | 6.25 | 6.25 | 6.25 |
| pBC147 | 15 | 0 | 15 | 10.1 | 0 |
| pcDNA 3.1 | 5 | 5 | 0 | 0 | 0 |
| puc18 | 10 | 1.25 | 3.75 | 2.4 | 0 |
Isolation of Noninteractor Mutants of Caspase 2.
We mutagenized Caspase 2 by using Mn2+-biased PCR as described (22, 23) with primers 5′-CTCTCAATTGGCCACCATGGCCGCTGACAGGGGACGCAG-3′ and 5′-GCTCCTCGCCCTTGCTCACCATTGTGGGAGGGTGTCCTGGGAAC-3′ and Taq polymerase for 40 cycles. We amplified enhanced GFP (EGFP) by using PfuTurbo polymerase with 5′-GTTCCCAGGACACCCTCCCACAATGGTGAGCAAGGGCGAGGAGC-3′ and 5′-CTCCGTCGACCTCGAGTTACTTGTACAGCTCGTCCATGC-3′. We fused the mutant Caspase 2 PCR product to the EGFP product by amplification with PfuTurbo polymerase using 5′-CTCTCAATTGGCCACCATGGCCGCTGACAGGGGACGCAG-3′ and 5′-CTCCGTCGACCTCGAGTTACTTGTACAGCTCGTCCATGC-3′ primers. We introduced the resulting PCR product into the plasmid by yeast recombination as above or by introducing MfeI/XhoI-cut fragment into the pJG4–5 (18) by ligation in vitro to create a small library of plasmids that encoded mutant Caspase 2. We introduced library DNA into yeast (EGY48) for two-hybrid analysis. We picked colonies that both appeared near-white on 5-bromo-4-chloro-3-indlyl β-d-galactopyranoside (Xgal) medium, representing presumptive noninteracting mutants, and that fluoresced green, representing missense rather than nonsense mutants in Caspase 2. We sequenced two such plasmids, which contained 11 and 15 point mutations respectively. To identify the critical mutations, we introduced three roughly equal-sized subfragments of the mutagenized ORFs into the wt Caspase 2, introduced those genes into pJG4–5 (see http://www.molsci.org/∼amendelsohn/ caspase-2-methods.html for complete details), and performed two-hybrid analysis on the resulting plasmids. For both plasmids, the middle segment of the Caspase 2 coding region contained the mutations that caused loss of interaction. We introduced individual mutations of this middle segment into wt Caspase 2 by site-directed mutagenesis, and introduced the Caspase 2 gene into pJG4–5 as described above. For mutant Y175N, we used primers 5′-CACACTTCCAGCTGGCGAATAGGTTGCAGTCTCGG-3′ and 5′-CCGAGACTGCAACCTATTCGCCAGCTGGAAGTGTG-3′, and for mutant A242P we used primers 5′-CAAGAGAAACTGCAGAATTTTCCACAGTTACCTGCACACC-3′ and 5′-GGTGTGCAGGTAACTGTGGAAAATTCTGCAGTTTCTCTTG-3′. We repeated the two-hybrid analysis as described and sequenced the positive clones. We identified the resulting mutations as Caspase 2 Y175N (522 T → A) and A242P (724 G → C).
Expression of Mutant Caspase 2 Proteins in Mammalian Cells.
We PCR amplified Caspase 2 carrying Y175N and A242P with 5′-CTCTCAATTGGCCACCATGGCCGCTGACAGGGGACGCAG-3′ and 5′-CTCCGTCGACCTCGAGTCATGTGGGAGGGTGTCCTGGG-3′ and introduced the MfeI/XhoI-cut PCR product into MfeI/XhoI-cut C-terminal myc-tag fusion vector pCDNA3.1.
We transfected HEK 293 cells with 6.25 μg of plasmid DNA containing either wt, Y175N, A242P, or no insert and 6.25 μg of either the HA fusion mammalian expression vector pBC147 or pBC147 driving the expression of wt Caspase 2, as described for Fig. 5. We grew cells for 24 h, and we scraped them from 100 mm dishes at 4°C in 400 μl of RIPA buffer for Western analysis. We added 125 μl of 4× sample lysis buffer (240 mM Tris, pH 6.8/240 mM SDS/40% glycerol/20% 2-mercaptoethanol), and heated at 100°C for 10 min in boiling water. We adjusted samples to equal protein concentrations and ran them on a 12.5% Laemmli SDS gel. After electrotransfer to PVDF [poly(vinylidene fluoride)] membranes, we probed Westerns with a 1:1000 dilution of 9E10 anti-Myc antibody for 1 h and then 1:150,000 of horseradish peroxidase-conjugated anti-mouse IgG for 30 min, and developed the blots with the Pierce Supersignal Western kit.
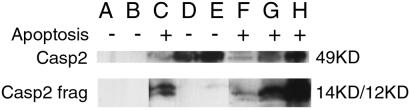
Figure 5.
Mutant Caspase 2s that do not interact with cyclin D3 are expressed at different levels, are not cleaved when expressed in HEK 293 cells, but are cleaved when coexpressed with wt Caspase 2. Immunoprecipitation Western probed with anti-Myc monoclonal antibody showing patterns of cleavage for mutant Caspase 2 in HEK 293 cells transfected with control Myc-fusion vector pcDNA3.1 and HA fusion vector pBC147 (lane A); pcDNA3.1 and pBC147-wt-Caspase 2 (lane B); pcDNA3.1-wt-Caspase 2 + pBC147 (lane C); pcDNA3.1-Y175N-Caspase 2 + pBC147 (lane D); pcDNA 3.1.-A242P-Caspase 2 + pBC147 (lane E); pcDNA3.1-wt-Caspase 2 + pBC147-wt-Caspase 2 (lane F); pcDNA3.1-Y175N-Caspase 2 + pBC147-wt-Caspase 2 (lane G); or pcDNA3.1-A242P-Caspase 2 + pBC-147-wt-Caspase 2 (lane H). + indicates that morphological evidence of apoptosis is present in the respective cell cultures; − indicates no evidence of apoptosis. The uncleaved Caspase 2 band runs at 49 kDa, as visualized in lanes D and E, in which the mutant Caspase 2 was coexpressed with a vector control. The Caspase 2 14 kDa and 12 kDa cleavage products are seen in lanes C (wt), F (wt), G (Y175N), and H (A242P). These bands correlate with the coexpression.
Results
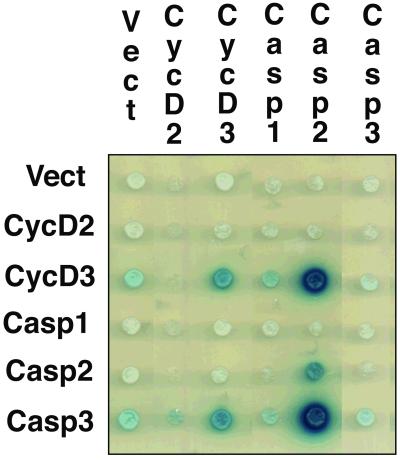
To survey possible connections between the cell cycle and apoptotic pathways, we performed interaction mating two-hybrid experiments (1). We used a panel of 680 defined LexA-fusion “bait” proteins (24) that contained proteins involved in cell proliferation, (including cyclins C, D1, D2, D3, and E, cyclin-dependent kinases 1, 2, 3, 4, 5, and 6, cyclin-dependent kinase inhibitors p16, p19, p21, and p27, and regulatory proteins Rb, p107, and p53), cell death proteins, numerous oncoproteins, and developmental regulatory proteins. We interrogated this panel with “prey” proteins known to be involved in apoptosis, including Caspase 1, Caspase 2, Bcl2, BclXL, Mcl1, and BclXS. Caspases 2 and 3 interacted with cyclin D3, but not with cyclin D2 (Fig. 1).
Figure 1.
Caspase 2 and Caspase 3 interact with cyclin D3 in yeast. Figure shows a modified yeast two-hybrid mating panel. Interactions are indicated by blue color on galactose-induced plates, corresponding to activation of the β-galactosidase reporter. Rows correspond to the baits and columns to the preys. Here, the dark blue color indicates interaction of cyclin D3 bait with Caspase 2 prey and the cyclin D3 prey with the Caspase 3 bait.
We verified that these caspase/cyclin interactions in the yeast nucleus also occurred in mammalian cells. Cyclin D3 activates cyclin-dependent kinases 2, 4, and 6 in the nucleus (25–28). Caspase 2, which was known to be present in the cytoplasm and mitochondria, has relatively recently been shown to reside in the nucleus (29, 30). On the other hand, Caspase 3 is predominately cytoplasmically localized, and we did not know whether it could enter the nucleus of mammalian cells (29, 30) for an interaction with cyclin D3 to occur. We expressed epitope-tagged versions of these proteins in HEK 293 cells. Fig. 2 shows that immunoprecipitation of Caspase 2–Myc fusion coprecipitated N-terminally HA-tagged cyclin D3, as judged by detection of HA-tagged cyclin D3 in the immunoprecipitate on a Western blot. Fig. 2 also shows that, as anticipated, a Caspase 3–Myc fusion was not precipitated with N-terminally HA-tagged cyclin D3.
Forced expression of Caspase 2 induces apoptosis in many cell lines (31). Forced expression causes conversion of inactive pro-Caspase 2 to cleaved active Caspase 2. Although the mechanism for this process is unknown, the extent of apoptosis is apparently proportional to the amount of active Caspase 2 and is cell-type specific (32). We tested whether coexpression of cyclin D3 would enhance Caspase 2-mediated apoptosis. We transfected HEK 293 cells with a construct that directed the synthesis of cyclin D3 from the CMV promoter and a GFP–G418 resistance gene product fusion (GFP-neo) from the mouse phosphoglycerate kinase promoter (pAcl4–cyclin D3), together with a construct that directed expression of Caspase 2-Myc from the CMV promoter (pcDNA3.1–caspase-2). We compared these transfected cells to cells transfected with an empty vector control that expresses GFP-neo (pACL-4) and the vector directing synthesis of Caspase 2 (pcDNA3.1). Dead cells displayed a distinctive small round or blebbed morphology and detached from the plates. About ≈59% of the cyclin D3/Caspase 2 cells exhibiting green fluorescence were dead, compared with ≈38% of the green fluorescing cells transfected with Caspase 2 and the vector control (Table 2). Cyclin D3 overexpression alone had no effect on the induction of apoptosis: only ≈4–5% of green fluorescing cells transfected with cyclin D3 and the empty expression vector (pcDNA3.1) were dead, a number comparable to the background cell death seen when cells were transfected with two control expression vectors. The extent of Caspase 2-dependent apoptosis potentiated by cyclin D3 is likely an underestimate. Coexpression of Caspase 2 and cyclin D3 consistently resulted in fewer cells expressing GFP than any of other combinations, presumably because of the general inhibition of transcription and translation associated with progression to the late stages of apoptosis (33, 34).
Table 2.
Directed synthesis of cyclin D3 and Caspase 2 results in increased cell death compared with directed synthesis of Caspase 2 alone
| Transfection | Percent death (±SD) |
|---|---|
| Cyclin D3/Caspase 2 | 59 (±13) |
| Caspase 2 | 38 (±6) |
| Cyclin D3 | 5 (±2) |
| Control | 4 (±2) |
We transfected HEK 293 cells with constructs directing the expression of cyclin D3 (pACL4-cyclinD3) and Caspase 2 (pcDNA3-caspase-2), Caspase 2 (pcDNA3-caspase-2), and a vector control (pACL-4), cyclin D3 (pACL4-cyclinD3) and a vector control (pcDNA3.1), or two expression vector controls (pcDNA3.1 and pACL4). Percent death is the fraction of GFP-expressing cells that appear dead under phase microscopy in two independent experiments. Green cells correspond to those expressing GFP and either cyclin D3 for pACL4–CyclinD3 or a noncoding insert for control vector pACL-4.
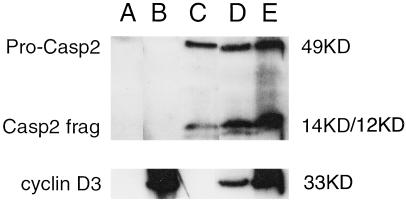
To investigate whether the observed increase in apoptosis caused by cyclin D3 was due to activation of Caspase 2, we coexpressed Caspase 2 and cyclin D3 and examined the extent of Caspase 2 cleavage by Western analysis (Fig. 3). Transfection of equimolar amounts of Caspase 2 and cyclin D3 cDNA-expression vectors resulted in an increase in the amount of cleaved Caspase 2 (Fig. 3, lane D), compared with transfection of the same amount wt Caspase 2 vector and control vector (Fig. 3, lane C). Transfection of a 3-fold molar excess of cyclin D3 with the same amount of Caspase 2 as before resulted in both more cleaved Caspase 2 and more uncleaved pro-Caspase 2 (Fig. 3, lane E). Thus, coexpression of cyclin D3 increased the amount of active Caspase 2. Coexpression of cyclin D3 did not increase the amount of Caspase 2 mRNA detected by quantitative reverse transcriptase–PCR analysis (data not shown). Whenever cyclin D3 was coexpressed with Caspase 2, the ratio of cleaved to uncleaved pro-Caspase 2 increased, these results suggest that cyclin D3 may help promote the cleavage of pro-Caspase 2. Moreover, these facts also suggest that cyclin D3 may stabilize Caspase 2, perhaps by means of a protein–protein interaction.
Figure 3.
Both cleaved (active) Caspase 2 and pro-Caspase 2 are expressed at higher levels when Caspase 2 is cotransfected with increasing amounts of cyclin D3 expression vector. Immunoprecipitation Westerns probed with either anti-Myc monoclonal antibody, showing patterns of cleavage for Caspase 2, or with anti-HA monoclonal antibody showing, cyclin D3 in HEK 293 cells transfected with controls HA-fusion vector pCDNA3.1 and Myc fusion vector pBC147 (lane A); pcDNA3.1 vector + pBC147-cyclin D3 (lane B); pCDNA3.1-Caspase 2 + pBC147 vector (lane C); pcDNA3.1-Caspase 2 + pBC147-cyclin D3 1:1 molar ratio (lane D); or pCDNA3.1-Caspase 2 + pBC147-cyclin D3 1:3 molar ratio (lane E). The same amount of pcDNA3.1-Caspase 2 was used for all transfections involving Caspase 2. Total DNA transfected was the same for all transfections. The uncleaved pro-Caspase 2 band runs at 49 kDa. The Caspase 2 14-kDa and 12-kDa cleavage products are seen in lanes C–E with increasing intensity. The cyclin D3 band appears at 33 kDa.
Isolation of Specific Noninteractors.
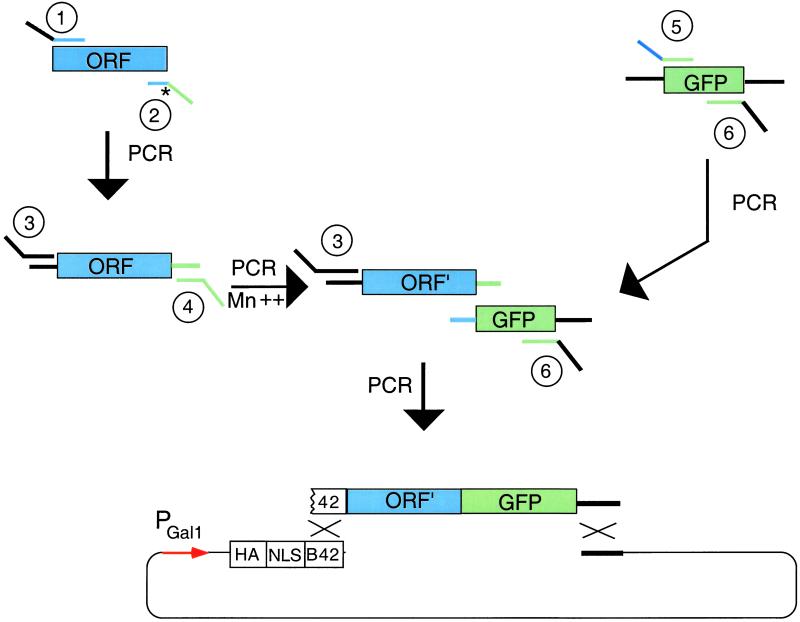
To test whether the cyclin D3-dependent increase in Caspase 2 cleavage and expression was caused by the interaction between Caspase 2 and cyclin D3, we isolated Caspase 2 mutants that no longer interacted with cyclin D3. For this experiment, we devised a general PCR mutagenesis/ligation approach to isolate noninteracting point mutations (Fig. 4). The method works as follows. First, PCR of the ORF (here, Caspase 2) removes a termination codon (*) and places amplifiable sequences upstream and downstream of the target sequence (Fig. 4, upper left). Primer 2 contains the GFP sequence for later PCR ligation. Second, PCR of the ORF with primers 3 and 4 in the presence of Mn2+ mutagenizes the ORF to ORF′ (Fig. 4, middle left). Primer 3 carries partial sequence of the B42 activation domain (see below) (35). Primer 4 (green) carries partial GFP-coding sequence. Third, GFP is amplified by PCR with primers 5 and 6 (Fig. 4, upper right). Primer 5 carries sequence (blue) that overlaps with the ORF and sequence (green) that overlaps with GFP. Primer 6 carries sequences that overlap GFP and the recombination target vector. Fourth, PCR creates a chimeric gene in which the 3′ end of the mutagenized ORF is fused to the GFP-coding sequence (Fig. 4, center). Primers 3 and 6 bear homology to the yeast recombination target, allowing yeast-mediated recombination in vivo.
Figure 4.
General method for isolating noninteracting missense mutants. ORF is PCR-amplified to remove its termination codon, mutagenized by Mn2+-doped PCR, PCR-ligated to GFP, and then introduced into a two-hybrid prey plasmid by recombination in vivo. Colonies of cells that carry missense mutant proteins fluoresce green, but may not interact with specific partner proteins as assayed by two-hybrid analysis.
In this method, we construct mutant recombinant plasmids by one of two means. We use yeast recombination to integrate homologous sequences on the PCR product into the cut vector to generate a prey plasmid. Alternatively, primers 3 and 6 contain restriction enzyme sites, thus allowing the PCR product cut with these enzymes to be introduced into cut vector by ligation in vitro (not shown). Both mutants described in this paper were isolated by using this alternative tactic. It is also possible (A.R.M., unpublished observation) to use primers 3 and 6 that carry sequences recognized by site-specific recombinases, so that the fragment can be introduced by site-specific recombination into appropriately engineered plasmids in vivo or in vitro.
In these experiments, we mutagenized Caspase 2 by PCR in the presence of Mn2+, fused it in frame with GFP derivative EGFP by PCR ligation, and introduced the PCR product into the pJG4–5 prey vector (18). We introduced these plasmids into yeast that contained a cyclin D3 bait and a pSH18–34 lexAop–lacZ reporter (Fig. 4). To isolate noninteractors, we looked for plasmids in which the ORF was uninterrupted (identified by green fluorescence), but in which interaction with the cyclin D3 partner was diminished (identified by lack of blue color on Xgal medium). We selected two mutants for further characterization by this method.
We sequenced the entire Caspase 2 coding region in these mutants. Because the sequencing revealed multiple point mutations (11 and 15 amino acid changes, respectively), we divided the ORFs into three roughly equal-sized restriction fragments, and introduced these segments individually into constructions in which the remainder of the Caspase 2 sequence was unmutagenized and observed their interaction by two-hybrid analysis. We found that for both mutants the middle segment of the coding region carried the mutation(s) that blocked interaction. We then introduced individual point mutations from the middle region into wild-type Caspase 2 by PCR-mediated site-directed mutagenesis (22, 23), and characterized the resulting Caspase 2 mutants by two-hybrid analysis. We identified the noninteractors as two point mutants Y175N (522 t → a) and A242P (724 g → c).
We expressed Y175N and A242P Caspase 2 mutants in HEK 293 cells under the control of the CMV promoter in pcDNA3.1. Expression of these proteins did not induce apoptosis, nor was apoptosis potentiated by coexpression of cyclin D3. Expression of these proteins did not cause any of the cell morphological changes associated with apoptosis. The Y175N protein was not cleaved, whereas A242P protein showed a small amount of cleavage when extracts from transfected cells were examined by Western analysis (Fig. 5, lanes D and E). Coexpression of cyclin D3 with the mutant Caspase 2 proteins had no effect on either their cleavage or their expression (data not shown). In these experiments, Y175N was expressed at levels comparable to wt, whereas A242P was expressed at higher levels than wt. The fact that neither mutant induced apoptosis is consistent with the fact that neither mutant produced large amounts of cleaved Caspase 2.
To determine whether the mutants remained capable of conversion to the active state by cleavage by wt Caspase 2, we coexpressed mutant Caspase 2 proteins with wt Caspase 2. Western analysis showed that both mutants were cleaved in the presence of wt Caspase 2 (Fig. 5, lanes G and H), suggesting that they were recognized as substrates by activated caspases in vivo.
Thus, these observations suggest that these mutations define residues important for Caspase 2 cleavage and activation. Because the mutants do not bind cyclin D3, and because cyclin D3 coexpression with wt Caspase 2 increases the amount of cleaved Caspase 2, these results suggest that physical interaction with cyclin D3 stimulates Caspase 2 cleavage and activation. However, because in the A242P mutant, low-level cleavage occurs in the absence of coexpressed D3, these results suggest that although cyclin D3 stimulates activation, it may not be required for it.
Discussion
Our results show that the cell cycle regulatory protein cyclin D3 interacts with the cell death enzyme Caspase 2 in yeast and in mammalian fibroblasts. Our results further show that cyclin D3 expression sensitizes cells to apoptosis induced by Caspase 2 overexpression by increasing the absolute amount of cleaved Caspase 2 and the amount that is present relative to the uncleaved form. Our results also show that cyclin D3 increases the amount of uncleaved Caspase 2 protein. Furthermore, our results show that Caspase 2 mutants that do not interact with cyclin D3 do not induce apoptosis when overexpressed. Taken together, these facts suggest that cyclin D3 stimulates activation of Caspase 2 by interacting with it.
These experiments demonstrate a physical and functional interaction between a cell cycle positive regulator and an apoptosis effector. How might this interaction potentiate apoptosis? One simple idea is that interaction with cyclin D3 stabilizes Caspase 2 by competing with protease(s) that destroy(s) Caspase 2, perhaps by binding near the sites recognized by the protease. The increased expression of wt Caspase 2 in the presence of cyclin D3 is consistent with this idea. In that view, mutations that abrogate binding of cyclin D3 might also abrogate binding of the destructive protein, which would mean that all noninteracting mutants should be more stable and express at higher levels than wt. Alternatively, if cyclin D3 and the protease(s) bind to overlapping sites, then we would expect some noninteracting mutants to increase stability and others to have no effect. The observations that A242P is expressed at a higher level than wt, and that Y175N is expressed at about the same level as wt, are consistent with this second idea, and suggest that cyclin D3 and the hypothetical protease might bind at an overlapping site on Caspase 2 that includes amino acid 242.
In addition to stabilizing Caspase 2, cyclin D3 expression promotes the cleavage of pro-Caspase 2 (Fig. 3). Potentiation of cleavage may be a consequence of increased expression levels, whereby cyclin D3 is tilting the balance away from the putative destructive protease toward proteases that cleave Caspase 2 into its active form. Alternatively, cyclin D3 might directly participate in the cleavage of pro-Caspase 2, for example by recruiting other apoptotic regulatory proteins (for example cyclin-dependent kinases or proteases), or by localizing Caspase 2 to a permissive cellular compartment, such as the nucleus, where cleavage can occur. Consistent with this latter hypothesis, Caspase 2 localized to the mitochondria remains inactive until released, pro-Caspase 2 microinjected into the cytoplasm is activated (36, 37), and we have been unable to detect Cdk activity in immunoprecipitates of Caspase 2 and associated proteins (not shown).
How might the cyclin D3/Caspase 2 interaction link cell proliferation to apoptosis? Our data are consistent with a two-step model in which cells first respond to a change in their cell cycle regulatory environment, such as activation of an oncogene, such as c-Myc, by releasing proapoptotic factors, such as Cytochrome c, from mitochondria (36, 37) into the cytoplasm. Note that there is evidence that pro-Caspase 2 is released from mitochondria whenever Cytochrome c is released (36, 37). Second, the normal activity of cyclin D3 would help activate Caspase 2. Heightened Caspase 2 activity would overcome the effects of the inhibitory proteins that normally inhibit caspase activity (such as inhibitor of apoptosis proteins; ref. 38), killing the inappropriately proliferating cells and thus protecting the organism.
These experiments confirm the power of yeast interaction mating panels to reveal hitherto unexpected connections between regulatory proteins (1). Perhaps more significantly, they describe a simple PCR mutagenesis/ligation/GFP/two-hybrid/yeast recombination approach to isolate informative missense mutants that are altered in their interaction with any particular protein. This method for isolating specific noninteractors does not require knowledge of the specific organism under genetic investigation, but only of widely used workhorse techniques and reagents, and it is clearly suitable for systematic application on a larger scale. The ability to generate informative missense mutants without knowledge of the specific system complements the recently realized ability to generate protein-based dominant reagents that block specific aspects of protein function (3, 4, 39, 40). We anticipate that both kinds of methods will eventually be used to determine the importance of individual interactions among the wild type and allelic variant products of entire genomes.
It would not be surprising to find similar regulatory interactions among other cyclins and caspases. Cyclin D3 and Caspase 7 interact in two-hybrid experiments (A.R.M., unpublished data), and it is possible that this interaction may also have functional significance. Whatever other connections between proliferation and apoptosis may exist, our current results define a protein–protein interaction in G1 that connects the pathways and may help determine whether proliferating cells live or die.
Acknowledgments
We thank Andrew Arnold for the human cyclin D3 gene, Alejandro Colman-Lerner for pACL-4, and George Gaitanaris for HEK 293 cells. During the early phases of this work, A.R.M. was supported by a postdoctoral fellowship from the American Cancer Society. Work on the interaction mating panel and interaction mutagenesis methods was funded by a National Human Genome Research Institute grant (to R.B.). Work at The Molecular Sciences Institute is funded by National Human Genome Research Institute and Defense Advanced Research Planning Agency.
Abbreviations
- GFP
green fluorescent protein
- EGFP
enhanced GFP
- Xgal
5-bromo-4-chloro-3-indlyl β-d-galactopyranoside
- Rb
human retinoblastoma protein
- CMV
cytomegalovirus
- wt
wild type
- HEK
human embryo kidney
- HA
influenza virus hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Finley R L, Jr, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 3.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 4.Geyer C R, Colman-Lerner A, Brent R. Proc Natl Acad Sci USA. 1999;96:8567–8572. doi: 10.1073/pnas.96.15.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartman P E, Roth J R. Adv Genet. 1973;17:1–105. doi: 10.1016/s0065-2660(08)60170-4. [DOI] [PubMed] [Google Scholar]
- 6.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 7.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, Glynn J M, Guilbert L J, Cotter T G, Bissonnette R P, Green D R. Science. 1992;257:212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Levine A J. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin X Q, Livingston D M, Kaelin W G, Jr, Adams P D. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitaker L L, Hansen M F. Oncogene. 1997;15:1069–1077. doi: 10.1038/sj.onc.1201277. [DOI] [PubMed] [Google Scholar]
- 12.Tiemann F, Hinds P W. EMBO J. 1998;17:1040–1052. doi: 10.1093/emboj/17.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araki S, Simada Y, Kaji K, Hayashi H. Biochem Biophys Res Commun. 1990;172:1081–1085. doi: 10.1016/0006-291x(90)91557-9. [DOI] [PubMed] [Google Scholar]
- 14.Haas-Kogan D A, Kogan S C, Levi D, Dazin P, T'Ang A, Fung Y K, Israel M A. EMBO J. 1995;14:461–472. doi: 10.1002/j.1460-2075.1995.tb07022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki A, Tsutomi Y, Akahane K, Araki T, Miura M. Oncogene. 1998;17:931–939. doi: 10.1038/sj.onc.1202021. [DOI] [PubMed] [Google Scholar]
- 16.Kato J. Front Biosci. 1999;4:D787–D792. doi: 10.2741/kato. [DOI] [PubMed] [Google Scholar]
- 17.Grutter M G. Curr Opin Struct Biol. 2000;10:649–655. doi: 10.1016/s0959-440x(00)00146-9. [DOI] [PubMed] [Google Scholar]
- 18.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 19.Motokura T, Keyomarsi K, Kronenberg H M, Arnold A. J Biol Chem. 1992;267:24412–24415. [PubMed] [Google Scholar]
- 20.Cohen B A, Colas P, Brent R. Proc Natl Acad Sci USA. 1998;95:14272–14277. doi: 10.1073/pnas.95.24.14272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sefton B M. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Struhl K, editors. New York: Wiley; 1996. p. 18.2.5. [Google Scholar]
- 22.Lin-Goerke J L, Robbins D J, Burczak J D. BioTechniques. 1997;23:409–412. doi: 10.2144/97233bm12. [DOI] [PubMed] [Google Scholar]
- 23.Cadwell R C, Joyce G F. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 24.Finley R L, Jr, Brent R. Annu Rev Genet. 1997;31:663–704. doi: 10.1146/annurev.genet.31.1.663. [DOI] [PubMed] [Google Scholar]
- 25.Xiong Y, Zhang H, Beach D. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 26.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 27.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J Y. Mol Cell Biol. 1994;14:2466–2476. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyerson M, Harlow E. Mol Cell Biol. 1994;14:2477–2486. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colussi P A, Harvey N L, Kumar S. J Biol Chem. 1998;273:24535–24542. doi: 10.1074/jbc.273.38.24535. [DOI] [PubMed] [Google Scholar]
- 30.Mancini M, Machamer C E, Roy S, Nicholson D W, Thornberry N A, Casciola-Rosen L A, Rosen A. J Cell Biol. 2000;149:603–612. doi: 10.1083/jcb.149.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Bergeron L, Cryns V, Pasternack M S, Zhu H, Shi L, Greenberg A, Yuan J. J Biol Chem. 1997;272:21010–21017. doi: 10.1074/jbc.272.34.21010. [DOI] [PubMed] [Google Scholar]
- 33.Delic J, Coppey-Moisan M, Magdelenat H. Int J Radiat Biol. 1993;64:39–46. doi: 10.1080/09553009314551091. [DOI] [PubMed] [Google Scholar]
- 34.de Belle I, Testolin L, Pandey S, Carson C, Walker P R, Armato U, Sikorska M. Biochem Cell Biol. 1994;72:639–648. doi: 10.1139/o94-084. [DOI] [PubMed] [Google Scholar]
- 35.Ma J, Ptashne M. Cell. 1988;55:443–446. doi: 10.1016/0092-8674(88)90030-x. [DOI] [PubMed] [Google Scholar]
- 36.Susin S A, Lorenzo H K, Zamzami N, Marzo I, Brenner C, Larochette N, Prevost M C, Alzari P M, Kroemer G. J Exp Med. 1999;189:381–394. doi: 10.1084/jem.189.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uehara T, Kikuchi Y, Nomura Y. J Neurochem. 1999;72:196–245. doi: 10.1046/j.1471-4159.1999.0720196.x. [DOI] [PubMed] [Google Scholar]
- 38.Goyal L. Cell. 2001;104:805–808. doi: 10.1016/s0092-8674(01)00276-8. [DOI] [PubMed] [Google Scholar]
- 39.Geyer C R, Brent R. Methods Enzymol. 2000;328:171–248. doi: 10.1016/s0076-6879(00)28398-5. [DOI] [PubMed] [Google Scholar]
- 40.Colman-Lerner A, Brent R. New Technologies for Life Sciences: A Trends Guide. London: Elsevier; 2000. pp. 56–60. [Google Scholar]