Abstract
The 1.2-kb DNA sequence element (5′HS4) at the 5′ end of the chicken β-globin locus has the two defining properties of an insulator: it prevents an “external” enhancer from acting on a promoter when placed between them (“enhancer blocking”) and acts as a barrier to chromosomal position effect (CPE) when it surrounds a stably integrated reporter. We previously reported that a single CTCF-binding site in 5′HS4 is necessary and sufficient for enhancer blocking. We show here that a 250-bp “core” element from within 5′HS4 is sufficient to confer protection against silencing of transgenes caused by CPE. Further dissection of the core reveals that 5′HS4 is a compound element in which it is possible to separate enhancer blocking and barrier activities. We demonstrate that full protection against CPE is conferred by mutant 5′HS4 sequences from which the CTCF-binding site has been deleted. In contrast, mutations of four other protein binding sites within 5′HS4 result in varying reductions in the ability to protect against CPE. We find that binding sites for CTCF are neither necessary nor sufficient for protection against CPE. Comparison of the properties of 5′HS4 with those of other CTCF-binding enhancer-blocking elements suggests that CPE protection is associated with maintenance of a high level of histone acetylation near the insulator, conferred by insulator binding-proteins other than CTCF.
Keywords: CTCF‖chromatin domain‖boundaries
Insulators are DNA sequence elements that protect transcribed regions from outside regulatory influences. They are present near chromatin domain boundaries or at sites where they prevent inappropriate activation of a promoter by a nearby heterologous enhancer. A growing number of insulators with varied binding sequences and associated proteins have been found in Drosophila, and a few have been found in vertebrates (1–3). Some time ago we identified, at the 5′ end of the chicken β-globin locus, an element (5′HS4) with the characteristic properties of an insulator (4). We applied two defining tests for insulator activity. In the first, the “enhancer-blocking” assay, we placed a 1.2-kb DNA sequence element containing 5′HS4 between an enhancer and promoter and showed that the action of the enhancer was impeded (4–6). This impedance did not occur when the insulator was inserted elsewhere. In the second assay, we showed that when double copies of this 1.2-kb sequence were placed on either side of an erythroid-specific reporter and stably integrated into an erythroid cell line, the expression of the reporter was uniform from one line to another (7). Expression was maintained after 80 days of incubation in the absence of selection. In contrast, expression was variable among uninsulated lines, and in most cases expression of uninsulated lines was extinguished in far less than 80 days. We concluded that this variability of expression and rapid extinction were manifestations of chromosomal position effects arising from the influence on the reporter of dominant regulatory elements flanking the sites of integration (7). The β-globin insulator was capable of acting as a barrier against these position effects, which is a second characteristic of insulator elements. Furthermore, in recent studies we showed that nucleosomes adjacent to the insulator site were highly acetylated in vivo in all cell types (8, 9).
In earlier papers we used the enhancer-blocking assay to analyze the large 1.2-kb insulator element (Fig. 1a). We found first that a 250-bp “core” containing 5′HS4 was effective in this assay (5, 6, 10), and then we further dissected the core element into five footprinted regions (FI–FV) and showed that only one (FII) was necessary for enhancer blocking (5). Purification of FII-binding proteins revealed that binding of the protein CTCF was responsible for its activity (5). Recent studies have shown that CTCF sites with enhancer-blocking properties are present within a number of important gene clusters (3, 5, 11). None of this information told us anything about the second property of the β-globin insulator: the ability to protect against position effects. We did not know whether this protection required the full 1.2-kb element, the 250-bp core, or perhaps only the CTCF-binding site.
Figure 1.
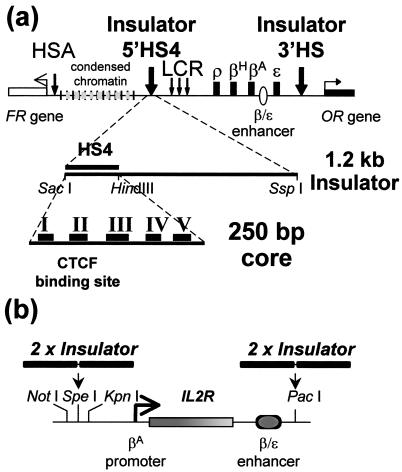
(a) The chicken β-globin domain, the 5′ insulator region, and the neighboring domains. The DNase I-hypersensitive site 5′HS4 (with both enhancer-blocking and barrier properties) marks the 5′ end of the globin locus. Upstream is a 16-kb-long condensed chromatin region, and beyond that is a folate receptor gene (22). At the 3′ end of the β-globin locus is another constitutive hypersensitive site, 3′HS, with enhancer-blocking properties, and beyond that is a gene for an odorant receptor (12). (b) Reporter used in assays for position effects (the pGI′ vector). Plasmids were stably transformed into 6C2 cells, which were analyzed for IL2R expression on the cell surface by flow cytometry (Materials and Methods).
To address this question, we again have begun to dissect the insulator, this time employing the position-effect assay (7). To our surprise, the ability to protect against position effects does not depend at all on the CTCF site. Rather, it seems to arise predominantly from the other footprinted regions within the core, with contributions coming from several independent binding sites for protein factors. Our results show that the chicken β-globin 5′ insulator is a complex element with separable enhancer-blocking and position-effect functions. Furthermore, comparison of the distribution of bound CTCF in vivo and earlier data on histone acetylation patterns makes it clear that acetylation is not correlated with CTCF binding and enhancer blocking but with the DNA sequences associated with barrier function and protection against position effect.
Materials and Methods
Recombinant Plasmid Constructs.
Several unique restriction sites were added to the pGEM-IL2R reporter. Downstream of the β/ɛ enhancer (7), we replaced the SalI site with a PacI restriction site. Upstream of the βA promoter, we introduced a polylinker (oligonucleotides: NSK-top, 5′-CGGCCGCACTAGTGGTAC-3′, and NSK-bot, 5′-CACTAGTGCGGGC-3) containing NotI, SpeI, and KpnI sites between the NarI and KpnI sites. We designated this new construct pGI′. The synthetic oligonucleotides used in PCR to generate FII and FII/FIII deletions in the 1.2-kb insulator fragment are: 1.2ΔFII, forward 5′-ccgttaattaaggGAGCTCACGGGGACAGCCCCCCCCCAAAGCCGCGAGCCGCCCGGGGCTCCG, 1.2ΔFII/III, forward, 5′-ccgttaattaggGAGCTCACGGGGACAGCCCCCCCCCAAAGCCCAGCGTGCGGGGACAGCCCG, and the reverse primer, PacI.2R, 5′-ccgttaattaaggAATATTCTCACTGACTCCGTCCTG. (Lower case letters indicate restriction sites.) For cloning into SpeI, SpeI linkers were ligated to the blunted PCR fragments. For cloning into PacI, PacI linkers were ligated to the blunted PCR fragments. To obtain the constructs pGI′–(1.2ΔFII)2S/P and pGI′–(1.2-ΔFII/III)2S/P, double-tandem copies of each appropriate fragment were cloned first into the SpeI site and then into the PacI site of pGI′. We screened for two copies of each insert by examining digestions of pGI′ with NotI–KpnI on agarose gels. The core insulator was cloned by the same strategy to obtain the pGI′–(CD) for the double core. All the deletions came from the pNI series of constructs (5), and in all the cases except for the FIII site alone the fragments were already cloned in two copies before being transferred to the pGI′ reporter. For 3′HS, the 730 base pairs tested come from the pJC3′HS-2 plasmid (12). The differentially methylated domain (DMD) and the BEAD-1 fragments were created by PCR using human K562 as a source of genomic DNA with the following primers: DMD F, aggcgcgccGGTACCTCGTGGACTCGGACTCCCAAATCA; DMD R, aggcgcgccAAGCTTTGTCACAGCGGACCCCAACCTATG; BEAD ascF, aggcgcgccGAATTCCAGAAATCTTTGATTTCAGATGCT; and BEAD ascR, aggcgcgccGGATCCCACTCTTAGCCATTATACTGCATTG. For the DMD we obtained 1,640-bp fragments and for BEAD-1 1,970-bp fragments. Both fragments containing the AscI site on both ends were cloned into the pNI plasmid (5) and then subcloned with the previous strategy in the SpeI and PacI sites of pGI′ but this time in only one copy on each side. All the final constructs were sequenced by using the primers 5′-CAGATGCGTAAGGAGAAAATACC-3′ and 5′-CTGATAACGGGGCAGGAGGGTGT-3′ located on each side of the new polylinker and the primers 5′-GCCCTTCCCAACAGTTGCGCAG-3′ and 5′-TAATACGACTCACTATAGGGAGA-3′ located on each side of the PacI site of the pGI′ reporter.
Cell Culture and Transformation.
6C2 cells, a preerythroid stable transformed chicken cell line, were grown in αMEM (Invitrogen/GIBCO) supplemented with 10% FCS/2% chicken serum/1 mM Hepes, pH 7.2/50 μM β2-mercaptoethanol/100 units of penicillin/0.1 mg of streptomycin per ml at a density of 1 × 106 cells per ml (13). The cells were transfected by electroporation with 1 μg of pREP7 fragment (see below), which carried the hygromycin resistance gene, and 1 μg of test constructions, linearized at the unique XmnI site interrupting the bla gene of the vector plasmid. This hygromycin resistance and reporter plasmid DNA ratio resulted in ≈50% of stable lines harboring single-copy integrants. The selectable marker plasmid pREP7 has the hygromycin resistance gene under the control of the thymidine kinase promoter (Invitrogen). It was excised for transfection by triple-restriction enzyme digestion (cleaving sites, XbaI, ClaI, and SacI), which removes the Epstein–Barr-virus (EBV) origin of replication and nuclear antigen regions, which are found also on this plasmid. The resulting 5.3-kb DNA fragment was gel-purified by using QiaEx II reagent (Qiagen, Chatsworth, CA).
For transfection, 1 × 107 cells were washed twice with Dulbecco's PBS, resuspended in 0.5 ml of PBS, and mixed with the DNA for electroporation (7). Cells then were plated in 2% methocel (Fluka) containing 2,000 units/ml of hygromycin (Calbiochem). After 2 weeks, individual colonies were picked and expanded in liquid medium containing 1,250 units/ml hygromycin. For extinction experiments the cells were maintained in 3 ml of αMEM medium without hygromycin. Screening for copy number of integrants was carried out by XbaI digestion, electrophoresis, and hybridization with an IL2R cDNA fragment. Single-copy integrant lines were identified with a single hybridization band of >2 kb or as multiple bands for multiple integrants.
Fluorescence-Activated Cell Sorter (FACS) Analysis.
Harvested cells were washed twice in Hanks' balanced salt solution (HBSS) complemented with 0.1% BSA and 0.1% sodium azide (HBSS+). Cells were incubated for 30 min on ice with 1 mg of mouse anti-IL2R monoclonal antibody (Upstate Biotechnology, Lake Placid, NY), washed twice with HBSS+ and a second incubation with 750 ng of FITC-conjugated goat anti-mouse IgG (Roche, Gipf-Oberfrick, Switzerland) for 30 min on ice and washed. Labeled cells were either analyzed by FAST Systems (Gaithersburg, MD) or on a FACScalibur flow cytometer (Becton Dickinson). FACS analysis was performed on days 0, 20, 40, 60, and 80 in absence of selection.
Chromatin Immunoprecipitation for CTCF.
Ten-day chicken erythrocytes were obtained by bleeding 10-day embryos from fertilized White Leghorn chicken eggs (Truslow Farms, Chestertown, MD). Formaldehyde chromatin fixation, purification, and sonication were carried out as described (8). Chromatin immunoprecipitations were carried out with 25 μl of CTCF antibody (obtained from Upstate Biotechnology) with formaldehyde-fixed chromatin from ≈1 × 107 cells. The data for histone H3 acetylated at Lys-9 and Lys-14 in 10-day chicken embryo erythrocytes by using MNase-digested chromatin are from ref. 8, which also describes Taqman probes and quantitative real-time PCR.
Results
The position-effect assay we developed used a human IL2R reporter driven by the chicken βA-globin promoter and the β/ɛ enhancer (refs. 7, 13, and 14; Fig. 1b). This construct was integrated stably into the genome of the chicken immature erythroid 6C2 cell line by cotransfection with the hygromycin resistance gene as a selectable marker (7, 13). Chromosomal position effect, monitored by flow cytometry (FACS) analysis, manifests as variability from line to line in the level of expression in the presence of hygromycin (day 0) and as decreases in expression over time (days 20–80) after removal of hygromycin selection. Both initial variability and time-dependent extinction are prevented if the reporter is surrounded with two copies of a 1.2-kb DNA element containing the chicken β-globin 5′ boundary element (7).
The Core Insulator with Multiple Footprinted Sites Protects Against Position Effect.
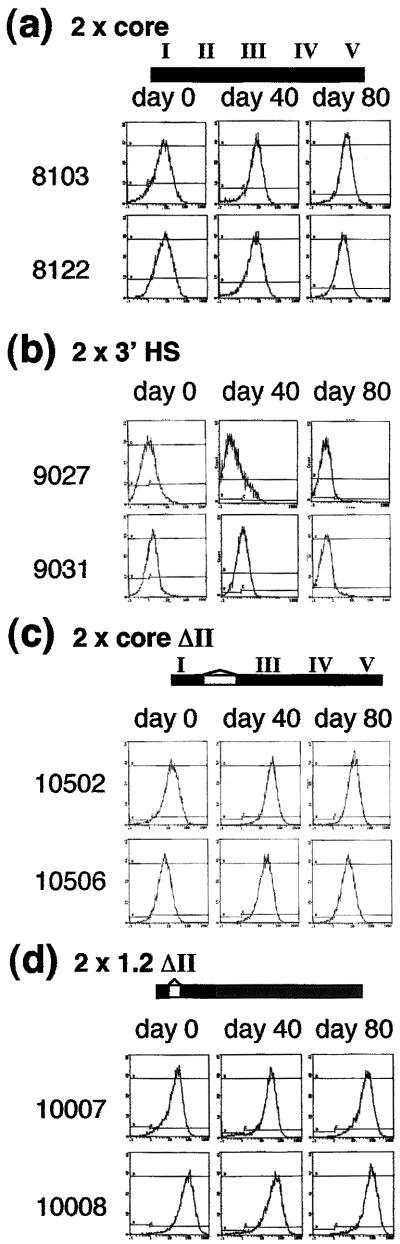
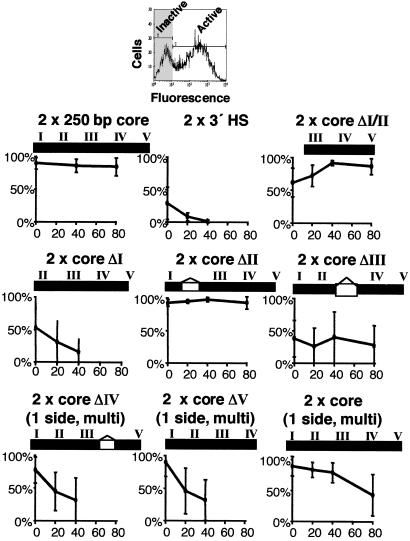
Because previous results had shown that the 250-bp core element was able to function in the enhancer-blocking assay, we began by testing its ability to protect against chromosomal position effects. Two copies of this element, containing footprinted sites FI–FV, were placed on either side of the reporter and integrated stably into 6C2 cells. The core element is as effective as the full 1.2-kb element (compare Fig. 2a with ref. 7). All lines exhibit high levels of recombinant IL2R expression after removal of hygromycin selection. Expression levels remain the same for at least 80 days in culture (see Figs. 6–21, which are published as supporting information on the PNAS web site, www.pnas.org, for expression data from all the lines tested).
Figure 2.
CTCF-binding sites are neither necessary nor sufficient for barrier activity. Two copies of test fragments were placed on each side of the IL2R reporter and stably integrated into 6C2 cells. Results of FACS analyses for IL2R expression at 0, 40, and 80 days after removal of hygromycin selection are shown; number of cells (y) versus intensity of antibody staining (x). A representative number of single-copy lines (numbered to the left of each set of panels) are shown unless otherwise stated. Data from all lines can be found online in Figs. 6–21. (a) A 250-bp core of the 5′HS4 insulator has full barrier activity. All lines express high levels of reporter IL2R at day 0; expression levels are maintained for at least 80 days in culture. The data are representative of the nine single-copy and eight multicopy lines tested (Figs. 6 and 7). (b) A 730-bp fragment of the 3′HS enhancer-blocking element does not have barrier activity. All lines (Fig. 8) lose recombinant IL2R expression by 20–40 days in culture. The lines shown are representative of the five single-copy and five multicopy lines tested (Fig. 8). (c and d) Deletion of the single binding site (FII) for CTCF has no effect on barrier activity of the 250-bp core or full 1.2-kb 5′HS4 insulators, respectively. The data are representative of the 10 single-copy and 12 multicopy core ΔII (Figs. 9 and 10) and the six single-copy and six multicopy 1.2ΔII lines tested (Figs. 11 and 12).
Other Enhancer-Blocking Elements Fail To Protect Against Position Effect.
Other boundary elements have been reported in the literature. We examined the DMD of the mouse Igf2 and H19 locus (15, 16) and the BEAD element of the human T-cell receptor α/δ locus (17). Both of these have been shown to possess enhancer-blocking activity and bind the protein CTCF (12, 18). Reporters were flanked with either one copy of a 1,604-bp DMD fragment or one copy of a 1,970-bp BEAD fragment. Neither of these elements is able to protect the IL2R reporter from chromosomal position effect (data not shown).
CTCF-Binding Sites Are Neither Necessary Nor Sufficient for Chromosomal Position-Effect Protection.
We next undertook a methodical dissection of the 250-bp core element. Because the CTCF-binding site FII had proven central to the enhancer-blocking activity of the core (5), we began by using a core element from which FII had been deleted. To our surprise (Fig. 2c) this deletion had no effect on barrier activity. We found that the same was true when FII was deleted from the full 1.2-kb insulator element (Fig. 2d). Because FII is the binding site for CTCF, these results indicate that CTCF does not play a role in barrier function.
We previously described the very strong enhancer-blocking properties of an element named (II/III)Q [also called (II/III-Ins)Q], containing four copies of a 90-bp sequence carrying footprint regions II and III (5, 6). The (II/III)Q element does not possess barrier activity (data not shown). We found that 90% of the lines carrying (II/III)Q were repressed for IL2R expression at day 0, and all were inactive by day 20 (data not shown). The rate of inactivation observed is more rapid than that for an uninsulated reporter, suggesting that the (II/III)Q element harbors silencing activity. Another sequence element marked by a DNase I-hypersensitive site and possessing enhancer-blocking properties has been found at the 3′ end of the β-globin locus (12). In contrast to the results with the 5′HS4 insulator element, two copies of a 730-bp 3′HS fragment were incapable of conferring any barrier to chromosomal position effect (Fig. 2b). It should be noted that although the 3′ sequence has a binding site for CTCF resembling that of FII, it does not appear to contain any of the motifs associated with binding to FI or FIII–FV. These results thus are consistent with the conclusion that CTCF does not play a role in barrier activity.
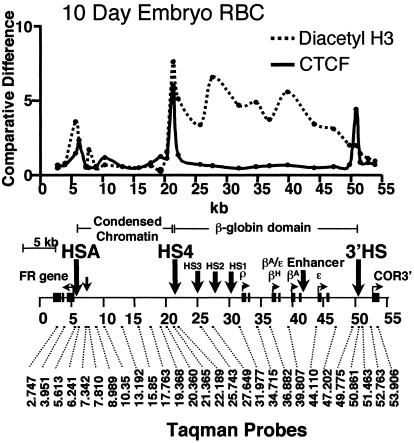
We wanted to understand better the relationship between CTCF binding, histone modifications, and barrier activity. Although FII has been shown to bind CTCF in vitro, there has been no direct evidence for its presence at the endogenous globin locus in vivo. We therefore carried out a chromatin immunoprecipitation analysis for the presence of CTCF across the entire β-globin locus in 10-day chicken embryonic erythrocytes. As shown in Fig. 3, CTCF is present both at the 5′HS4 insulator site and at the 3′HS enhancer-blocking site at the other end of the β-globin locus in vivo. When we compare the CTCF-binding pattern with that of histone acetylation over the β-globin locus (Fig. 3), we note that a peak of acetylation is present over the 5′HS4 insulator but not over the 3′HS region marked by the second CTCF site. Thus we tentatively attribute the acetylation peak to some or all of the proteins bound at FI and FIII–V and not to the presence of CTCF, which may provide a clue to the mechanism of barrier action at the β-globin locus (see Discussion).
Figure 3.
A comparison of enrichment for CTCF and histone H3 acetylated on Lys-9 and Lys-14 (K9 and K14) at locations across the chicken β-globin locus in 10-day chicken erythrocytes after chromatin immunoprecipitation and quantitative PCR analysis (see Materials and Methods). Locations of PCR primer sets are shown below on a map of the chicken β-globin neighborhood drawn to approximate scale. The data from chromatin preparations using anti-CTCF (solid line) or anti-acetyl K9 and K14 histone H3 (dotted line) antibodies are shown. The differences in DNA site enrichment represent the ratio of the immunoprecipitated DNA divided by the input DNA with all points for anti-acetyl K9 and K14 histone H3 divided by two for better comparison
Dissection of the Core Insulator.
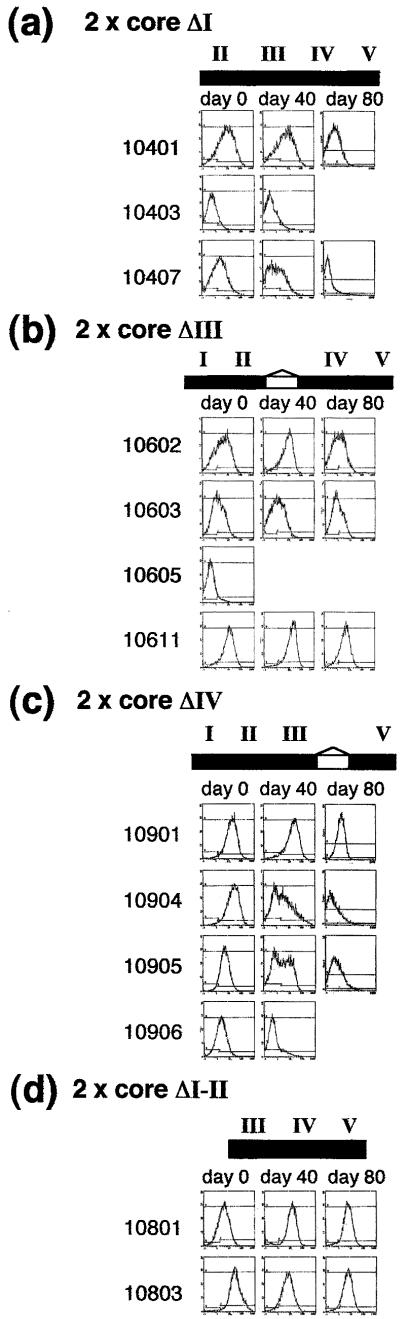
We next deleted each of the other binding sites within the core. Deletions of FI, FIII, FIV, or FV did cause a decrease in the barrier activity of the 5′HS4 core insulator (Figs. 4 and 19). Deletion of FIII in combination with FII also caused a decrease in barrier activity of the full 1.2-kb 5′HS4 insulator (Figs. 13 and 14). Results for core ΔFV were similar to those for ΔFIV (Fig. 10 and 19). Although single footprint deletions all had deleterious effects, the deletion of FI and FII together (core ΔI–II), leaving only FIII, IV, and V, resulted in an element capable of effective chromosomal position-effect protection (Fig. 4d). Thus the contributions of individual elements are not necessarily additive.
Figure 4.
Deletion of binding sites for proteins other than CTCF disrupt barrier activity of the core insulator. The results of FACS analyses for recombinant IL2R expression at 0, 20, 40, and 80 days after removal of hygromycin selection are shown as described for Fig. 2. (a) Deletion of the FI protein-binding site disrupts barrier activity. The lines show variable expression at day 0. Most lines lose IL2R expression by day 40. The data are representative of three single-copy and nine multicopy lines tested (Fig. 15). (b) Deletion of the FIII protein-binding site disrupts barrier activity. All lines show variable expression at day 0 but maintain levels of recombinant IL2R expression at least to day 80. The data are representative of the 12 single-copy and eight multicopy lines tested (Figs. 16 and 17). (c) Deletion of the FIV protein-binding site disrupts barrier activity. All multicopy lines display high levels of recombinant IL2R expression at day 0 but become silenced with time in culture. The data are representative of the 10 multicopy lines tested (Fig. 18). Deletion of the FV protein-binding site had a similar affect on the activity of the core (10 multicopy lines, Fig. 19). (d) The FI and FII protein-binding sites together are dispensable for barrier activity. A minimal fragment containing only FIII, IV, and V has near full barrier activity. All lines maintain high levels of expression for at least 40 days. Some extinction is observed in multicopy lines after 80 days. The data shown are representative of the nine single-copy and 10 multicopy lines tested (Figs. 20 and 21).
The ΔFIV or ΔFV constructions contained double copies of the mutant insulator element only on one side of the IL2R reporter. For that reason we studied only lines containing multiple-copy inserts so that with the exception of an end element each reporter is still surrounded by double copies. This approach was validated by control experiments (Fig. 5) carried out with double copies of the wild-type core only on one side, which showed that expression in multicopy lines was largely maintained after 20 days in culture. Although there were small decreases in expression levels in a few lines, the behavior is quite different from the rapid loss of expression seen with the ΔFIV or ΔFV constructions.
Figure 5.
Summary of the barrier activities of wild-type and mutant insulator fragments. (Upper) A selected line that exhibits a biphasic expression pattern. Cells not expressing recombinant IL2R (shaded gray) are gated based on a control where no secondary antibody was used. (Lower) Percentages of cells expressing IL2R from all lines were averaged at each time point and plotted. Raw FACS plots used for this quantitation are shown in Figs. 6–21.
The data for these experiments are summarized in Fig. 5, which makes it clear that although each of these deletions decreases protection, there are differences among them. Most single-copy lines carrying mutant ΔFI insulators are silent at day 0, and all expressing lines rapidly become extinct. The ΔFIII mutants also exhibit variable expression levels, mostly low, at day 0. In contrast to the ΔFI mutants, however, expression levels of the ΔFIII lines persist to day 80. Conversely, the ΔFIV or ΔFV multicopy mutants are mostly active at day 0 but rapidly extinguish to become largely inactive by 20 days, with a further increase in the inactive lines by day 40.
Discussion
The insulator element at the 5′ end of the chicken β-globin locus shares with many insulators identified in Drosophila two properties: positional enhancer-blocking activity and the ability to act as a barrier against chromosomal position effect. This element was identified originally and studied as a 1.2-kb DNA fragment containing the constitutive hypersensitive site HS4. Later work focusing on enhancer-blocking activity showed that a 250-bp core element containing the hypersensitive site is fully functional, and that a single binding site for the protein CTCF, FII, is necessary and sufficient to block the action of an enhancer on a promoter when placed between them (5, 6, 10).
We repeated this approach in attempting to dissect the elements in the 1.2-kb insulator responsible for barrier activity. We found that, as in the earlier assay, we could confer complete position-effect protection with two copies of the core element on either side of the reporter, but the CTCF-binding site FII was dispensable for this barrier activity. More directly, we found that a single copy of a CTCF-binding site, present in the 3′HS element for example, seems to contribute neither silencing nor barrier activity. Furthermore, the (II/III)Q construction seems to silence expression relative to what is observed with an uninsulated reporter (see below). It thus became clear that barrier activity must involve other sequence elements within the core. A series of deletions based on information from the DNase I footprinting patterns over the element showed that individual removal of FI, FIII, FIV, or FV affected barrier activity in somewhat different ways; multiple deletions revealed that at least some of the contributions from individual elements can be affected by their neighbors such that there is interdependence between the elements.
The effects of these deletions fall into three classes: (i) FI deletion causes both variable expression at day 0 and rapid shutdown over time, (ii) single deletions of FIV or V have less effect at day 0 but reduce protection against extinction over time, and (iii) deletion of FIII on the other hand causes variability of expression at day 0, but its removal has little effect on the rate of long-term extinction when FI, FIV, and FV are present. This result suggests that there may be separate mechanisms affecting barrier action against initial silencing, mediated by FIII, and long-term repression, conferred by the other footprinted regions, which in part may explain the necessity for a compound insulator element.
More complicated results are obtained with double deletions. For example FIII, FIV, and FV together are sufficient to afford complete protection against silencing. Because the mutants with FI alone deleted are impaired in barrier activity, it is possible that the FI regions may counteract the suppressive effects of the double copies of FII on each side of the reporter. The protein bound to FII is CTCF, which has been reported in some contexts to interact with Sin3A, known to recruit a histone deacetylase that might contribute to silencing (21). It is also known (Fig. 3; ref. 8; see below) that the sequences surrounding the endogenous 5′HS4 are a site of very strong histone acetylation in chicken cells. Perhaps this is the basis of the counteracting effect of FI on FII.
Other loci have been identified in which only the CTCF site is present. These sites have enhancer-blocking activity, but the additional elements necessary for barrier activity appear to be absent (12, 17, 18, 20). Why is the β-globin 5′HS4 insulator more complex? One major difference in its environment may be the presence of ≈16 kb of condensed chromatin immediately upstream, marked by high levels of CpG DNA methylation (22) and constitutively low levels of histone acetylation (8). The data presented in Fig. 3 show that there are high concentrations of CTCF both at 5′HS4 and at the 3′ end of the β-globin domain in vivo. In contrast, as also shown, there is a peak of constitutive histone acetylation only at 5′HS4, which indicates that CTCF is not responsible for the maintenance of high acetylation levels and strongly suggests that the other components of 5′HS4, lacking at the 3′ end, are the source of the acetylation signals. These signals thus may be connected with barrier activity. We showed recently that there is a high level of histone H3 Lys-9 methylation over the 16-kb condensed chromatin region (9), and we suggest that high acetylation levels at HS4 prevent the further advance of this modification and consequently the encroachment of condensed chromatin on the active globin locus. Although CTCF is responsible for enhancer-blocking activity, other proteins bound to HS4 may serve to recruit histone acetyltransferases that give rise to barrier activity (see ref. 9). We note that such protection may not be necessary at the 3′ end of the chicken β-globin locus, which is not located near the same kind of constitutive condensed chromatin (12).
Our results have the practical consequence of reducing to two copies of the 250-bp core the size of the minimum sequence required for complete barrier action (Fig. 2). Other recent studies have exploited the properties of the core element in experiments involving adeno-associated virus (AAV) vectors (23). Earlier work in a number of laboratories has demonstrated the usefulness of the full 1.2-kb β-globin insulator element in protecting reporters from chromosomal position effect in many transgenic animal (24–28) and cell (7, 29–31) lines. Both plasmid and retroviral vectors have been used in these experiments. The use of the much smaller core element should simplify modification of such vectors and make possible some constructions where the total length of the insert must be limited. The data presented here suggest that eventually it may be possible to reduce the size of the effective element even further.
Supplementary Material
Abbreviations
- FACS
fluorescence-activated cell sorter
- DMD
differentially methylated domain
References
- 1.Gerasimova T I, Gdula D A, Gerasimov D V, Simonova O, Corces V G. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 2.Bell A, Boyes J, Chung J, Pikaart M, Prioleau M-N, Recillas F, Saitoh N, Felsenfeld G. Cold Spring Harbor Symp Quant Biol. 1998;63:509–514. doi: 10.1101/sqb.1998.63.509. [DOI] [PubMed] [Google Scholar]
- 3.Bell A C, West A G, Felsenfeld G. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 4.Chung J H, Whiteley M, Felsenfeld G. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 5.Bell A, West A G, Felsenfeld G. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 6.Recillas-Targa F, Bell A C, Felsenfeld G. Proc Natl Acad Sci USA. 1999;96:14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pikaart M J, Recillas-Targa F R, Felsenfeld G. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litt M D, Simpson M, Recillas-Targa F, Prioleau M-N, Felsenfeld G. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litt M D, Simpson M, Gaszner M, Allis C D, Felsenfeld G. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 10.Chung J H, Bell A C, Felsenfeld G. Proc Natl Acad Sci USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filippova G N, Thienes C-P, Penn B-H, Cho D H, Hu Y J, Moore J M, Klesert T R, Lobanenkov V V, Tapscott S J. Nat Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 12.Saitoh N, Bell A C, Recillas-Targa F, West A G, Simpson M, Pikaart M, Felsenfeld G. EMBO J. 1999;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyes J, Felsenfeld G. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 14.Reitman M, Felsenfeld G. Proc Natl Acad Sci USA. 1988;85:6267–6271. doi: 10.1073/pnas.85.17.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorvaldsen J L, Duran K L, Bartolomei M S. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hark A T, Tilghman S M. Nucleic Acids Res. 1998;12:1979–1985. [Google Scholar]
- 17.Zhong X P, Krangel M S. Proc Natl Acad Sci USA. 1997;94:5219–5224. doi: 10.1073/pnas.94.10.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. Nature (London) 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 19.Bell A C, Felsenfeld G. Nature (London) 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 20.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qui C F, Wolffe A, Ohlsson R, Lobanenkov V V. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 21.Lutz M, Burke L J, Barreto G, Goeman F, Greb H, Arnold R, Schultheiss H, Brehm A, Kouzarides T, Lobanenkov V, Renkawitz R. Nucleic Acids Res. 2000;28:1707–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prioleau M-N, Nony P, Simpson M, Felsenfeld G. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue T, Yamaza H, Sakai Y, Mizuno S-I, Ohno M, Hamasaki N, Fukumaki Y. J Hum Genet. 1999;44:152–162. doi: 10.1007/s100380050133. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, DeMayo F J, Tsai S-Y, O'Malley B W. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- 25.Potts W, Tucker D, Wood H, Martin C. Biochem Biophys Res Commun. 2000;273:1015–1018. doi: 10.1006/bbrc.2000.3013. [DOI] [PubMed] [Google Scholar]
- 26.Boeda B, Weil D, Petit C. Hum Mol Genet. 2001;10:1581–1589. doi: 10.1093/hmg/10.15.1581. [DOI] [PubMed] [Google Scholar]
- 27.Ciana P, Di Luccio G, Belcredito S, Pollio G, Vegeto E, Tatangelo L, Tiveron C, Maggi A. Mol Endocrinol. 2001;15:1104–1113. doi: 10.1210/mend.15.7.0658. [DOI] [PubMed] [Google Scholar]
- 28.Taboit-Dameron F, Malassagne B, Viglietta C, Puissant C, Leroux-Coyau M, Chereau C, Attal J, Weill B, Houdebine L M. Transgenic Res. 1999;8:223–235. doi: 10.1023/a:1008919925303. [DOI] [PubMed] [Google Scholar]
- 29.Rivella S, Callegari J A, May C, Tan C W, Sadelain M. J Virol. 2000;74:4679–4687. doi: 10.1128/jvi.74.10.4679-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emery D W, Yannaki E, Tubb J, Stamatoyannopoulos G. Proc Natl Acad Sci USA. 2000;97:9150–9155. doi: 10.1073/pnas.160159597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinwaerder D S, Lieber A. Gene Ther. 2001;7:556–567. doi: 10.1038/sj.gt.3301139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.