Abstract
Double-stranded RNA (dsRNA) triggers homology-dependent posttranscriptional gene interference (RNAi) in a diverse range of eukaryotic organisms, in a process mechanistically related to viral and transgene-mediated cosuppression. RNAi is characterized by the conversion of long dsRNA into ≈21–25-nt small interfering RNAs (siRNA) that guide the degradation of homologous mRNA. Many of the genes required for siRNA production and target mRNA degradation are widely conserved. Notably, members of the Argonaute-like gene family from Arabidopsis, Caenorhabditis elegans, Drosophila, and Neurospora have been genetically and/or biochemically identified as components of the RNAi/cosuppression pathway. We show here that mutations in the Drosophila Argonaute1 (AGO1) gene suppress RNAi in embryos. This defect corresponds to a reduced ability to degrade mRNA in response to dsRNA in vitro. Furthermore, AGO1 is not required for siRNA production in vitro nor can the introduction of siRNA bypass AGO1 mutants in vivo. These data suggest that AGO1 functions downstream of siRNA production.
Many species, across a wide phylogenetic range, respond to aberrant/foreign RNA by degrading endogenous mRNA in a sequence-specific manner (1). This phenomenon, broadly referred to as posttranscriptional gene silencing (PTGS), can be triggered by the introduction of double-stranded RNA (dsRNA) [RNA interference (RNAi)], transformation with sense transgenes (cosuppression/quelling), or viral infection (2). RNAi acts as a cellular defense against parasitic nucleic acids and provides a fortuitous technique for biologists to reduce or eliminate a gene activity (3). RNAi-like mechanisms are also involved in the production of small noncoding RNAs that control developmental timing (4, 5). A better understanding of RNAi may then shed light on genome defense and endogenous developmental pathways.
The molecular mechanisms underlying RNAi are beginning to be elucidated. dsRNA is processed into small double-stranded fragments of 21–25 nucleotides, called small interfering RNA (siRNA; refs. 6–8), by the enzyme Dicer (9–11). These siRNA are then incorporated into a protein complex, called RNA-induced silencing complex (RISC), which degrades homologous mRNA (8, 12). The siRNA signal can also be amplified, possibly by an RNA-dependent RNA polymerase (13, 14). siRNA seems to be the crucial mediator of sequence specificity targeting mRNA for degradation (6, 15–17).
RNAi seems to be an ancient and evolutionarily conserved process. The Argonaute gene family was first defined by the Arabidopsis Argonaute gene (AGO1; ref. 18) and encodes related proteins of unknown molecular function in plants, animals, and fungi. Members of the Argonaute gene family, including rde-1 in Caenorhabditis elegans (19), qde-2 in Neurospora (20), AGO1 in Arabidopsis (21), and piwi in Drosophila (22), are genetically defined as being required for PTGS. The Drosophila Argonaute2 (AGO2) protein was biochemically identified as a component of the RISC complex (23). Thus far, AGO2 is the only identified RISC component. The conservation of genes involved in PTGS extends beyond members of the Argonaute family. The Dicer enzyme, whose activity was identified in Drosophila (9), has homologs in plants (24), C. elegans (4, 10, 11), and mammals (5). Also, RNA-dependent RNA polymerases have been implicated in PTGS in plants (25), Neurospora (26), and C. elegans (27). Thus, homologous proteins may have similar functions in PTGS throughout a diverse range of eukaryotic organisms.
Based on the evolutionary conservation of genes involved in PTGS, we have begun to address the roles of members of the Drosophila Argonaute gene family. We have identified five Argonaute-like genes encoded in the Drosophila genome. For comparison, the C. elegans genome encodes 23 Argonaute-like genes. Three of the Drosophila genes are maternally expressed, but zygotic expression in the embryo is limited to the presumptive gonad. Thus, they likely have tissue-specific functions. The other two genes, Argonaute1 (AGO1) and AGO2, are expressed broadly throughout embryonic development and are better candidates to function in RNAi in embryos. We chose to examine the role AGO1 plays in RNAi in the Drosophila embryo. We show that AGO1 mutant embryos have a reduced RNAi response when injected with either long dsRNA or siRNA. We also show that AGO1 is required for the degradation of targeted mRNA in vitro but is not required for the Dicer-mediated cleavage of dsRNA. We propose that AGO1 functions in RNAi and, specifically, functions downstream of siRNA production.
Materials and Methods
Drosophila Strains.
Flies were maintained with standard procedures. P element insertions in AGO1, l(2)k08121, and l(2)k16601 were each out-crossed to w1118 and isogenized. The lethality of l(2)k08121 was reverted by precise excision of the P element. In addition, the lethality of l(2)k08121 was rescued in a strain containing heat shock Gal4 and UAS-AGO1 transgenes (28). The lethality of the strain, therefore, is caused by the insertion of the P element in AGO1. All lethal mutations were balanced over a CyO chromosome carrying an armadillo (arm)-GFP transgene that is expressed throughout the embryo.
RNA Synthesis and Injections.
For dsRNA production, regions of the even-skipped (eve), fushi tarazu (ftz), and white genes were amplified with oligonucleotides that add 5′ T7 promoter sequences (available on request) by using pEve, pGEM F1 (29), and pUAST (28) as templates, respectively. The resulting PCR products were each used as templates for in vitro transcription reactions by using the Ambion (Austin, TX) MEGAscript kit following the manufacturer's instructions with or without the addition of 20 μCi of [α-32P]UTP (Amersham Pharmacia Biotech). The reactions were boiled and allowed to anneal >12 h at room temperature. The resulting RNA was analyzed by agarose gel electrophoresis to verify that it was double stranded and of the appropriate size. dsRNA (21 bp) with 2 base 3′ overhangs (siRNA) corresponding to the eve gene (siEVE-2 AACUCCUUGAACGGCAGCCGC) was purchased from Dharmacon (Lafayette, CO). The siRNA was 5′-phosphorylated with T4 polynucleotide kinase (NEB, Beverly, MA). Approximately 100 pl of dsRNA or siRNA (1 μg/μl in H2O) was injected into syncitial blastoderm embryos. After 40–48 h at 18°C, the surviving embryos were scored for the number of ventral denticle belts. For consistency, any embryo with 7 or 8 visible denticle belts was scored as wild type and embryos with 4, 5, or 6 belts were scored as eve. The number of eve embryos may be under-represented, because embryos with a weak phenotype (i.e., only missing a single denticle belt) would have been scored as wild type.
For mRNA production, regions of the eve and white genes were amplified with a forward oligonucleotide with a 5′ T7 transcription start and a reverse primer that adds 25 A residues to the 3′ end. The PCR products were then used as templates for in vitro transcription reactions with the Ambion mMessage kit plus the addition of 20 μCi of [α-32P]UTP. The RNA products were verified by agarose gel electrophoresis.
In situ probes were generated by in vitro transcription with a linearized expressed sequence tag clone as a template. Embryo collections, hybridizations, and detection were performed as described by the Berkeley Drosophila Genome Project (www.fruitfly.org/about/methods/RNAinsitu.html) with minor modifications.
Extract Preparation and in Vitro RNAi Reactions.
Extracts used for in vitro RNAi reactions were prepared as described in Tuschl et al. (12) and Zamore et al. (7) with some modifications. In short, embryos were collected for 4 h and then aged for an additional 12 h at 25°C to allow for zygotic expression of the arm-GFP transgene on the CyO balancer chromosome. Homozygous AGO1k08121 embryos were sorted from their AGO1k08121/CyO-armGFP and CyO-armGFP/CyO-armGFP siblings following the procedure of Furlong et al. (30). Approximately 6,000 embryos were homogenized in 2 ml of lysis buffer (100 mM potassium acetate/30 mM Hepes, pH 7.4/2 mM magnesium acetate/5 mM DTT/0.5 mg/ml Pefabloc SC) with a glass tissue grinder, centrifuged for 25 min at 14,500 × g at 4°C, and flash-frozen in 10-μl aliquots. The total protein concentration in the extracts was determined with a colorimetric Bio-Rad assay.
The cleavage of dsRNA was assessed by incubating 200 ng of labeled dsRNA in a 20-μl reaction containing 5 μg of extract protein for increasing times at 25°C. Reactions were stopped by the addition of formamide tracking dye. Sequence-specific degradation of mRNA was assessed by adding 200 ng of unlabeled dsRNA to a 20-μl reaction containing 5 μg of extract protein. After a 10-min incubation at 25°C, 5 ng of labeled mRNA was added. The reactions were stopped following the procedure of Tuschl et al. (12). All reactions were analyzed on a 12% denaturing polyacrylamide gel followed by autoradiography. RNA molecular weight markers were made by T1 RNase (Ambion) digestion of labeled eve RNA.
Results
The Argonaute Gene Family in Drosophila.
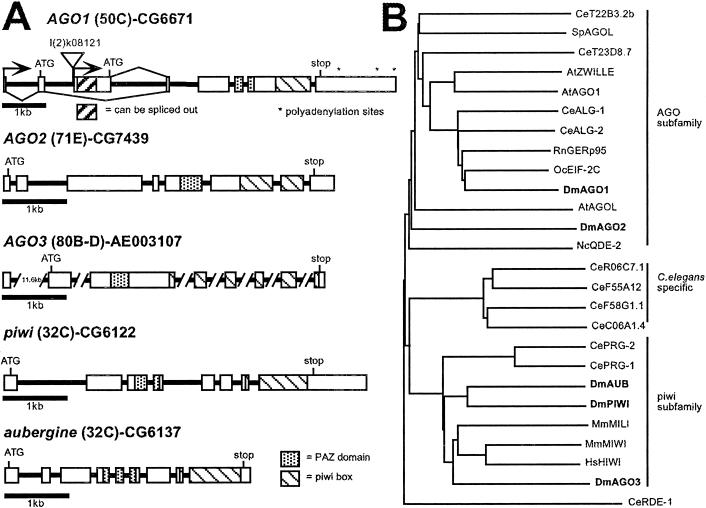
Searches of the Drosophila genome and expressed sequence tag (EST) databases reveal five separate transcribed members of the Argonaute gene family (Fig. 1A). Argonaute-like genes are defined as encoding two conserved domains: a well conserved region of approximately 300 amino acids, called the PIWI box, near the C-terminal region of the ORF and a more centrally located and less well conserved region of 110 amino acids called the PAZ domain (ref. 31; Fig. 1A). The molecular functions of Argonaute-like proteins are not known; however, two of the genes, piwi and aubergine (aub)/sting, are necessary for proper germ-line development (32–35). AGO1 is required for viability and embryonic neural growth (36). The AGO2 protein has been implicated in the degradation of target mRNA in response to dsRNA (23). A fifth member, Argonaute3 (AGO3), has not yet been characterized (Fig. 1A). Additional information on the EST clones can be found in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
Figure 1.
The Argonaute gene family. (A) Genomic organization and cytological location of the five Drosophila Argonaute-like genes. Exons are shown as boxes and introns are shown as solid lines. The PAZ and PIWI domains are defined by Cerutti et al. (31). The AGO1 gene has two transcriptional start sites, resulting in two different ORFs differing in the first 65 amino acids. The AGO1 P element insertion l(2)k08121 is shown. l(2)k16601, an independent insertion in AGO1, is located 8 base pairs downstream from the l(2)k08121 insertion site. AGO3 is located near the centromere at 80B-D. Only one of the seven introns has been completely sequenced. The curated gene name, CG, is given for all except AGO3. The GenBank accession no. for a genomic scaffold sequence that contains AGO3 is listed instead. (B) Phylogenetic grouping of the Drosophila Argonaute-like proteins and representatives from other organisms. The tree was constructed with the full-length protein sequences with CLUSTALX and a BLOSUM protein weight matrix. Dm, Drosophila melanogaster; Ce, C. elegans; At, Arabidopsis thaliana; Sp, Schizosaccharomyces pombe; Mm, Mus musculus; Hs, Homo sapiens; Oc, Oryctolagus cuniculus; Rn, Rattus norvegicus. The Drosophila sequences are shown in bold.
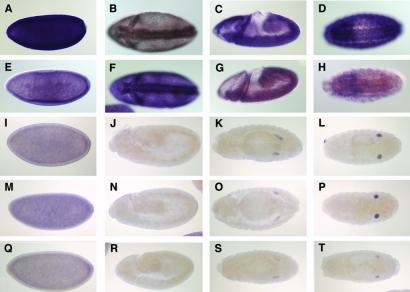
Based on sequence alignments, the five genes can be subdivided into two subcategories (Fig. 1B). Interestingly, the embryonic RNA expression patterns of the five Drosophila genes reflect this sequence-based grouping as well. AGO1 and AGO2 are maternally deposited and have fairly ubiquitous embryonic expression patterns (Fig. 2 A–H). They are, however, more strongly expressed in the ventral and cephalic furrows. Later, AGO1 is also up-regulated in the developing nervous system. piwi, aub, and AGO3, which are similar in sequence, are also expressed maternally but unlike AGO1 and AGO2, their expression disappears by embryonic stages 10–12. Zygotic transcription is then restricted to the presumptive gonad (Fig. 2 I–T), suggesting they have tissue-specific roles during embryo development and are less likely to a part of the general RNAi machinery that is assumed to be ubiquitous.
Figure 2.
Embryonic expression patterns of the five Drosophila Argonaute-like genes. (A–D) AGO1, (E–H) AGO2, (I–L) AGO3, (M–P) piwi, and (Q–T) aub. (A, E, I, M, and Q) Stage 4–5 embryos. (B and F) Stage 6 embryos initiating gastrulation. (C and G) Stage 8 embryos. (D and H) Stage 15–16. (J, N, and R) Stage 10–12 embryos. (K, O, and S) Stage 14. (L, P, and T) Stage 15–16. Anterior is to the left. Lateral views in A, C, E, G, I, J, M, N, Q, and R. Ventral view in B, D, F, and H. Dorsal view in K, L, O, P, S, and T.
AGO1 Mutants Are RNAi-Defective.
With the availability of loss-of-function mutations in AGO1, we were able to examine its role as a possible component of the RNAi machinery in Drosophila embryos. Mutations in AGO1 result in late embryonic/early larval lethality and have defects in the central and peripheral nervous system (36). Precise excision of the l(2)k08121 P element reverted lethality, as did a heat shock-driven AGO1 cDNA (data not shown). l(2)k08121, an insertion near the transcription start site of two of the AGO1 isoforms (Fig. 1A), was previously shown to be a strong allele (36). Most of our analysis was done in this out-crossed insertion line we refer to as AGO1k08121.
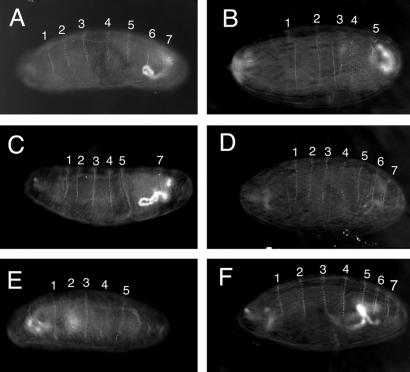
We tested for RNAi in vivo by assaying the ability of dsRNA corresponding to the eve gene to produce an eve phenotype when injected into wild-type and AGO1 mutant embryos. AGO1k08121 was maintained as a heterozygous stock balanced over a CyO-armGFP chromosome. One-quarter of the embryos produced in this stock are AGO1k08121 homozygous mutants, as identified by the absence of zygotic GFP expression. The remaining three-quarters of the embryos, which express GFP and represent both heterozygous and CyO-armGFP homozygous embryos, are referred to as AGO1+. Wild-type embryos, wild-type embryos injected with control dsRNA corresponding to the white gene (Fig. 3A), and uninjected AGO1k08121 mutant embryos have eight ventral denticle belts. By comparison, eve mutant embryos or wild-type embryos injected with eve dsRNA have a visible reduction in the number of denticle belts (ref. 29; Fig. 3B). eve dsRNA was injected into AGO1k0812 and AGO1+ embryos. AGO1k08121 zygotic mutants were less responsive than AGO1+ embryos to eve dsRNA (Fig. 3 C and D and Table 1). Specifically, only 8% of AGO1k08121 zygotic mutants had fewer than 7 denticle belts whereas 38% of their GFP-expressing AGO1+ siblings exhibited an eve phenotype. We also observed a similar reduction in RNAi-induced embryonic phenotypes (12% vs. 34%) by using dsRNA corresponding to the ftz gene (Table 1). AGO1k16601 zygotic mutants, resulting from an independent P element insertion 8 nucleotides downstream of the l(2)k08121 insertion site, also exhibited a reduced response to eve dsRNA (7% vs. 28%). It should be noted that RNAi is not completely inhibited in AGO1k08121 mutants, as a small percentage of eve embryos are observed after injection with eve dsRNA (see Discussion). However, the percentage of embryos exhibiting an RNAi-induced phenotype is clearly reduced in AGO1 mutants compared with wild type.
Figure 3.
AGO1 mutant embryos have a reduced response to eve dsRNA. Representative embryonic phenotypes of wild-type, GFP-expressing AGO1+, and AGO1k08121 mutant embryos injected with dsRNA are shown. (A) w1118 embryo injected with white dsRNA showing no alteration in the number of ventral denticle belts. (B) w1118 embryo injected with eve dsRNA. Denticle belt 4 is fused with 5, and 6 is missing. (C) AGO1+ embryo injected with eve dsRNA. Belt 6 is missing. (D) AGO1k08121 embryo injected with eve dsRNA showing no alteration in the number of denticle belts. (E) AGO1+ embryo injected with eve siRNA. Belt 4 is partially formed whereas belt 6 is missing. (F) AGO1k08121 embryo injected with eve siRNA showing no alteration in the number of denticle belts. The denticle belts corresponding to each abdominal segment are labeled, except 8, which is not in the plane of focus.
Table 1.
Effects of dsRNA injected into Drosophila embryos
| Genotype* | Injected RNA | Wild type† | eve† | Expected‡ |
|---|---|---|---|---|
| w1118 | evedsRNA | 44 | 66 (60%) | |
| w1118 | evesiRNA | 156 | 54 (26%) | |
| CyO-armGFP/Adv | evedsRNA | 55 | 32 (37%) | |
| Adv/Adv | evedsRNA | 30 | 10 (25%) | |
| CyO-armGFP/AGO1k08121 | evedsRNA | 91 | 56 (38%) | |
| AGO1k08121 | evedsRNA | 55 | 5 (8%) | 23 |
| CyO-armGFP/AGO1k16601 | evedsRNA | 31 | 12 (28%) | |
| AGO1k16601 | evedsRNA | 13 | 1 (7%) | 4 |
| CyO-armGFP/AGO1k08121 | ftzdsRNA | 39 | 20 (34%) | |
| AGO1k08121 | ftzdsRNA | 17 | 2 (12%) | 6 |
| AGO1k08121 revertant | evedsRNA | 34 | 17 (33%) | |
| AGO1k08121[hsAGO1] | evedsRNA | 32 | 13 (29%) | |
| CyO-armGFP/AGO1k0812 | evesiRNA | 157 | 61 (28%) | |
| AGO1k08121 | evesiRNA | 75 | 1 (1%) | 21 |
| w1118 | whitedsRNA | 61 | 0 | |
| CyO-armGFP/AGO1k08121 | whitedsRNA | 39 | 0 | |
| AGO1k08121 | whitedsRNA | 12 | 0 |
Zygotic genotype. Genotypes listed as CyO-armGFP/AGO1k08121 were scored as GFP-positive and represent a mixed population including AGO1k08121/CyO-armGFP and CyO-armGFP/CyO-armGFP embryos.
Number of embryos with wild-type or eve phenotype after injection. Embryos were scored as eve if they had 6 or fewer ventral denticle belts. Embryos were scored as wild type if they had 7 or 8 denticle belts (see Materials and Methods). Percentages are given in parentheses.
The expected number of eve embryos based on the percent penetrance from the above row assuming AGO1 mutants have no affect on RNAi.
We performed a number of control experiments to confirm that the decrease in RNAi-induced phenotypes was a specific effect of AGO1 reduction-of-function. Of the embryos homozygous for a l(2)k08121 revertant chromosome that were injected with eve dsRNA, 33% exhibited an eve phenotype (Table 1). Similarly, AGO1k08121 embryos containing the heat shock-driven AGO1 transgene that rescues lethality also had a near wild-type response to dsRNA (29%). Therefore, the P element insertion in AGO1 causes the reduced RNAi response exhibited in AGO1k08121 embryos.
AGO1 Mutants Are Defective in Degrading Targeted mRNA.
As a first step toward understanding the molecular function of AGO1, we used an in vitro extract to narrow down the step(s) in RNAi that are affected in AGO1 mutants. Previous in vitro analysis of RNAi in Drosophila relied on extracts, prepared from either early syncitial embryos (12) or S2 cells (8), which were capable of processing dsRNA into siRNA and then degrading mRNA in a sequence-specific manner. We prepared extracts from older 12–16-h cellular embryos, when the zygotic expression of the armGFP clearly distinguishes CyO-containing embryos from AGO1k08121 embryos, by following a procedure similar to Zamore et al. (7).
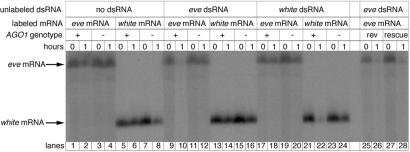
We assessed the ability of extracts prepared from AGO1k08121 and GFP-expressing embryos, laid by an AGO1k08121/CyO-armGFP balanced stock, to degrade target mRNA in response to dsRNA. mRNA incubated in the extracts without dsRNA for 1 h were only modestly degraded (Fig. 4, lanes 1–8). Extracts prepared from AGO1+ embryos and preincubated with eve dsRNA efficiently degrade eve mRNA (lanes 9 and 10). Similarly, white dsRNA could target white mRNA for degradation (lanes 21 and 22). This process is sequence-specific because eve dsRNA does not affect the stability of white mRNA nor does white dsRNA affect the stability of eve mRNA (lanes 13 and 14 and 17 and 18), suggesting that our embryo extracts faithfully reproduce RNAi in vitro. Extracts prepared from AGO1k08121 embryos, however, did not degrade mRNA when preincubated with a homologous dsRNA (lanes 11 and 12 and 23 and 24). The ability to degrade mRNA in a sequence specific manner was restored in extracts prepared from the revertant line (lanes 25 and 26) and in extracts prepared from the line where the AGO1k08121 insertion is rescued by the expression of an AGO1 cDNA (lanes 27 and 28). The reduced RNAi observed in living AGO1k08121 embryos correlates with the lack of dsRNA-triggered mRNA degradation in vitro.
Figure 4.
Degradation of targeted mRNA in response to dsRNA is reduced in AGO1 mutants. Gel of uniformly 32P-labeled mRNA that was incubated in extracts prepared from AGO1k08121 (−) and GFP-expressing AGO1+ (+) embryos. Before the addition of labeled mRNA, the extracts were incubated with or without unlabeled dsRNA. Samples collected just after the addition of mRNA (0) or after a 1-h incubation (1) are in neighboring lanes. There is some nonspecific degradation of the mRNA after 1 h in the embryo extracts as seen in the “no dsRNA” controls. However, the amount of mRNA is greatly reduced in (+) extracts preincubated with homologous dsRNA. mRNA is unaffected in extracts prepared from AGO1 mutants. Extracts prepared from a viable revertant line derived from l(2)k08121 (rev) are able to degrade targeted mRNA. Similarly, extracts prepared from AGO1k08121 embryos rescued with an AGO1 cDNA (rescue) degrade mRNA targeted with homologous dsRNA.
AGO1 Acts Downstream of siRNA Production.
To determine whether AGO1 is required for the initial Dicer-mediated cleavage of dsRNA into siRNA, we injected synthetic eve siRNA into embryos laid by w1118 and AGO1k08121/CyO-armGFP females and scored them for the number of denticle belts. siRNA was capable of producing an eve phenotype in wild-type embryos, albeit at a lower penetrance than longer dsRNA (26% vs. 60% for w1118 and 28% vs. 38% for GFP-expressing embryos; Fig. 3 E and F and Table 1). This lower penetrance may be the result of incomplete phosphorylation of the 5′ ends of the siRNA, which is required for efficient degradation of mRNA (38–40) or may reflect a lower potency of the siRNA. AGO1k08121 mutants, however, produced only 1% eve embryos after injection with eve siRNA. Therefore, AGO1k08121 mutants have a reduced response to synthetic siRNA, suggesting that AGO1 functions downstream of siRNA production.
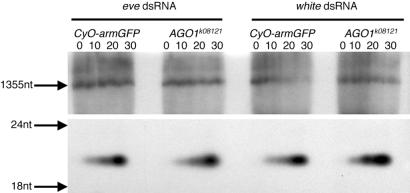
We then assayed the ability of extracts from AGO1k08121 mutant embryos and their AGO1+, GFP-expressing siblings to process dsRNA into short ≈21-nt fragments. Extracts prepared from both AGO1k08121 zygotic mutants and AGO1+ embryos were able to cleave dsRNA corresponding to both the eve and white genes into ≈21-nt fragments (Fig. 5). The rate of siRNA production was similar in both AGO1k08121 and AGO1+ extracts. Taken together, these experiments indicate that wild-type AGO1 activity is required after the Dicer-mediated production of siRNA.
Figure 5.
Production of ≈21-nt fragments is unaffected in AGO1 mutants. Gel of uniformly 32P-labeled dsRNA, corresponding to either the eve or white gene, which was incubated in extracts from GFP-expressing AGO1+ embryos and AGO1k08121 mutant embryos for 0, 10, 20, and 30 min. Upper is the top part of the gel showing the full-length input RNA. Lower shows the lower part of the same gel showing that both eve dsRNA and white dsRNA were cleaved into ≈21-nt fragments.
Discussion
A Family of Argonaute-like Genes in Drosophila.
The Drosophila genome encodes five Argonaute-like proteins. Consistent with their embryonic expression patterns, piwi and aub are defined by mutations that affect aspects of germ-line development. piwi is required to maintain germ-line stem cells perhaps by regulating a somatically derived stem cell promoting signal (33). Recently, piwi has also been shown to affect transgene-mediated cosuppression in Drosophila (22). aub is required for embryo patterning, by regulating oskar and gurken translation, and for pole cell formation (32, 35). aub is also required for Su(Ste)-mediated suppression of Stellate in the testis (34). Interestingly, this regulation seems to involve an RNAi-like mechanism (41). The related AGO3 gene has an embryonic expression pattern very similar to piwi and aub, suggesting that it may have a role in gonad and/or germ-line development as well.
AGO1 is an essential gene that plays a role in neuronal growth (36). The related AGO2 protein was identified as part of the RISC protein complex that degrades targeted mRNA in response to dsRNA (23). Both AGO1 and AGO2 transcripts are maternally deposited and expressed throughout embryonic development. Consistent with its mutant phenotype, AGO1 is up-regulated in the embryonic nervous system (36). Unlike piwi, aub and AGO3, AGO1, and AGO2 are expressed outside the gonad and thus may have more general somatic functions.
Possible Roles for AGO1 in RNAi.
Based on its sequence similarity to the C. elegans rde-1, the Neurospora qde-2, and the Arabidopsis Argonaute genes, AGO1 is a likely candidate to function in RNAi. This idea is also supported by its sequence similarity to the Drosophila AGO2 and its wide embryonic expression pattern. With both in vivo embryo injections and in vitro assays, we have shown that AGO1 zygotic mutants are compromised for RNAi.
AGO1 mutant embryos still exhibit some RNAi activity, however. There are several possible explanations for this observation. First, AGO1k08121 may not be a null allele. The P element is inserted near the transcriptional start site for two of the isoforms and in the second intron of a third isoform (Fig. 1A), leaving the possibility that a functional mRNA could be produced; however, there is a strong reduction of all AGO1 transcripts in the l(2)k08121 allele (36). Secondly, because AGO1 and AGO2 have similar expression patterns and both may function in RNAi, they may share functional redundancy. Partial redundancy has been demonstrated for two related Argonaute-like genes during C. elegans and Arabidopsis development (4, 18, 37). An interesting and untested idea is that AGO1 and AGO2 may have some redundancy during early embryogenesis but then later tissue-specific differences in their expression patterns would uncover the lethality associated with AGO1 mutants. This model would be consistent with the continued up-regulation of AGO1 mRNA, especially in the ventral nerve cord, near the end of embryogenesis. Third, the maternal contribution of AGO1 mRNA might provide some level of AGO1 activity, although it cannot support wild-type levels of RNAi.
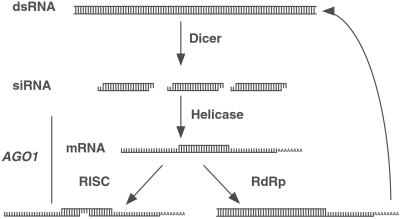
AGO1 is not required for siRNA production. This model is supported by the cleavage of dsRNA into ≈21fragments in AGO1 mutants and the inability to bypass AGO1 function by injection of synthetic siRNA. Therefore, AGO1 functions at some step(s) between siRNA production and target mRNA degradation (Fig. 6).
Figure 6.
Proposed step(s) during RNAi where AGO1 may function. RNAi is initiated by the Dicer-mediated cleavage of dsRNA into ≈21-nt siRNA. siRNA is then used to target mRNA for degradation by the RISC complex. In another step, which may or may not be mediated by RISC, siRNA can act as a primer for second-strand synthesis of RNA complementary to the targeted mRNA. The newly dsRNA may then become a substrate for Dicer. Based on our results, we proposed that AGO1 functions downstream of siRNA production. Our experiments do not address whether AGO1 is specific for mRNA degradation or whether it is required for the RNA-dependent RNA polymerase (RdRp) amplification of the siRNA.
However, there are still several possible steps in which AGO1 may function (38). Recent studies of C. elegans rde-1 mutants have shown that in vitro extracts are capable of processing dsRNA into siRNA, but siRNA was greatly reduced in vivo. This work suggested that wild-type rde-1 activity is required to stabilize siRNA (42, 43). Thus, AGO1/rde-1 may protect siRNA from a nuclease. AGO1 may help to incorporate siRNA into a functional RISC or, like AGO2, is itself a component of RISC. Such activity could include maintaining the required 5′ phosphate group on siRNA, acting as a scaffold to assemble the multiprotein complex, unwinding siRNA/strand selection for base pairing to target mRNA, or acting as a component of the nuclease that cleaves mRNA. These possible functions for AGO1 are not exclusive. For example, siRNA could be stabilized by incorporation into RISC. However, the biochemical activity or in vivo partners of the AGO1 protein are not known.
Developmental Roles of AGO1?
AGO1 mutants are late embryo/early larval lethal and exhibit defects in the embryonic nervous system (36). This finding is not surprising because other components of the RNAi pathway have been shown to function during development. Dicer/dcr-1 and the rde-1 homologs alg-1 and alg-2 are required for the production of the small temporal RNA (stRNA) in C. elegans (4, 5). stRNAs, encoded by the lin-4 and let-7 genes (44–49), are 21–22-nt single-stranded RNAs that function by base pairing to the 3′ untranslated regions and inhibiting translation of genes that control developmental timing. stRNAs are initially made as ≈70-nt primary transcripts that can fold into a hairpin structure. The double-stranded stem portion of the RNA is cleaved and processed into the functional 21–22-nt stRNA. Recent studies have uncovered a large and diverse population of endogenous microRNA (50–52) that share many of the characteristics of lin-4/let-7. These observations raise the possibility that small RNA represents a common mode of gene regulation and their production/usage requires a mechanism similar to RNAi.
The neuronal defect in AGO1 mutants is particularly intriguing. Lai and Posakony (53) have proposed that short regions of RNA:RNA duplex formation between the 3′ untranslated regions (UTRs) of the proneural genes and members of the Enhancer of split complex E(spl) may represent a level of gene regulation during neurogenesis. Most recently, it was noted that several Drosophila microRNAs have regions of complementarity to negative regulatory elements in the 3′ UTRs of multiple members of the Bearded and E(spl) complexes (54). Because members of these gene families regulate development of the nervous system, it would be very interesting if the developmental defects in AGO1 mutants are the results of a defect in microRNA processing or use.
Supplementary Material
Acknowledgments
We thank Mike Brodsky (Univ. of Massachusetts, Worcester) and Kathy Sullivan (Univ. of California, Berkeley) for the CyO-armGFP stock. Richard Carthew (Northwestern University, Evanston, IL) kindly provided the even-skipped (pEve) and fushi tarazu (pGEM F1) clones. For use of and assistance with the embryo sorter, we especially thank Eileen Furlong and Matthew Scott. Amy Beaton, Audrey Huang, Eric Lai, Andrea Page-McCaw, and Mark Running provided helpful comments on this manuscript. Todd Laverty and Amy Beaton maintained the P element fly stocks. Michael Muse provided valuable technical assistance. The Berkeley Drosophila Genome Project supplied numerous expressed sequence tag clones. This work was supported by the Howard Hughes Medical Institute and by a Helen Hay Whitney Fellowship (to R.W.W.).
Abbreviations
- dsRNA
double-stranded RNA
- RNAi
RNA interference
- siRNA
small interfering RNA
- PTGS
posttranscriptional gene silencing
- RISC
RNA-induced silencing complex
References
- 1.Hammond S M, Caudy A A, Hannon G J. Nat Rev Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 2.Cogoni C, Macino G. Proc Natl Acad Sci USA. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Grishok A, Pasquinelli A E, Conte D, Li N, Parrish S, Ha I, Baillie D L, Fire A, Ruvkun G, Mello C C. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 5.Hutvagner G, McLachlan J, Pasquinelli A E, Balint E, Tuschl T, Zamore P D. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton A J, Baulcombe D C. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 7.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 8.Hammond S M, Bernstein E, Beach D, Hannon G J. Nature (London) 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature (London) 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Knight S W, Bass B L. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ketting R F, Fischer S E J, Bernstein E, Sijen T, Hannon G J, Plasterk R H A. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuschl T, Zamore P D, Lehmann R, Bartel D P, Sharp P A. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipardi C, Wei Q, Paterson B M. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- 14.Sijen T, Fleenor J, Simmer F, Thijssen K L, Parrish S, Timmons L, Plasterk R H, Fire A. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 15.Parrish S, Fleenor J, Xu S, Mello C, Fire A. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir S M, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 18.Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabara H, Sarkissian M, Kelly W G, Fleenor J, Grishok A, Timmons L, Fire A, Mello C C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 20.Catalanotto C, Azzalin G, Macino G, Cogoni C. Nature (London) 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- 21.Fagard M, Boutet S, Morel J B, Bellini C, Vaucheret H. Proc Natl Acad Sci USA. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pal-Bhadra M, Bhadra U, Birchler J A. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 23.Hammond S M, Boettcher S, Caudy A A, Kobayashi R, Hannon G J. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 24.Jacobsen S E, Running M P, Meyerowitz E M. Development (Cambridge, UK) 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- 25.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe D C. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 26.Cogoni C, Macino G. Nature (London) 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 27.Smardon A, Spoerke J M, Stacey S C, Klein M E, Mackin N, Maine E M. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 28.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 29.Kennerdell J R, Carthew R W. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 30.Furlong E E, Profitt D, Scott M P. Nat Biotechnol. 2001;19:153–156. doi: 10.1038/84422. [DOI] [PubMed] [Google Scholar]
- 31.Cerutti L, Mian N, Bateman A. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- 32.Wilson J E, Connell J E, Macdonald P M. Development (Cambridge, UK) 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- 33.Cox D N, Chao A, Baker J, Chang L, Qiao D, Lin H. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt A, Palumbo G, Bozzetti M P, Tritto P, Pimpinelli S, Schafer U. Genetics. 1999;151:749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris A N, Macdonald P M. Development (Cambridge, UK) 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- 36.Kataoka Y, Takeichi M, Uemura T. Genes Cells. 2001;6:313–325. doi: 10.1046/j.1365-2443.2001.00427.x. [DOI] [PubMed] [Google Scholar]
- 37.Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton M K. Development (Cambridge, UK) 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- 38.Nykanen A, Haley B, Zamore P D. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 39.Boutla A, Delidakis C, Livadaras I, Tsagris M, Tabler M. Curr Biol. 2001;11:1776–1780. doi: 10.1016/s0960-9822(01)00541-3. [DOI] [PubMed] [Google Scholar]
- 40.Elbashir S M, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aravin A A, Naumova N M, Tulin A V, Vagin V V, Rozovsky Y M, Gvozdev V A. Curr Biol. 2001;11:1017–27. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 42.Tijsterman M, Ketting R F, Okihara K L, Sijen T, Plasterk R H. Science. 2002;295:694–697. doi: 10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- 43.Parrish S, Fire A. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- 44.Lee R C, Feinbaum R L, Ambros V. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 45.Wightman B, Ha I, Ruvkun G. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 46.Ha I, Wightman B, Ruvkun G. Genes Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 47.Olsen P H, Ambros V. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 48.Reinhart B J, Slack F J, Basson M, Pasquinelli A E, Bettinger J C, Rougvie A E, Horvitz H R, Ruvkun G. Nature (London) 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 49.Pasquinelli A E, Reinhart B J, Slack F, Martindale M Q, Kuroda M I, Maller B, Hayward D C, Ball E E, Degnan B, Muller P, et al. Nature (London) 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 50.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 51.Lau N C, Lim L P, Weinstein E G, Bartel D P. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 52.Lee R C, Ambros V. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 53.Lai E C, Posakony J W. Cell. 1998;93:1103–1104. doi: 10.1016/s0092-8674(00)81454-3. [DOI] [PubMed] [Google Scholar]
- 54.Lai E C. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.