Abstract
Immune cells are activated during cellular responses to antigen by two described mechanisms: (i) direct uptake of antigen and (ii) extraction and internalization of membrane components from antigen-presenting cells. Although endocytosis of microbial antigens by pattern recognition molecules (PRM) also activates innate immunity, it is not known whether this involves extraction and internalization of microbial surface components. Epithelial cells on mucosal surfaces use a variety of receptors that are distinct from the classical endocytic PRM to bind and internalize intact microorganisms. Nonclassical receptor molecules theoretically could act as a type of endocytic PRM if these molecules could recognize, bind, extract, and internalize a pathogen-associated molecule and initiate cell signaling. We report here that the interaction between the cystic fibrosis transmembrane conductance regulator (CFTR) and the outer core oligosaccharide of the lipopolysaccharide (LPS) in the outer membrane of Pseudomonas aeruginosa satisfies all of these conditions. P. aeruginosa LPS was specifically recognized and bound by CFTR, extracted from the organism's surface, and endocytosed by epithelial cells, leading to a rapid (5- to 15-min) and dynamic translocation of nuclear transcription factor NF-κB. Inhibition of epithelial cell internalization of P. aeruginosa LPS prevented NF-κB activation. Cellular activation depended on expression of wild-type CFTR, because both cultured ΔF508 CFTR human airway epithelial cells and lung epithelial cells of transgenic-CF mice failed to endocytose LPS and translocate NF-κB. CFTR serves as a critical endocytic PRM in the lung epithelium, coordinating the effective innate immune response to P. aeruginosa infection.
Although numerous bacterial cell-surface molecules have been identified as ligands for eukaryotic cell receptors (1), it has not been determined whether the ligand–receptor interaction serves solely to promote bacterial binding and ingestion by eukaryotic cells. The hypothesis that bacterial ligands can be extracted and internalized by epithelial cells to serve a signaling function for the host has not been tested. Studies using purified pathogen-associated molecular pattern (PAMP) molecules have suggested that recognition of conserved PAMPs—such as bacterial lipopolysaccharide (LPS) and peptidoglycan—by pattern recognition molecules (PRM) of the innate immune system represents a key component of host resistance to infection. Almost all studies published to date on PRM activation of cells used purified PAMPs such as LPS, CpG-rich DNA, peptidoglycan, etc., without evidence that these are released naturally from microbes, particularly during the early stages of infection.
Early recognition of a pathogen by epithelial cells on a mucosal surface likely involves binding of the whole microbial cell rather than binding of released PAMP. Eukaryotic cell extraction of PAMPs could serve as an amplification mechanism to stimulate an effective innate immune response. Vesy et al. showed that LPS-binding protein and phospholipid transfer protein can release LPS from isolated Gram-negative bacterial outer membrane fragments and transfer the LPS to human high-density lipoproteins (2) but did not report on extraction of LPS from the membranes of intact bacteria. Interactions between T lymphocytes and antigen-presenting cells can lead to endocytosis of membrane components and subsequent cellular activation (3). Therefore, we investigated whether extraction of LPS from the surface of Pseudomonas aeruginosa by the cystic fibrosis transmembrane conductance regulator (CFTR), which specifically binds to the outer core oligosaccharide of this organism's LPS, could result in responses that initiate innate immunity.
Methods
Cell Lines.
Respiratory epithelial cell lines used were CFT1-ΔF508 cells (three alleles of human ΔF508 CFTR) and CFT1-LCFSN cells that express a functional wild-type (wt) human CFTR gene in this cellular background (4).
In Vivo Mouse Experiments.
Wt C3H/HeN mice were obtained commercially. Noninbred, homozygous ΔF508 cftr mice (5) and doubly transgenic S489X cftrFABPhuCFTR mice expressing no murine CFTR protein, but expressing wt human CFTR protein in the gastrointestinal tract from the CFTR gene under the control of the mouse fatty acid binding protein promoter (6), were bred in our facility (7). Breeding and mouse genotyping and phenotyping were performed as described (7).
Mice were anesthetized as described (7) for intratracheal injection of LPS and intranasal application of whole bacteria (8), and lung tissues were processed for histology. The slides were deparaffinized, rehydrated, and incubated with rabbit antibody to human (p65) NF-κB (Santa Cruz Biotechnology) for 90 min at 37°C and then incubated with Alexa Fluor 594 dye-labeled goat anti-rabbit IgG and with Alexa Fluor 488 dye directly conjugated to mouse anti-CFTR mAb CF3 (5) for 90 min at 37°C. 4′,6-Diamidino-2-phenyindole dihydrochloride was added for the final 1 h as a nuclear stain. The slides then were visualized by using confocal microscopy.
Bacterial Strains, LPS Purification, and Fluorescent Labeling of LPS.
Wt P. aeruginosa strain PAO1-V (7) and two nonmucoid, LPS-smooth clinical isolates from CF patients obtained early in the course of infection were used. Escherichia coli TG1 also was used in some experiments. Wt LPS and LPS lacking the outer-core oligosaccharide were purified as described (9). LPS was labeled with FITC as described (10). Purity, fluorescent labeling, and immunological reactivity of FITC-LPS were confirmed by UV scan, SDS/PAGE, and immunoblot analysis.
Preparation of Liposomes and Planar Bilayers.
Unilamellar membranes were formed by freeze-thawing and extrusion (11) to incorporate different densities of FITC-LPS into the liposomes. Planar bilayers were formed on round, glass cover slips (25 mm diameter) as described (12).
Confocal and Fluorescence Microscopy.
LPS uptake by epithelial cells was measured by using a Meridian Ultima laser cytometer system, including an argon laser (Coherent Radiation, Palo Alto, CA; excitation wavelength, 488 nm) and a ×40 oil-immersion objective (numerical aperture = 1.3). The ability of FITC-LPS to diffuse laterally in the planar bilayers was confirmed as described previously (13). For nonconfocal XY images, the size of the scanned field was 90 × 90 μm and fluorescence counts were measured at 1-μm intervals. To view directly the intracellular uptake of fluorescence, confocal XZ and XY planes also were scanned by using a field of 36 × 36 μm, and fluorescence counts were measured at 0.4-μm intervals. Each point was sampled 20 times with 4-μs laser pulses, and the fluorescence counts were averaged before the next point was scanned. The laser power at the microscope stage was about 1 mW, and the beam diameter was about 2 μm.
Additional confocal images were obtained by using a Bio-Rad MRC-1024 microscope interfaced with a krypton–argon laser or with a Tsunami mode-locked, titanium–sapphire multiphoton laser (Spectra-Physics) as described (7), except that in some cases, the TO-PRO-3 nuclear stain was used to visualize the nucleus.
NF-κB Activation Analysis.
An Effectene transfection kit (Qiagen, Chatsworth, CA) was used to transfect cultured human cells with 0.35 μg of NF-κB-Luciferase Mercury profiling plasmid (CLONTECH) for 43 h at 37°C. Cells then were washed with F12 and stimulated for 15 min at 37°C with 6 μl of a 5% solution of liposomes containing either no LPS or wt P. aeruginosa LPS. Cells then were washed, incubated for 4 h in F12, washed again, and lysed, and the cell extracts were harvested. Luciferase activity was analyzed by using a Luciferase assay kit (Promega); the luminescence of each sample was measured for 10 s with an Optocomp I Luminometer (MGM Instruments, Hamden, CT).
FACS Analysis.
Sonicated (5 min) wt P. aeruginosa FITC-LPS (8 μg/ml in F12) was incubated for 2 h at 37°C with 50 nM of peptide SYDPDNKEER, representing amino acids 108–117 of the first CFTR extracellular domain, or with peptide KDPNYDERSE, a scrambled version of peptide 108–117. LCFSN or ΔF508 cells (2 × 105) in F12 medium were gently pelleted in polypropylene tubes, resuspended in 400 μl of medium only or the FITC-LPS/peptide mix, and incubated for 1–2 h at 37°C. Cells were washed three times with PBS to remove unbound LPS and then fixed in 4% paraformaldehyde. Flow cytometry using the 488-nm excitation line was performed as described previously (7).
Fusion of LPS-FITC into the P. aeruginosa Membrane.
P. aeruginosa strain PAO1-V was grown overnight at 37°C and adjusted to 109 colony-forming units/ml with a spectrophotometer. Twenty microliters of a 5% FITC-LPS liposome solution was added to 4 ml of bacteria. This mixture was rotated gently for 24 h at 4°C. Bacterial cells were counterstained with 5 μM SYTO-17 dye (Molecular Probes) for 30 min at room temperature. Bacteria were washed eight times before use to remove unbound dye and unincorporated LPS.
Immunoelectron Microscopy.
CFT1-LCFSN cells were incubated with P. aeruginosa PAO1-V for 4 h. Unbound bacteria were washed away with PBS. Staining for LPS was carried out as described by using normal or immune serum and Protein A gold particles for visualization by electron microscopy (14). Images were taken at a magnification of ×25,000.
Statistical Analysis.
ANOVA and the Fisher probable least significant differences posthoc test were used on multiple comparison data. Differences in cellular labeling using FACS were determined by using Kolmogorov–Smirnov statistical analysis.
Results
In Vivo Epithelial Cell Response to P. aeruginosa Infection.
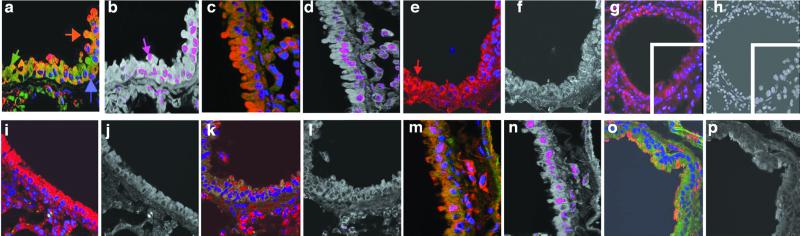
Mice were infected either intranasally with 2 × 107 whole bacterial cells or intratracheally with liposomes containing 10 μg of LPS from wt P. aeruginosa. Both treatments induced rapid NF-κB nuclear translocation in lung epithelial cells of mice expressing wt CFTR. Translocation was detected as early as 5 min after infection (not shown), peaked around 15 min postinfection (Fig. 1 a–d), and could no longer be detected by 45 min postinfection (not shown). Little to no detectable NF-κB nuclear translocation was seen in lung cells from transgenic ΔF508 cftr mice (Fig. 1 e– h), although immunofluorescence microscopy confirmed the presence of P. aeruginosa in the lungs of these mice (not shown). There was no translocation of NF-κB after intratracheal injection of P. aeruginosa into doubly transgenic S489X cftrFABPhuCFTR mice, which lacked expression of murine CFTR in any tissue but expressed human CFTR in the gastrointestinal tract (ref. 6; Fig. 1 i and j). Infection of wt and ΔF508 cftr mice with clinical isolates of P. aeruginosa produced the same result as infection with P. aeruginosa PAO1-V (one example, Fig. 1 k– n). E. coli, whose LPS lacks the CFTR ligand, failed to elicit NF-κβ nuclear translocation in the lung epithelium of mice expressing wt CFTR (Fig. 1 o and p).
Figure 1.
NF-κB nuclear translocation in vivo in epithelial cells in small airways sectioned from lungs of wt and transgenic ΔF508 CF mice 15 min after intranasal infection with whole bacteria or intratracheal infection with LPS in liposomes. Sections were stained for NF-κB (red channel, arrow, e), CFTR (green channel, arrow, a), and nuclear DNA (blue channel, arrow, a). Yellow to orange color (arrow, a) represents overlap of cytoplasmic CFTR and NF-κB. Colocalization of translocated NF-κB with nucleic acid is shown as a magenta color (arrow, b) produced from the overlap of the blue and red fluorescence signals. (×400.) (a) Wt lungs + P. aeruginosa LPS. (b) Colocalization depiction of a. (c) Wt lungs + P. aeruginosa bacteria. (d) Colocalization depiction of c. (e) ΔF508 CF lungs + P. aeruginosa LPS. (f) Colocalization depiction of e. (g) ΔF508 CF lungs + P. aeruginosa bacteria. (h) Colocalization depiction of g. (Insets) Higher-power view (×800) of section of lung airway from ΔF508 CF lungs. (i) FABP CF lungs + P. aeruginosa bacteria. (j) Colocalization depiction of i. (k) ΔF508 CF lungs + P. aeruginosa clinical isolate. (l) Colocalization depiction of k. (m) Wt lungs + P. aeruginosa clinical isolate. (n) Colocalization depiction of m. (o) Wt lungs + E. coli bacteria. (p) Colocalization depiction of o.
In Vitro Responses of Human Epithelial Cells.
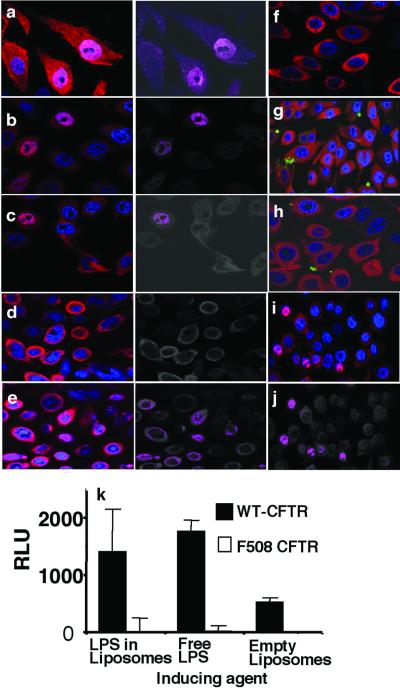
Similar results were obtained by using cultured human lung epithelial cells that were isogenic for expression of either the ΔF508 or the wt CFTR alleles. For some experiments, P. aeruginosa LPS was incorporated into liposomes before exposure of the cells to LPS. In other experiments, P. aeruginosa LPS was labeled with FITC. Wt-CFTR human cells showed rapid nuclear translocation of cytoplasmic NF-κB after exposure to either P. aeruginosa cells, P. aeruginosa LPS in liposomes, or free LPS (Fig. 2 a–c). Previous studies have demonstrated that incubation of P. aeruginosa cells with CFTR peptide 108–117 inhibits bacterial uptake (4, 15) and activation of epithelial cell src-like tyrosine kinases p60Src and p59Fyn (15), although this treatment also increases adherence of the bacteria to epithelial cells. Here, CFTR peptide 108–117 inhibited NF-κB translocation induced by P. aeruginosa cells (Fig. 2d) or by P. aeruginosa LPS in liposomes (not shown), whereas a scrambled version of this peptide did not (Fig. 2e). Importantly, treatment of P. aeruginosa cells or LPS with CFTR peptide 108–117 did not prevent binding of the bacteria or liposomes to epithelial cells. Thus, bacterial binding to epithelial cells, in and of itself, did not induce NF-κB nuclear translocation.
Figure 2.
In vitro translocation of NF-κB to the nucleus of cultured human epithelial cells expressing either ΔF508 or wt CFTR. Sections were stained for NF-κB (red) and nuclear DNA (blue). FITC-labeled LPS is green. Translocation of NF-κB into the nucleus is shown as a magenta color produced from overlap of the blue and red fluorescence signals in the images to the immediate right of a–e. (a) Wt CFTR cells incubated with P. aeruginosa whole bacteria. (b) Wt CFTR cells incubated with P. aeruginosa LPS in liposomes. (c) Wt CFTR cells incubated with free P. aeruginosa LPS. (d) Wt CFTR cells incubated with P. aeruginosa and CFTR peptide 108–117. (e) Wt CFTR cells incubated with P. aeruginosa and a scrambled version of CFTR peptide 108–117. (f) Wt CFTR cells incubated with E. coli LPS in liposomes. (g) ΔF508 CFTR cells incubated with wt P. aeruginosa FITC-LPS in liposomes. (h) Wt CFTR cells incubated with FITC-labeled P. aeruginosa incomplete core LPS (lacks CFTR ligand) in liposomes. (i) ΔF508 CFTR cells treated with 10% glycerol overnight to induce cell-surface expression of the ΔF508 CFTR protein, then incubated with wt P. aeruginosa LPS. (j) Colocalization depiction of i. (k) Induction of NF-κB luciferase activity, measured as relative light units (RLU), in transfected wt or ΔF508 CFTR cells exposed to P. aeruginosa LPS in liposomes or as free LPS. No activity above background was seen when ΔF508 CFTR cells were exposed to P. aeruginosa LPS.
E. coli LPS in liposomes did not elicit NF-κB translocation in epithelial cells expressing wt CFTR (Fig. 2f). Because NF-κB translocation in response to E. coli LPS requires the cellular proteins CD14, MD-2, and toll-like receptor 4, these results indicated that at least one of these molecules was not present or functional in this system. Human airway epithelial cells that expressed only ΔF508 CFTR alleles translocated little to no NF-κB to the nucleus after exposure to FITC-labeled P. aeruginosa LPS (Fig. 2g) or to whole P. aeruginosa cells (not shown). Because P. aeruginosa cells are flagellated and express a high content of CpG DNA, neither of these ligands for toll-like receptors 5 and 9 were active in this system. FITC-labeled incomplete-core LPS, which lacks the CFTR-binding oligosaccharide but contains the lipid A core, did not elicit NF-κβ translocation (Fig. 2h). Using a reporter plasmid to measure NF-κB activation, only the cells with wt-CFTR produced detectable luciferase activity after stimulation with either free P. aeruginosa LPS or LPS in liposomes (Fig. 2k). Thus, the rapid epithelial cell NF-κB translocation induced by P. aeruginosa cells or LPS depended on binding of P. aeruginosa complete-core LPS to wt CFTR protein.
Treatment of ΔF508 CFTR cells with 10% glycerol promotes trafficking of the mutant protein from intracellular compartments into the plasma membrane (16). Human airway epithelial cells expressing only the ΔF508 CFTR allele and treated with 10% glycerol overnight showed substantial nuclear translocation of NF-κB 15 min after addition of wt P. aeruginosa LPS in liposomes (Fig. 2 i and j). The ΔF508 CFTR protein thus was capable of mediating rapid NF-κB nuclear translocation when the mutant protein was properly chaperoned in the cell.
CFTR-Dependent Extraction of LPS from Lipid Bilayers.
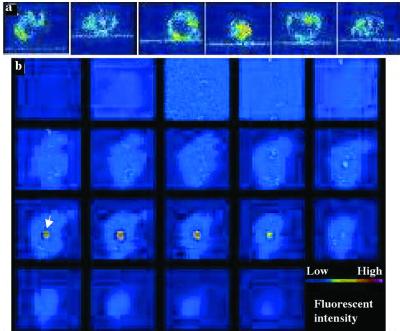
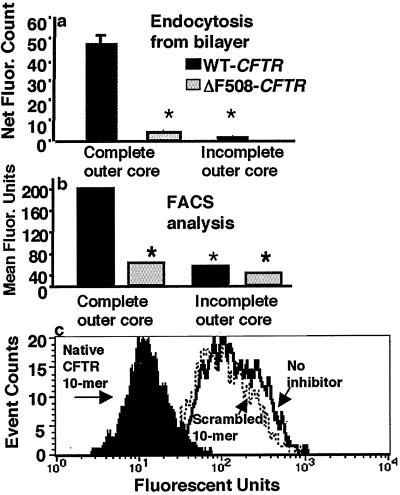
Cell lines with wt or ΔF508 CFTR were incubated on a glass-supported planar bilayer membrane (13) reconstituted with FITC-labeled wt P. aeruginosa LPS, and the interaction between the unilamellar membrane and the epithelial cell was observed by using a Meridian Ultima interactive laser cytometer (Meridian Instruments, Lansing, MI). Scans through the XZ planes showed that wt CFTR cells rapidly internalized the FITC-labeled LPS (Fig. 3a). Scans through the XY planes, from below the planar bilayer to the top of the epithelial cell, confirmed the cellular uptake of LPS (Fig. 3b). FITC-labeled LPS from a mutant P. aeruginosa strain that lacks the bacterial oligosaccharide ligand for CFTR was not taken up by wt epithelial cells, and ΔF508 CFTR epithelial cells were unable to extract wt LPS from the planar bilayer (not shown). Quantitative measurements confirmed that epithelial cells with the wt CFTR allele took up significantly more fluorescent wt LPS from the planar bilayer than did cells with the ΔF508 CFTR allele (Fig. 4a). FACS analysis of epithelial cell endocytosis of liposomal FITC-labeled LPS confirmed the findings obtained with FITC-labeled LPS that had been presented in planar bilayers (Fig. 4b). Specific uptake of wt LPS by cells expressing wt CFTR was inhibited by incubation of the LPS with CFTR peptide 108–117 but not by incubation with a scrambled version of this peptide (Fig. 4c).
Figure 3.
Extraction and internalization of wt P. aeruginosa LPS from a unilamellar lipid bilayer. (a) Confocal XZ scans of six different wt CFTR cells taking up FITC-LPS from a planar lipid bilayer. (b) Nineteen confocal XY scans, from the bottom to the top of the cell at 1-μm intervals, of a single cell taking up FITC-LPS. The first image plane was ≈2 μm below the plane of the lipid bilayer containing P. aeruginosa FITC-LPS. White arrow indicates internalized FITC-LPS in one panel.
Figure 4.
Quantitative analysis of LPS extraction and uptake from a planar lipid bilayer. (a) Comparative endocytosis, by wt and ΔF508 CFTR cells, of wt P. aeruginosa FITC-LPS that had been reconstituted in a planar lipid bilayer. Wt CFTR cells endocytosed only a minimal amount of P. aeruginosa FITC-LPS with an incomplete core (lacks CFTR ligand). (b) Flow cytometer analysis of epithelial cell endocytosis of P. aeruginosa FITC-LPS. Maximal endocytosis required both wt CFTR epithelial cells and wt P. aeruginosa FITC-LPS with a complete outer core. Bars represent means and error bars represent standard errors (not visible because of small size for some bars). Asterisks (*) indicate a significant difference of P < 0.001 by ANOVA and Fisher probable least significant differences. (c) Inhibition of LPS uptake by incubation of wt CFTR cells with CFTR peptide 108–117 (black histogram). A scrambled version of CFTR peptide 108–117 (dashed line) showed no inhibition compared with the control (solid line). Incubation of LPS with the native peptide significantly inhibited uptake by epithelial cells (P < 0.001, Kolmogorov–Smirnov statistical analysis).
CFTR-Dependent Extraction of LPS from the Bacterial Surface.
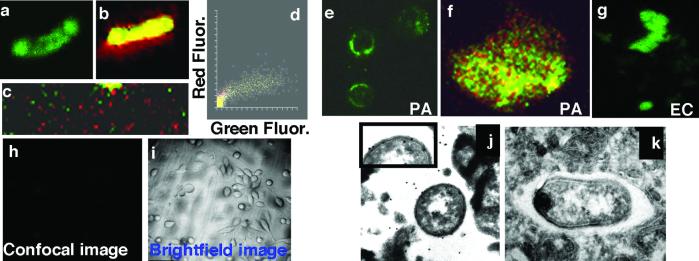
CFTR-dependent extraction of LPS from the P. aeruginosa outer membrane, followed by NF-κB nuclear translocation and cellular activation, could be the molecular basis for an effective innate immune response that is lacking in patients with two mutant CFTR alleles. Previous studies have shown that, after ingestion of Salmonella enterica into a cytosolic vacuole of epithelial cells, the LPS dissociates from the bacterial surface (17). The epithelial cell components or mechanisms involved in LPS dissociation have not been described, however. Here, FITC-labeled LPS was reconstituted into the bacterial outer membrane by fusing LPS-containing liposomes with P. aeruginosa cells and washing the bacteria extensively to remove unbound LPS (Fig. 5 a–d). FITC-labeled LPS was extracted from the reconstituted P. aeruginosa cells by epithelial cells expressing wt CFTR; such cells showed a pattern of diffuse cellular LPS uptake (Fig. 5e). Higher magnification views showed cells with diffuse, green LPS interspersed among punctate red-stained, intact bacterial cells (Fig. 5f). Epithelial cells expressing wt CFTR also were able to extract FITC-labeled P. aeruginosa LPS from E. coli bacteria reconstituted with P. aeruginosa LPS (Fig. 5g). Epithelial cells expressing only the ΔF508 CFTR allele did not take up P. aeruginosa FITC-LPS from cells reconstituted with this labeled lipopolysaccharide (Fig. 5 h and i).
Figure 5.
Epithelial cells extract LPS from bacterial cells. (a– c) Confocal images of wt FITC-LPS reincorporated into the outer membrane of live P. aeruginosa cells. (a) FITC fluorescence (green channel) of wt FITC-LPS in the membrane of P. aeruginosa strain PAO1-V. (b) PAO1-V cells with reincorporated wt FITC-LPS were counterstained with SYTO-17 (red), and cells were visualized in both the red and green channels. (c) Low-power (×400) view of a field of PAO1-V cells with reincorporated wt-FITC-LPS. (d) Colocalization box plot showing distributions of intensity of red and green fluorescent (Fluor) stains. (e–g) Cultured wt CFTR human epithelial cells extracted LPS from live bacterial cells. (e) Wt P. aeruginosa FITC-LPS as extracted from the outer membrane of PAO1-V by epithelial cells (×400) and visualized as a diffuse, green stain. (f) High-magnification (×1,000) scan of a cell from e, showing diffuse, green LPS and punctate, red P. aeruginosa cells. (g) Wt P. aeruginosa FITC-LPS reincorporated into E. coli was extracted from the outer membrane, as visualized by the diffuse green staining. (h and i) Wt P. aeruginosa FITC-LPS reincorporated into PAO1-V cells was not extracted from the outer membrane by epithelial cells expressing ΔF508 CFTR. Confocal (h) and bright-field (i) images, as indicated (×100). (j and k) Extraction and endocytosis of P. aeruginosa LPS in vacuoles of wt CFTR epithelial cells. (j) Gold particles (20 nm) demonstrated the presence of LPS on the osmiophilic coat of ingested P. aeruginosa PAO1-V as well as in a space in close proximity to the bacterium. Inset provides an enlarged view of a portion of the bacterial surface to demonstrate LPS molecules detected by the immunogold stain. (k) Control using normal rabbit serum. (×25,000.)
Immunoelectron microscopy was used to verify that cells expressing wt CFTR could promote extraction of LPS from intact whole P. aeruginosa cells. Substantial binding of anti-LPS antibody was observed within the cytoplasmic vacuole of epithelial cells, identifying both free LPS molecules and LPS molecules on the surface of P. aeruginosa cells that had been ingested into the vacuole (Fig. 5j). No such binding was seen when using normal rabbit serum (Fig. 5k). Thus, wt CFTR was required for extraction of P. aeruginosa LPS from the bacterial surface, although it is not clear whether the CFTR protein itself carried out the extraction or whether other cofactors also were needed.
Discussion
The recently completed human genome sequence predicts only 30,000–40,000 genes (18), perhaps twice that of the nematode Caenorhabditis elegans (18). Thus, a number of mechanisms, including alternative splicing of mRNA (18), use of small genetic elements to produce variable proteins such as antibodies and T cell receptors (19), and multifunctionality of individual proteins, are important for carrying out the variety of physiologic activities in a human organism. A protein with multiple functional phenotypes likely would use several of its intrinsic properties to generate these different phenotypes. Some of the multifunctional properties of CFTR have been characterized, including its ability to regulate other ion channels, to interact with other proteins through PDZ domains, and to traffic through the plasma membrane (20). Cell surface receptor trafficking and signaling are key mechanisms by which information about extracellular conditions is transmitted to the inside of the cell to activate appropriate cellular responses. Our results indicate that CFTR has acquired the ability to function as an endocytic PRM by using a small, 10-aa portion of an extracellular loop to bind to and remove the outer-core oligosaccharide of P. aeruginosa LPS and signal airway epithelial cell activation. CFTR that lacks the amino terminus required for binding to P. aeruginosa LPS still functions as a chloride ion channel (21), indicating that the amino terminus of CFTR is dispensable for this function. Thus, the multiple properties of CFTR allow for functionally disparate phenotypes to be mediated by one molecule.
CFTR functions as an endocytic PRM not only in epithelial cells in culture but also in lung tissues of wt, but not transgenic CF, mice. Previous work has shown that CFTR binding to P. aeruginosa results in bacterial internalization and elimination via cellular desquamation (4, 7, 22). The findings here extend the role of CFTR in innate immunity to P. aeruginosa infection by showing that the CFTR–P. aeruginosa interaction also leads to rapid NF-κB nuclear translocation, a process that occurs in an intact, living tissue as well as cultured cells. Li et al. (23) showed a commercial P. aeruginosa LPS preparation of undefined purityactivated NF-κB in cultures of cells with wt CFTR but did not define the CFTR dependence of this activation or perform an analysis in living tissue.
Some in vitro studies have suggested that P. aeruginosa does not enter polarized epithelial cells via the apical surface, where the CFTR protein is localized in the plasma membranes of cells in uninfected tissue (24, 25). It is clear, however, that the cells used in these studies had a much higher transepithelial resistance (>1,000 ohms) than the resistance of about 350–400 ohms measured in intact tissue (26). When polarized cells are treated with agents that disrupt tight junctions sufficiently to bring the resistance into a more physiologic range, then P. aeruginosa readily binds to and invades these cells via CFTR. P. aeruginosa infection also promotes CFTR trafficking to the nonapical surface of polarized epithelial cells via rapid translocation from cytoplasmic stores (27). Therefore, our finding of an in vivo role for CFTR in NF-κB activation because of P. aeruginosa infection validates the more detailed in vitro findings in cultured cells.
Controversy exists over the role of NF-κB translocation and associated activation of inflammatory cytokine production in the pathogenesis of CF lung disease. Some clinical studies detect inflammatory cytokines such as IL-8 that are likely to be controlled by NF-κB activation in the lungs of young, apparently uninfected CF patients (28, 29). Other studies have not found any differences in cytokine levels between uninfected CF and control subjects (30). One study indicated that alveolar macrophages are the principal source of IL-8 (31). Burns et al. (32) recently have shown that P. aeruginosa infection occurs early but intermittently in almost all CF patients by the age of 3, suggesting that studies detecting elevated IL-8 but no infection could have been measuring a response to a recently resolved infection. Some reports have indicated that transformed cells with nonfunctional CFTR show constitutive NF-κB nuclear translocation and associated production of IL-8 (33, 34) whereas others have not (35). Bedard et al. (36) finds no evidence for elevated production of IL-6, IL-8, or granulocyte/macrophage colony-stimulating factor when using primary epithelial cell cultures from the lungs of CF patients. Finally, Scheid et al. (37) report no difference in the basal levels of NF-κB translocation and IL-8 production when comparing CF and non-CF primary nasal epithelial cells in culture, wt and transgenic CF mouse tissues, and wt and CF cell lines, although more prolonged interaction between CF cells and P. aeruginosa does lead to NF-κB activation. Our observations reported here are consistent with the conclusion that CF epithelial cells do not constitutively translocate NF-κB to the nucleus.
Genetic deficiencies and polymorphisms in PRM genes recently have been appreciated to have a role in susceptibility and response to infectious pathogens (38). CF patients lack functional CFTR and are hypersusceptible to lung infections with a variety of bacterial pathogens; P. aeruginosa is the predominant pathogen that is responsible for most of the morbidity and mortality in CF. Our finding that CFTR is a PRM for P. aeruginosa supports the observations of others that genetic polymorphisms in PRM underlie disease susceptibility. The rapid and dynamic NF-κB nuclear translocation induced in wt lung epithelial cells by CFTR-promoted extraction of P. aeruginosa LPS appears to be a key molecular mechanism of innate immunity to this pathogen. This response is lacking in CF patients. Overall, the lack of proper PRM activity because of missing or nonfunctional CFTR compromises the ability of the lung epithelium to orchestrate effective innate immunity to P. aeruginosa infection.
Acknowledgments
We thank Michelle Lowe at the Confocal and Multiphoton Imaging Core Facility, Brigham and Women's Hospital, Gloria Meluleni and Fadie Coleman, who bred the transgenic cftr mice, and Ervin Meluleni for histopathology. This work was supported by National Institutes of Health Grants HL58398, HL32854, HL15157, and HL04277 and by an Institutional Interdisciplinary grant from Brigham and Women's Hospital. M.M.L. was a recipient of a fellowship from the Canadian Lung Association/Bayer. T.H.S. was supported by the Walter-Marget-Vereinigung zur Foerderung der Klinischen Infektiologie e.V.
Abbreviations
- CFTR
cystic fibrosis transmembrane conductance regulator
- LPS
lipopolysaccharide
- wt
wild type
- PAMP
pathogen-associated molecular pattern
- PRM
pattern recognition molecules
References
- 1.Schilling J D, Mulvey M A, Hultgren S J. Urology. 2001;57:56–61. doi: 10.1016/s0090-4295(01)01130-x. [DOI] [PubMed] [Google Scholar]
- 2.Vesy C J, Kitchens R L, Wolfbauer G, Albers J J, Munford R S. Infect Immunol. 2000;68:2410–2417. doi: 10.1128/iai.68.5.2410-2417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang I, Huang J F, Kishimoto H, Brunmark A, Peterson P A, Jackson M R, Surh C D, Cai Z, Sprent J. J Exp Med. 2000;191:1137–1148. doi: 10.1084/jem.191.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pier G B, Grout M, Zaidi T S. Proc Natl Acad Sci USA. 1997;94:12088–12093. doi: 10.1073/pnas.94.22.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pier G B, Grout M, Zaidi T, Meluleni G, Mueschenborn S S, Banting G, Ratcliff R, Evans M J, Colledge W H. Nature (London) 1998;392:79–82. doi: 10.1038/30006. [DOI] [PubMed] [Google Scholar]
- 6.Zhou L, Dey C R, Wert S E, Duvall M D, Frizzell R A, Whitsett J A. Science. 1994;266:1705–1708. doi: 10.1126/science.7527588. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder T H, Reiniger N, Meluleni G, Grout M, Coleman F T, Pier G B. J Immunol. 2001;166:7410–7418. doi: 10.4049/jimmunol.166.12.7410. [DOI] [PubMed] [Google Scholar]
- 8.Allewelt M, Coleman F T, Grout M, Priebe G P, Pier G B. Infect Immunol. 2000;68:3998–4004. doi: 10.1128/iai.68.7.3998-4004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coyne M J, Russell K S, Coyle C L, Goldberg J B. J Bacteriol. 1994;176:3500–3507. doi: 10.1128/jb.176.12.3500-3507.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haas C J, van Leeuwen H J, Verhoef J, van Kessel K P, van Strijp J A. J Immunol Methods. 2000;242:79–89. doi: 10.1016/s0022-1759(00)00207-6. [DOI] [PubMed] [Google Scholar]
- 11.Pohl P, Saparov S M, Borgnia M J, Agre P. Proc Natl Acad Sci USA. 2001;98:9624–9629. doi: 10.1073/pnas.161299398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dustin M L, Golan D E, Zhu D M, Miller J M, Meier W, Davies E A, van der Merwe P A. J Biol Chem. 1997;272:30889–30898. doi: 10.1074/jbc.272.49.30889. [DOI] [PubMed] [Google Scholar]
- 13.Dustin M L, Ferguson L M, Chan P Y, Springer T A, Golan D E. J Cell Biol. 1996;132:465–474. doi: 10.1083/jcb.132.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenney D, Pouliot K L, Wang Y, Murthy V, Ulrich M, Doring G, Lee J C, Goldmann D A, Pier G B. Science. 1999;284:1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
- 15.Esen M, Grassme H, Riethmuller J, Riehle A, Fassbender K, Gulbins E. Infect Immunol. 2001;69:281–287. doi: 10.1128/IAI.69.1.281-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato S, Ward C L, Krouse M E, Wine J J, Kopito R R. J Biol Chem. 1996;271:635–638. doi: 10.1074/jbc.271.2.635. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-del Portillo F, Stein M A, Finlay B B. Infect Immunol. 1997;65:24–34. doi: 10.1128/iai.65.1.24-34.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 19.Glusman G, Rowen L, Lee I, Boysen C, Roach J C, Smit A F, Wang K, Koop B F, Hood L. Immunity. 2001;15:337–349. doi: 10.1016/s1074-7613(01)00200-x. [DOI] [PubMed] [Google Scholar]
- 20.Kunzelmann K. News Physiol Sci. 2001;16:167–170. doi: 10.1152/physiologyonline.2001.16.4.167. [DOI] [PubMed] [Google Scholar]
- 21.Flotte T R, Afione S A, Conrad C, McGrath S A, Solow R, Oka H, Zeitlin P L, Guggino W B, Carter B J. Proc Natl Acad Sci USA. 1993;90:10613–10617. doi: 10.1073/pnas.90.22.10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pier G B, Grout M, Zaidi T S, Olsen J C, Johnson L G, Yankaskas J R, Goldberg J B. Science. 1996;271:64–67. doi: 10.1126/science.271.5245.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J D, Feng W, Gallup M, Kim J H, Gum J, Kim Y, Basbaum C. Proc Natl Acad Sci USA. 1998;95:5718–5723. doi: 10.1073/pnas.95.10.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleiszig S M, Evans D J, Do N, Vallas V, Shin S, Mostov K E. Infect Immunol. 1997;65:2861–2867. doi: 10.1128/iai.65.7.2861-2867.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee A, Chow D, Haus B, Tseng W, Evans D, Fleiszig S, Chandy G, Machen T. Am J Physiol. 1999;277:L204–L217. doi: 10.1152/ajplung.1999.277.1.L204. [DOI] [PubMed] [Google Scholar]
- 26.Barker P M, Boucher R C, Yankaskas J R. Am J Physiol. 1995;268:L270–L277. doi: 10.1152/ajplung.1995.268.2.L270. [DOI] [PubMed] [Google Scholar]
- 27.Zaidi T S, Lyczak J, Preston M, Pier G B. Infect Immunol. 1999;67:1481–1492. doi: 10.1128/iai.67.3.1481-1492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cantin A. Amer J Respir Crit Care Med. 1995;151:939–941. doi: 10.1164/ajrccm.151.4.7697269. [DOI] [PubMed] [Google Scholar]
- 29.Abman S H, Ogle J W, Harbeck R J, Butlersimon N, Hammond K B, Accurso F J. J Pediatr. 1991;119:211–217. doi: 10.1016/s0022-3476(05)80729-2. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong D S, Grimwood K, Carzino R, Carlin J B, Olinsky A, Phelan P D. Br Med J. 1995;310:1571–1572. doi: 10.1136/bmj.310.6994.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan T Z, Wagener J S, Bost T, Martinez J, Accurso F J, Riches D W H. Amer J Respir Crit Care Med. 1995;151:1075–1082. doi: 10.1164/ajrccm/151.4.1075. [DOI] [PubMed] [Google Scholar]
- 32.Burns J L, Gibson R L, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith A L, Ramsey B W. J Infect Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 33.Eidelman O, Srivastava M, Zhang J, Leighton X, Murtie J, Jozwik C, Jacobson K, Weinstein D L, Metcalf E L, Pollard H B. Mol Med. 2001;7:523–534. [PMC free article] [PubMed] [Google Scholar]
- 34.DiMango E, Ratner A J, Bryan R, Tabibi S, Prince A. J Clin Invest. 1998;101:2598–2605. doi: 10.1172/JCI2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pizurki L, Morris M A, Chanson M, Solomon M, Pavirani A, Bouchardy I, Suter S. Am J Pathol. 2000;156:1407–1416. doi: 10.1016/S0002-9440(10)65009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bedard M, McClure C D, Schiller N L, Francoeur C, Cantin A, Denis M. Am J Respir Cell Mol Biol. 1993;9:455–462. doi: 10.1165/ajrcmb/9.4.455. [DOI] [PubMed] [Google Scholar]
- 37.Scheid P, Kempster L, Griesenbach U, Davies J C, Dewar A, Weber P P, Colledge W H, Evans M J, Geddes D M, Alton E W. Eur Respir J. 2001;17:27–35. doi: 10.1183/09031936.01.17100270. [DOI] [PubMed] [Google Scholar]
- 38.Bax W A, Cluysenaer O J, Bartelink A K, Aerts P C, Ezekowitz R A, van Dijk H. Lancet. 1999;354:1094–1095. doi: 10.1016/s0140-6736(99)02563-5. [DOI] [PubMed] [Google Scholar]