Abstract
HIV-1 entry into cells involves formation of a complex between gp120 of the viral envelope glycoprotein (Env), a receptor (CD4), and a coreceptor, typically CCR5. Here we provide evidence that purified gp120JR-FL–CD4–CCR5 complexes exhibit an epitope recognized by a Fab (X5) obtained by selection of a phage display library from a seropositive donor with a relatively high broadly neutralizing serum antibody titer against an immobilized form of the trimolecular complex. X5 bound with high (nM) affinity to a variety of Envs, including primary isolates from different clades and Envs with deleted variable loops (V1, -2, -3). Its binding was significantly increased by CD4 and slightly enhanced by CCR5. X5 inhibited infection of peripheral blood mononuclear cells by a selection of representative HIV-1 primary isolates from clades A, B, C, D, E, F, and G with an efficiency comparable to that of the broadly neutralizing antibody IgG1 b12. Furthermore, X5 inhibited cell fusion mediated by Envs from R5, X4, and R5X4 viruses. Of the five broadly cross-reactive HIV-1-neutralizing human monoclonal antibodies known to date, X5 is the only one that exhibits increased binding to gp120 complexed with receptors. These findings suggest that X5 could possibly be used as entry inhibitor alone or in combination with other antiretroviral drugs for the treatment of HIV-1-infected individuals, provide evidence for the existence of conserved receptor-inducible gp120 epitopes that can serve as targets for potent broadly cross-reactive neutralizing antibodies in HIV-1-infected patients, and have important conceptual and practical implications for the development of vaccines and inhibitors.
Keywords: AIDS‖antibody‖vaccines‖inhibitors
Binding of the HIV-1 envelope glycoprotein (Env, gp120-gp41) to CD4 and coreceptors initiates a series of conformational changes that are the heart of the fusion machinery leading to viral entry (1, 2). The elucidation of the nature of the Env conformational changes is not only a clue to the mechanism of HIV-1 entry but may also provide new tools for the development of inhibitors and vaccines (3–5). It has been proposed that the interaction of coreceptor molecules with the Env–CD4 complex leads to intermediate Env conformations that may include structures conserved among various HIV-1 isolates that could be used as vaccines (6, 7). Currently, there are only four well characterized monoclonal antibodies with broadly neutralizing activity—the anti-gp120 Abs b12 (8) and 2G12 (9), and the anti-gp41 Abs 2F5 (10) and 4E10/Z13 (11). None of these antibodies recognizes receptor-inducible epitopes. The high-affinity binding antibody b12, which interacts with the CD4 binding site on gp120 and is able to neutralize a variety of primary HIV-1 isolates, was identified by selection of human phage display libraries against gp120 (8).
We hypothesized that the use of purified Env-CD4-coreceptor complexes as the selecting antigen for human phage display libraries might provide a new approach for elucidation of the nature of the intermediate Env structures and for development of broadly neutralizing antibodies. Some of the transient Env conformations on the pathway to entry might be associated with such complexes and used for selection of human monoclonal antibodies, which in turn could be helpful for characterization of HIV-1 entry intermediates. These antibodies could be broadly neutralizing if their epitopes include conserved intermediate structures that are exposed during entry. In addition, the use of complexed coreceptor molecules may prevent selection of antibodies against the coreceptor binding site on gp120 that may be minimally accessible following attachment of native Env to cell-surface-associated CD4; such antibodies as 17b and CG10 are only weakly neutralizing against primary isolates (12). Here we report the identification of human antibody Fab, X5, isolated from a phage display library by using a trimolecular complex of gp120, the receptor CD4, and the coreceptor CCR5 as the screening agent. The epitope of this antibody is inducible by CD4 and its exposure is slightly enhanced by the major HIV-1 coreceptor CCR5. The antibody potently neutralizes R5, X4, and R5X4 viruses, including primary isolates from different clades.
Materials and Methods
Cells, Viruses, Plasmids, Soluble CD4 (sCD4), gp120, gp140, and Monoclonal Antibodies (mAbs).
3T3 cells expressing CD4 and CCR5 were a gift from D. Littman (New York University). Cf2Th cells expressing high concentrations of CCR5 were a gift from J. Sodroski (Dana Farber Institute, Boston); the parental cells were purchased from American Type Culture Collection and used as a negative control. The stable cell line TF228 expressing LAI Env was a gift from Z. Jonak (SmithKline Beecham, Philadelphia) through R. Blumenthal (National Cancer Institute–Frederick). The CEM cells expressing CCR5 (CEM-CCR5) were a gift from J. Moore (Cornell University, New York). The T-cell line H9 was obtained from the Centralized Facility for AIDS Reagents (CFAR), U.K. Peripheral blood mononuclear cells (PBMCs) were isolated and pooled from three wild-type CCR5 donors. All HIV isolates were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (ARRRP). Recombinant vaccinia viruses used for the reporter gene fusion assay have been described (13). Plasmids used for expression of various Envs were obtained through the ARRRP from B. Hahn (University of Alabama, Tuscaloosa, AL). Two-domain sCD4 was obtained from the ARRP. Purified gp12089.6 and gp14089.6 were produced by recombinant vaccinia virus (gift of R. Doms, University of Pennsylvania, Philadelphia) with a combination of lentil lectin affinity chromatography and size exclusion chromatography. Recombinant gp140 from the primary isolates 92UG037.8, 92HT593.1, 93MW965.26, and 93ZR001.3, and gp120 from 93ZR001.3 were prepared similarly by using clones obtained from the ARRRP. Recombinant gp120JR-FL was a gift from A. Schultz and N. Miller (National Institute of Allergy and Infectious Diseases, Bethesda). The single-chain fusion protein gp120Bal–CD4 (14) was a gift from T. Fouts (Institute of Human Virology, Baltimore). gp120JR-FL and gp140JR-FL with deleted variable loops and the disulfide bond stabilized 140HXB2 have been described (15, 16). The anti-gp120 mAbs 17b and 48d were previously derived by a method described in ref. 17 and characterized in ref. 18; 23e and 21c are newly derived and characterized (S.-H. Xiang, N. Doka, R. K. Choudhary, J. Sodroski, and J.R., unpublished data). The anti-gp120–CD4-complex-specific mAb CG10 has been described (19). Various other anti-gp120 mAbs have been described: 19b (20); F91, A32, G3–136, and G3–519 (21); b12 (8); 2G12 (9). The mAb MAG45 was a gift from C.-Y. Kang (IDEC Pharmaceuticals, San Diego). The anti-CCR5 mAb 5C7 was a gift from L. Wu (Millenium Pharmaceuticals, Cambridge, MA).
Production, Purification, and Quantification of gp120–CD4–CCR5 Complexes.
Purified complexes between gp120, CD4, and CCR5 were produced by a previously described methodology (22) but modified to allow production of larger amounts. NIH 3T3 transfectants (109 in 100 ml) expressing high levels of CD4 and CCR5 were washed twice with cold (4°C) PBS, then pelleted by centrifugation and resuspended in 100 ml of lysis buffer (1% Brij97, 5 mM iodoacetamide, added immediately before use, 150 mM NaCl, 20 mM Tris (pH 8.2), 20 mM EDTA, and protease inhibitors) at 4°C for 40 min with gentle mixing. The nuclei were pelleted by centrifugation at 4°C. The anti-CCR5 antibody 5C7 at 2 μg/ml and protein G-Sepharose beads (Sigma) (1 ml) prewashed with PBS were added to the cell suspension and incubated at 4°C for 14 h. The beads were then washed five times with 100 ml of ice cold lysis buffer and incubated with gp120JR-FL at 5 μg/ml in 20 ml of lysis buffer for 1 h at 4°C. They were again washed five times with 100 ml of cold lysis buffer and once with cold PBS, incubated with 0.2% formaldehyde overnight, and finally washed twice with cold lysis buffer. They contained approximately 10 μg of CD4, 10 μg of CCR5, and 20 μg of gp120 as quantified by calibrating amounts of soluble CD4, detergent-solubilized CCR5 (dsCCR5), and gp120. For quantification of CD4 and gp120, two duplicate samples each containing 0.1% of the total amount of bead-associated gp120–CD4–CCR5 complexes were used. They were eluted by adding 4× sample buffer for SDS/PAGE gel, kept overnight at 37°C and run on a 10% SDS/PAGE gel simultaneously with calibrating amounts (1, 3, 10, 30, and 100 ng) of soluble four-domain CD4 or gp120. Quantification of CCR5 was performed similarly, but the calibrating amounts of dsCCR5 were themselves precalibrated with a 38-aa residue N-terminal CCR5 peptide by using rabbit polyclonal anti-CCR5 antibodies against this peptide provided by H. Golding (Food and Drug Administration, Bethesda). The samples were electrophoretically transferred to nitrocellulose membranes. The membranes were blocked with 20 mM Tris⋅HCl (pH 7.6) buffer containing 140 mM NaCl, 0.1% Tween-20, and 5% nonfat powdered milk. For Western blotting these membranes were incubated with anti-CD4, anti-gp120, or anti-CCR5 antibodies, then washed, incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies, and developed by using the supersignal chemiluminescent substrate from Pierce (Rockford, IL). The images were acquired by a Bio-Rad phosphorimager. The signal from the calibrating molecules was integrated for each band and plotted on a calibration curve for the signal vs. amount dependence. The amounts of CD4, CCR5, and gp120 were then calculated by interpolation using calibration curves. DsCCR5 was produced by a previously described methodology (23).
Phage Display Library Screening.
A phage library (IgG1κ) from a seropositive individual with a relatively high cross-clade neutralizing titer (FDA2) (24, 25) was used. Phage (50 μl) were preadsorbed on protein G beads in PBS for 1 h at 37°C. Unbound phage were recovered by centrifugation at 4°C and then incubated with protein G beads associated with gp120–CD4–CCR5 complexes for 2 h at 4°C under gentle agitation. Beads were washed ten times with PBS containing 0.5% Tween. Phage were eluted from the beads by incubation with 50 μl of 0.1 M HCl-glycine (pH 2.2) solution containing BSA at 1 mg/ml for 10 min at room temperature. The solution was neutralized with 3 μl of 2 M Tris-base. XL1-Blue E. coli cells were reinfected and panning repeated for total of five rounds (26).
Preparation of Soluble Fab Fragments.
Phagemid DNA was isolated from the panned library and digested with SpeI and NheI to remove the gene III fragment and self-religated as described (27). The Δgene III-phagemid library was used to transform XL1-Blue E. coli. Sixty individual clones were grown up. The corresponding Fabs were obtained by lysing the cell pellet. Cells were frozen in a dry ice-ethanol bath for 5 min followed by thawing in a 37°C water bath. This process was repeated four times and the cell debris was pelleted by centrifugation at 15,000 rpm for 15 min at 4°C. Soluble Fabs were produced as described (26). Protein G columns were used for purification.
ELISA Analysis of Fab Supernatants.
ELISA wells were coated overnight at 4°C with 50 μl of gp120 (10 μg/ml in PBS), blocked in 100 μl of 3% BSA/PBS for 1 h at 20°C. After five washes with 0.05% Tween 20/PBS washing buffer (WB), wells were incubated with 50-μl Fab supernatants for 1 h at 20°C. After ten washes with WB, 50 μl of a 1:1,000 dilution of alkaline phosphatase-conjugated goat anti-human IgG F(ab′)2 was added and incubated for 1 h at 20°C. Following ten washes with WB, the assay was developed at 20°C for 30 min with p-nitrophenyl phosphate substrate (Sigma) and monitored at 405 nm. Positive clones showed absorbance values >1. The CDR3 region of the heavy chains from positive clones were sequenced using the SeqGz primer (5′-GAAGTAGTCCTTGACCAG-3′).
ELISA Binding Assays.
The ELISA D7324 capture assay has been described (28). The amount of captured gp120s and gp140s was evaluated by using Ig from HIV-infected patients (HIVIG). The amount of bound X5 was detected by Fab-specific alkaline phosphatase conjugate (Accurate Antibodies, Westbury, NY); IgG Fc of the bound antibodies was detected with goat anti-mouse or anti-human IgG Fc alkaline phosphatase conjugate and quantitated by a colorimetric assay that measures the optical density at 490 nm (OD 490 nm). In another ELISA assay gp120(gp140) or sCD4–gp120(gp140) were coated directly on 96-well Nunc-Immuno Maxisorp surface plates (Nalge Nunc) by incubation of 0.1 ml of solution containing 100 ng of the protein at 4°C overnight. Plates were treated with 4% nonfat milk (Bio-Rad) to prevent nonspecific binding, then washed with TBS (50 mM Tris⋅HCL (pH 7.5)/150 mM NaCl) containing 0.1% Tween-20. DsCCR5 was diluted in cymal lysis buffer (1% Cymal-5/100 mM (NH4)2SO4/20 mM Tris/10% glycerol) to 1 μg/ml and incubated at 4°C overnight. Unbound molecules of dsCCR5 were washed and X5 was added. Bound X5 or biotinylated X5 was detected by anti-human F(ab′)2-HRP or streptavidin-HRP, respectively, and quantitated by a colorimetric assay that measures the optical density at 405 nm (OD 405 nm). Biotinylated X5 was prepared by incubation with 2 mM biotin [prepared from solid NHS-LS-Biotin (Pierce, CA) dissolved at 200 mM in DMSO as stock solution] on wet ice for 1 h. The biotinylation was quenched with 20 mM glycine on ice for 15 min.
Flow Cytometry Assay.
The flow cytometry measurements were performed with FACSCalibur (Becton Dickinson) by using CellQuest software, and the mean fluorescence intensity from the acquisition of 10,000 gated events from cells with antibodies bound to cell-surface-associated Env was used as a measure of binding.
Cell–Cell Fusion Assays.
The β-gal reporter gene cell–cell fusion assay has been described (13). Fusion induced by sCD4 (29) was performed by incubation of 105 293 cells expressing Env with sCD4 (1 μg/ml) at 37°C for 30 min before mixing with 105 293 cells expressing CXCR4 or CCR5 (after infection with recombinant vaccinia viruses) for 2 h at 37°C. The inhibitory effect of X5 was evaluated by mixing the effector cells with X5 for 30 min at 37°C and then performing the fusion assay. β-Gal activity was quantified by a colorimetric assay that measures the optical density at 595 nm (OD 595).
HIV Neutralization Assays.
The peripheral blood mononuclear cell (PBMC)-based neutralization assay was performed essentially as described (30). Briefly, serial 2-fold dilutions of Abs in 50 μl were incubated with an equal volume of virus containing 100 TCID50 for 1 h at 37°C and added to 100 μl of PHA-activated PBMC (5 × 105/ml). The calculated neutralization titers refer to the Ab concentration present during this incubation step. After overnight incubation, the cells were washed twice with tissue culture medium. On day 4, 100 μl of medium was replaced with fresh tissue culture medium. Triplicate samples were taken on days 4 and 7, treated with 1% Empigen (Calbiochem), and tested for p24 Ag content by using an in-house ELISA, as described (31). When the values for the p24 concentration at day 7 were saturated, data for day 4 were used for analysis. The pseudotype virus neutralization assay was performed in triplicate by using infection with a luciferase reporter HIV-1 Env pseudotyping system as described (32).
Results
Selection of a Phage Fab (X5) with High Affinity for gp120–CD4–CCR5 Complexes.
An anti-CCR5 mAb was used for preparation of the gp120–CD4–CCR5 complexes to allow only those CD4 molecules constitutively associated with CCR5 (22) to bind gp120, thus ensuring the interaction of most of the gp120 molecules with CCR5. The specific interaction of dsCCR5 with gp120 in the trimolecular gp120–CD4–CCR5 complex was also tested by an experiment where gp120 displaced RANTES bound to dsCCR5 (X.X. and D.S.D., unpublished work). For panning we used complexes containing about 10 μg of CD4, 10 μg of dsCCR5, and 20 μg of gp120JR-FL corresponding to molar ratios of 1:1:1. After five rounds of panning, one phage Fab was selected. The CDR3 sequence of the heavy chain of this Fab was unique—it was not found either in the laboratory of D.R.B. or reported in the GenBank database. Phage-displayed X5 exhibited binding activity to protein G cross-linked to Sepharose beads with an apparent affinity (equilibrium dissociation constant) of 1.4 nM, which was 15-fold lower than the affinity for gp120JR-FL–sCD4 complexes (0.09 nM).
Binding of X5 to gp120 and gp140 from Different Isolates Is Significantly Increased by CD4 and Slightly Enhanced by dsCCR5.
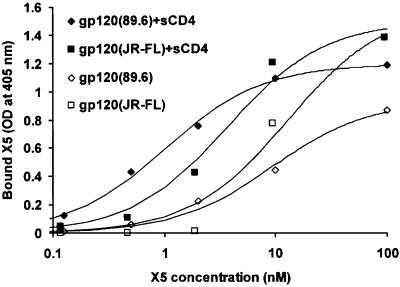
X5 bound gp120 from the primary isolate 89.6 with an affinity (equilibrium dissociation constant) of 9.4 nM, which was increased 10-fold to 1 nM after binding of sCD4 to gp120 as measured by an ELISA assay (Fig. 1). Similarly, the affinity of X5 binding to gp120JR-FL was increased from 10 nM to 2 nM after addition of sCD4 (Fig. 1). Binding of X5 to gp120 from the dual tropic primary isolate 93ZR001.3 (Clade D) and gp120HXB2 was also increased significantly by CD4 (data not shown). X5 also bound a single-chain fusion protein, where gp120Bal and CD4 are intramolecularly associated (14), at a level similar to that for the sCD4–gp120JR-FL complex (data not shown).
Figure 1.
Binding isotherms of X5 to gp120 and gp120–sCD4. Gp120 and gp120–sCD4 complexes were coated directly on 96-well plates and washed, and X5 was added at the indicated concentrations. Bound X5 was detected by anti-human F(ab′)2-HRP and represented as optical density at 405 nm. The background was estimated by the amount of X5 bound to BSA and subtracted. The data were fitted to the Langmuir adsorption isotherm [B/Bmax = X5/(Kd + X5), where B is the amount of bound X5, Bmax is the maximal amount of bound X5, X5 is its bulk concentration, and Kd is the equilibrium dissociation constant]. The continuous lines represent fitting of the data for X5 binding to gp120 [data represented by empty squares (JR-FL) and diamonds (89.6)] and sCD4–gp120 (data represented by solid symbols).
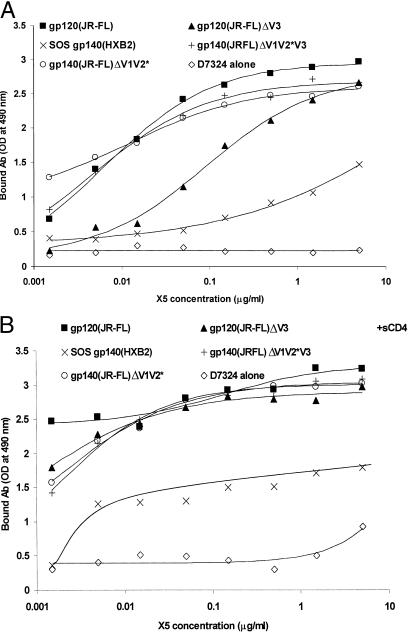
X5 bound a cleaved disulfide bond stabilized gp140HXB2 (16), and the binding was increased by sCD4 (Fig. 2). Binding of X5 to gp14089.6 and its complex with sCD4 was somewhat lower but comparable to the binding to gp12089.6 and sCD4–gp12089.6. Recombinant gp140 from primary isolates from different clades (92UG037.8, Clade A, R5; 92HT593.1, Clade B, R5X4; 93MW965.26, Clade C, R5; 93ZR001.3, Clade D, R5X4) complexed with sCD4 bound X5 (data not shown). The binding was at levels similar or lower than to 89.6 gp140 complexed with sCD4; for two isolates (92UG037.8 and 92HT593.1) binding to gp140 in the absence of sCD4 was significantly lower than to gp14089.6. X5 also bound gp120 with deleted V3 loop, and gp140 with deleted V1 and V2 or V1, V2, and V3 loops (15) in a CD4-dependent manner (Fig. 2).
Figure 2.
Binding of X5 to gp140 (120)s lacking variable loops and disulfide bond stabilized gp140. Wild-type gp120JR-FL, gp120 with deleted V3 loop (gp120JR-FLΔV3), and gp140 with deleted V1 and V2 loops (gp140JR-FLΔV1V2*) or deleted V1, V2, and V3 loops (gp140JR-FLΔV1V2*V3) (15), as well as cleaved disulfide bond stabilized gp140HXB (SOS gp140HXB2) (16), were captured by the antibody D7324 to 96-well plates and X5 added at the indicated concentrations in the absence (A) or presence (B) of sCD4 (2 μg/ml). The amount of bound X5 was detected by Fab-specific alkaline phosphatase conjugate and represented as optical density at 490 nm.
The amount of bound X5 to gp120JR-FL–sCD4 complexes and to single-chain gp120Bal–CD4 molecules was slightly (on average 1.3-fold) enhanced in presence of dsCCR5 (data not shown). Although small, this increase was statistically significant (P = 0.005 as calculated by a t test); to ensure that the enhancement was not due to artifacts and was reproducible, this experiment was performed six times during a 3-month period, using different aliquots of dsCCR5 either freshly prepared or stored for up to 1 week. In addition, the native conformation of dsCCR5 was tested by using the conformationally dependent anti-CCR5 mAb 2D7 and sCD4–gp120 complexes, which bound specifically to dsCCR5 (data not shown; ref. 23). Finally, in another set of experiments the enhancing effect of dsCCR5 was partially reversed by adding RANTES or MIP-1β (data not shown). The results of these control experiments suggested that the enhancement of the X5 epitope by dsCCR5 was specific and reproducible. X5 did not bind denatured gp12089.6, suggesting discontinuity of its epitope. Therefore, X5 binds to a conserved conformational gp120 epitope that is significantly affected by CD4 and slightly by CCR5 but differs from their binding sites.
X5 Binding to Cell-Surface-Associated Oligomeric Env.
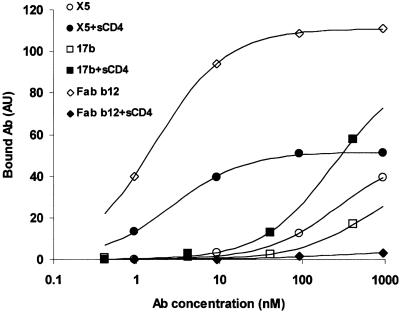
X5 bound oligomeric, fusion-active gp120-gp41MN expressed at the surface of HIV-1MN-infected H9 cells (Fig. 3). For this experimental system, the X5 affinity (2.7 nM) in the presence of sCD4 (20 μg/ml) was comparable to that of the CD4bs-specific mAb Fab b12 (1.7 nM) and significantly higher than the affinity of 17b, which was previously reported to exhibit an increased affinity to the gp120–CD4 complex (33). These results suggest that the X5 epitope is exhibited on native Env structures.
Figure 3.
Binding of X5, Fab b12, and 17b to cell-surface-associated oligomeric gp120-gp41. The T-cell line H9 was infected with the TCLA X4 HIV-1MN isolate for 10 days. At this time postinfection there was no detectable CD4 remaining at the cell surface, no syncytium formation in the culture, but strong Env expression detected using gp120-specific mAbs and flow cytometry. These H9 cells were preincubated with sCD4 (20 μg/ml) or buffer alone for 1 h at 20°C, then incubated with Fab X5, 17b, or Fab b12 at the indicated concentrations. The amount of bound antibodies was measured by flow cytometry and represented in arbitrary units. The continuous lines represent fitting of the binding data by using the Langmuir adsorption isotherm as described in the legend of Fig. 1.
X5 Competition with Known Antibodies to gp120.
To further characterize the X5 epitope, we measured X5 competition with anti-gp120 mAbs in the presence or absence of sCD4. The major results are (Table 1): (i) X5 competed to varying degrees with the antibodies 17b, CG10, 48d, 23e, and 21c, which bind CD4-inducible epitopes. (ii) X5 binding to gp120 was enhanced by the A32 mAb, which binds a CD4-inducible epitope; in turn X5 binding enhanced the exposure of the A32 epitope (data not shown). (iii) X5 competed to some extent with mAbs to the CD4 binding site such as IgG1b12 and F91. (iv) X5 did not compete with mAbs against other regions of gp120 including the V3 (19b) and V2 (G3–519) loops, C1 (MAG45, C11), C3/V4 (2G12), C4 (G3–136), and C5 (C11). (v) The competition pattern was not significantly dependent on the Env used, at least for the Envs from the two isolates investigated in detail (89.6 and JR-FL). These results suggest that the X5 epitope is likely located in close proximity to the coreceptor and CD4 binding sites.
Table 1.
Competition of anti-gp120 mAbs with X5 for gp120JR-FL and gp12089.6
| mAb | Epitope | Env | Competition
|
|
|---|---|---|---|---|
| −sCD4 | +sCD4 | |||
| 17b | CD4i | 89.6 | + | ++ |
| 17b | CD4i | JR-FL | + | ++ |
| CG10 | CD4i | 89.6 | 0 | ++ |
| CG10 | CD4i | JR-FL | 0 | +++ |
| A32 | CD4i(C1–C4) | 89.6 | −−− | − |
| A32 | CD4i(C1–C4) | JR-FL | −− | −− |
| 48d | CD4i | 89.6 | + | + |
| 23e | CD4i | 89.6 | + | + |
| 21c | CD4i | 89.6 | + | + |
| F91 | CD4bs | JR-FL | + | NA |
| IgG1b12 | CD4bs | JR-FL | + | NA |
| 19b | V3 | JR-FL | 0 | 0 |
| G3-136 | V2 | JR-FL | 0 | 0 |
| MAG45 | C1 | JR-FL | 0 | 0 |
| C11 | C1–C5 | JR-FL | 0 | 0 |
| 2G12 | C3/V4 | JR-FL | 0 | 0 |
| G3–519 | C4 | JR-FL | 0 | 0 |
| X5 | CD4i | 89.6 | +++ | +++ |
Gp120JR-FL was captured by the anti-gp120 Ab D7324 and incubated with X5 (5 μg/ml) and various antibodies at different concentrations in absence or presence of sCD4 (2 μg/ml), and IgG Fc of the bound antibodies was detected with goat anti-mouse or anti-human IgG Fc alkaline phosphatase conjugate. Gp12089.6 was directly coated on 96-well plates and incubated with biotinylated X5 (0.5 μg/ml) and various antibodies, and the amount of bound X5 was detected by using streptavidin-HRP. +, Denotes some competition; ++, significant competition; +++, complete displacement; −, various degrees of enhancement; 0, lack of measurable effects; NA, not applicable.
Inhibition of HIV-1 Infection and Env-Mediated Cell Fusion.
To determine the breadth and potency of HIV-1 neutralization by X5 we measured its ability to inhibit infection of peripheral blood mononuclear cells (PBMCs) by primary isolates from different clades in comparison with the potent broadly neutralizing antibody b12 (Table 2). X5 neutralized all tested primary isolates with a potency that was generally comparable to or in some cases better than IgG1 b12. The potency was particularly noteworthy given that X5 was assessed as an Fab fragment; its potency may improve as a whole IgG1 molecule. This molecule is currently being generated. X5 was also able to neutralize several representative R5 (JR-FL and Bal), X4 (NL4–3), and X4R5 (89.6) viruses at IC50 in the range of 0.1–10 μg/ml in another assay based on a luciferase reporter HIV-1 Env pseudotyping system (data not shown).
Table 2.
Neutralization of HIV infections of PBMCs by X5 and b12
| Isolate | Clade | IC50 μg/ml
|
IC90 μg/ml
|
n
|
|||
|---|---|---|---|---|---|---|---|
| Fab X5 | IgG1 b12 (Fab b12) | Fab X5 | IgG1 b12 (Fab b12) | Fab X5 | IgG1 b12 (Fab b12) | ||
| RW009 | A | 11 | >200 | 56 | >200 | 1.3 | 0.47 |
| MN | B | 35 | 0.033 | 96 | 0.42 | 2.2 | 0.86 |
| JR-FL | B | 14 | 0.017 | 57 | 3.4 | 1.6 | 0.41 |
| JR-CSF | B | 4.2 | 4.3 | 21 | 6.7 | 1.4 | 5.0 |
| SF2 | B | 6.4 | 2.8 | 29 | 25 | 1.4 | 1 |
| SF162 | B | 8.7 | 2.0 | 16 | 4.1 | 3.7 | 3.0 |
| Bal | B | 3.7 | 4.2 | 20 | 23 | 1.3 | 1.3 |
| BR025 | C | 0.29 | 15 | 22 | >200 | 0.51 | 0.39 |
| UG024 | D | 42 | 31 (69) | 65 | 60 (560) | 5 | 3.3 (1) |
| BR029 | F | 6.5 | 27 | 77 | >200 | 0.89 | 0.34 |
| RU570 | G | 125 | 4.4 | 230 | 9.6 | 3.7 | 2.8 |
| G3 | G | 10 | 15 | 32 | 61 | 1.9 | 1.6 |
Serial 2-fold dilutions of X5, IgG1 b12, or Fab b12 (results shown in parentheses) in 50 μl were incubated with an equal volume of virus containing 100 TCID50 for 1 h at 37°C and added to 5 × 105/ml PHA-stimulated PBMCs in 100 μl of medium. The calculated neutralization titers refer to the Ab concentration present during this incubation step. After overnight incubation, the cells were washed twice with medium, triplicate samples were taken, and p24 concentration measured using an in-house ELISA, as described (31). The data were fitted with a Hill-type function I = Kn/(Kn + CAbn), where I is the extent of infection defined as the p24 concentration in presence of Ab of concentration C divided by the p24 concentration in the absence of Ab, K is 50% inhibitory concentration (IC50), and n is parameter of fitting. IC90 was calculated from this formula as 9(1/n)K.
X5 inhibited cell–cell fusion mediated by Envs of primary isolates from different clades with a potency comparable to that of IgG b12 as measured by a reporter gene assay (Table 3). X5 almost completely inhibited sCD4-induced fusion mediated by Envs from NL4–3, HXB2, 89.6, JR-FL, ADA, Bal, and SF162 at very low (100 ng/ml) concentrations; inhibition of fusion mediated by X4 Envs was less efficient (71–83% inhibition) compared with inhibition of R5 Env-mediated fusion (96–100%) (data not shown). These results suggest that X5 is a potent, broadly HIV-1-neutralizing antibody.
Table 3.
Inhibition of cell–cell fusion by X5 and b12
| Envelope | Clade | Antibody
|
|
|---|---|---|---|
| X5 | b12 | ||
| UG037.8 | A | 29 | 24 |
| RW020.5 | A | 56 | 58 |
| US715.6 | B | 35 | 28 |
| HT593.1 | B | 20 | 39 |
| US005.11 | B | 24 | 47 |
| 89.6 | B | 100 | 100 |
| BR025.9 | C | 62 | 22 |
| ZR001.3 | D | 44 | 39 |
| TH022.4 | E | 17 | 34 |
| BR029.2 | F | 42 | 24 |
| BR019.4 | F/B | 62 | 63 |
| UG975.1 | G | 25 | 41 |
105 293 cells, transfected with plasmids encoding various HIV Envs under the control of T7 promoter and infected with recombinant vaccinia virus encoding T7 polymerase gene, were preincubated with X5 or IgG b12 at 100 μg/ml for 30 min at 37°C, and then mixed with 105 CEM-CCR5 cells infected with recombinant vaccinia virus encoding β-galactosidase gene. The extent of cell fusion was quantified colorimetrically 2 h after mixing the cells. The data are averages of duplicate samples and presented as % of fusion inhibition.
Discussion
The major result of this study is the identification of a human monoclonal antibody Fab, X5, that binds to a conserved epitope on gp120 induced by CD4 that is different from the coreceptor binding site and is slightly enhanced by CCR5 binding. X5 neutralizes a broad range of HIV-1 isolates. To our knowledge, this is the first report of an antibody selected for binding to purified gp120–CD4-coreceptor complexes; compared with the four known potent broadly neutralizing antibodies (b12, 2F5, 2G12, and 4E10/Z13), it is the only one against a receptor-inducible epitope. The use of a coreceptor complexed with gp120–CD4 allowed for selection of an antibody to an epitope that differs from the coreceptor binding site and is somewhat more exposed after CCR5 binding. Interestingly, an Fab almost identical to X5 was also selected by panning the FDA2 library against proteoliposomes (34) associated with gp160YU2, suggesting that the X5 epitope can become available in differing environments. The characterization of epitopes such as that of X5 may provide valuable information for the nature of the initial conformational changes of the Env required for its fusogenic activity.
It has been demonstrated (35) that binding of CD4 to the Env induces conformational changes as measured by the increased binding of antibodies to the V3 loop of gp120 and its enhanced cleavage by an exogenous protease. Subsequent studies (36, 37), including the solution of the crystal structure of a trimolecular CD4–gp120–17b complex, provided a wealth of information about the gp120 conformation in presence of CD4. However, the conformational changes induced by coreceptors, which are most likely responsible for driving the fusion process to completion, are less well characterized. It was proposed that the interactions of the coreceptors with Env and possibly with CD4 may help in the relocation of the fusion peptide from the interior of gp41 to a position close to the surface of the host membrane (6). The existence of major conformational changes specifically induced by coreceptor interactions with the Env–CD4 complex was demonstrated by using fluorescent dyes as indicators of exposure of hydrophobic regions of the Env or membrane destabilization (38, 39). Models of fusion intermediate states containing coiled coils derived from gp41 sequences have also been proposed, and experimental evidence has been provided for their existence (3, 40–46). However, the nature of the conformational changes leading to these intermediate structures remains unclear. The new antibody identifies an epitope that may play a role in the coupling of the gp120 conformational changes to those in gp41. One can speculate that the X5 binding to the CD4–gp120–CCR5 complex interferes with those conformational changes in gp120 that are required to transduce an activation signal for gp41 to undergo the major conformational changes leading to fusion.
The X5 epitope is conserved and partially overlaps the epitope of the antibody 17b, which binds a CD4 inducible epitope that partially overlaps the coreceptor binding site on gp120. Antibody 17b neutralizes weakly TCLA HIV-1 and does not neutralize primary isolates (37). The X5 epitope is significantly different in that it is outside the CCR5 binding site. Its precise localization is currently under investigation, but it is likely located at close proximity to the coreceptor and CD4 binding sites, as also indicated by the X5 competition with b12, which binds to the CD4 binding site on gp120 (47). X5 binds with higher affinity to gp120 than 17b and neutralizes various HIV-1, including primary isolates with potency comparable to that of IgG1 b12. The crystal structure of gp120 complexed with CD4 and the Fab 17b reveals an orientation of the 17b epitope toward the target cell membrane (36). It was also shown recently that access to the CD4-induced coreceptor-binding domain on gp120 is largely blocked at the fusing cell membranes and is unlikely to represent a target for neutralizing antibodies (48). Thus one might speculate that the X5 epitope is oriented differently than the 17b epitope and the coreceptor binding site, and it is easier to access in agreement with its potent HIV-neutralizing activity. The precise mechanism of HIV-1 inhibition by X5 is likely to involve blocking of post-receptor binding events and is currently under investigation.
The identification of X5 also shows that antibodies against conserved epitopes whose exposure is enhanced by interaction with receptor and coreceptor molecules may exist in infected individuals. Importantly, the results imply that certain epitopes, distinct from the coreceptor binding site, can be recognized by the immune system and elicit broadly HIV-1-neutralizing antibodies. This result may have important conceptual consequences for the development of vaccines able to induce broadly cross-reactive antibodies. In addition, X5 could be possibly used as entry inhibitor alone or in combination with other antiretroviral drugs for the treatment of HIV-1-infected individuals.
Acknowledgments
We wish to thank J. Sodroski, J. Moore, E. Berger, R. Doms, T. Fouts, L. Wu, G. Quinnan, and N. Miller for generous gifts of reagents. This study was supported by Collaborative Project Award from the Center for Cancer Research, National Cancer Institute, and by the National Institutes of Health Intramural AIDS Targeted Antiviral Program (D.S.D.). D.R.B. and J.R. acknowledge financial support from the National Institutes of Health (Grants AI33292 and AI24030, respectively). M.M. was supported by a grant from the Philippe Foundation (New York and Paris). The gp140 constructs from the primary isolates 92UG037.8, 92HT593.1, 93MW965.26, and 93ZR001.3 were prepared by Agnes Jones-Trower, using clones obtained from the National Institutes of Health AIDS Research and Reference Reagent Program; their development was supported by National Institutes of Health Grant AI42599 (to C.C.B.).
Abbreviations
- sCD4
soluble CD4
- dsCCR5
detergent-solubilized CCR5
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Doms R W. Virology. 2000;276:229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 2.Dimitrov D S. Cell. 2000;101:697–702. doi: 10.1016/s0092-8674(00)80882-x. [DOI] [PubMed] [Google Scholar]
- 3.Chan D C, Kim P S. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 4.Sodroski J G. Cell. 1999;99:243–246. doi: 10.1016/s0092-8674(00)81655-4. [DOI] [PubMed] [Google Scholar]
- 5.Moore J P, Stevenson M. Nat Rev Mol Cell Biol. 2000;1:40–49. doi: 10.1038/35036060. [DOI] [PubMed] [Google Scholar]
- 6.Dimitrov D S. Nat Med. 1996;2:640–641. doi: 10.1038/nm0696-640. [DOI] [PubMed] [Google Scholar]
- 7.LaCasse R A, Follis K E, Trahey M, Scarborough J D, Littman D R, Nunberg J H. Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]
- 8.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W, Sawyer L S, Hendry R M, Dunlop N, Nara P L, et al. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 9.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conley A J, Kessler J A, II, Boots L J, Tung J S, Arnold B A, Keller P M, Shaw A R, Emini E A. Proc Natl Acad Sci USA. 1994;91:3348–3352. doi: 10.1073/pnas.91.8.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwick M B, Labrijn A F, Wang M, Spenlehauer C, Saphire E O, Binley J M, Moore J P, Stiegler G, Katinger H, Burton D B, Parren P W. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nussbaum O, Broder C C, Berger E A. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouts T R, Tuskan R, Godfrey K, Reitz M, Hone D, Lewis G K, DeVico A L. J Virol. 2000;74:11427–11436. doi: 10.1128/jvi.74.24.11427-11436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders R W, Schiffner L, Master A, Kajumo F, Guo Y, Dragic T, Moore J P, Binley J M. J Virol. 2000;74:5091–5100. doi: 10.1128/jvi.74.11.5091-5100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binley J M, Sanders R W, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma D J, Maddon P J, Olson W C, et al. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson J E, Holton D, Pacheco-Morell S, Liu J, McMurdo H. AIDS Res Hum Retroviruses. 1990;6:567–579. doi: 10.1089/aid.1990.6.567. [DOI] [PubMed] [Google Scholar]
- 18.Thali M, Furman C, Ho D D, Robinson J, Tilley S, Pinter A, Sodroski J. J Virol. 1992;66:5635–5641. doi: 10.1128/jvi.66.9.5635-5641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gershoni J M, Denisova G, Raviv D, Smorodinsky N I, Buyaner D. FASEB J. 1993;7:1185–1187. doi: 10.1096/fasebj.7.12.7690724. [DOI] [PubMed] [Google Scholar]
- 20.Moore J P, Trkola A, Korber B, Boots L J, Kessler J A, McCutchan F E, Mascola J, Ho D D, Robinson J, Conley A J. J Virol. 1995;69:122–130. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao X, Wu L, Stantchev T S, Feng Y R, Ugolini S, Chen H, Shen Z, Riley J L, Broder C C, Sattentau Q J, et al. Proc Natl Acad Sci USA. 1999;96:7496–7501. doi: 10.1073/pnas.96.13.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirzabekov T, Bannert N, Farzan M, Hofmann W, Kolchinsky P, Wu L, Wyatt R, Sodroski J. J Biol Chem. 1999;274:28745–28750. doi: 10.1074/jbc.274.40.28745. [DOI] [PubMed] [Google Scholar]
- 24.Vujcic L K, Quinnan G V., Jr AIDS Res Hum Retroviruses. 1995;11:783–787. doi: 10.1089/aid.1995.11.783. [DOI] [PubMed] [Google Scholar]
- 25.Quinnan G V, Zhang P F, Fu D W, Dong M, Alter H J. AIDS Res Hum Retroviruses. 1999;15:561–570. doi: 10.1089/088922299311088. [DOI] [PubMed] [Google Scholar]
- 26.Barbas C F, Burton D R, Scott J K, Silverman G J. Phage Display: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 27.Barbas C F, III, Kang A S, Lerner R A, Benkovic S J. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binley J M, Klasse P J, Cao Y, Jones I, Markowitz M, Ho D D, Moore J P. J Virol. 1997;71:2799–2809. doi: 10.1128/jvi.71.4.2799-2809.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salzwedel K, Smith E D, Dey B, Berger E A. J Virol. 2000;74:326–333. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trkola A, Pomales A B, Yuan H, Korber B, Maddon P J, Allaway G P, Katinger H, Barbas C F, III, Burton D R, Ho D D. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 32.Connor R I, Chen B K, Choe S, Landau N R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 33.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirzabekov T, Kontos H, Farzan M, Marasco W, Sodroski J. Nat Biotechnol. 2000;18:649–654. doi: 10.1038/76501. [DOI] [PubMed] [Google Scholar]
- 35.Sattentau Q J, Moore J P. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poignard P, Saphire E O, Parren P W, Burton D R. Annu Rev Immunol. 2001;19:253–274. doi: 10.1146/annurev.immunol.19.1.253. [DOI] [PubMed] [Google Scholar]
- 38.Jones P L, Korte T, Blumenthal R. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- 39.Dimitrov A S, Xiao X, Dimitrov D S, Blumenthal R. J Biol Chem. 2001;276:30335–30341. doi: 10.1074/jbc.M103788200. [DOI] [PubMed] [Google Scholar]
- 40.Carr C M, Kim P S. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 41.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 42.Wild C, Dubay J W, Greenwell T, Baird T, Jr, Oas T G, McDanal C, Hunter E, Matthews T. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 44.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 45.Skehel J J, Wiley D C. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 46.LeDuc D L, Shin Y K. Biosci Rep. 2000;20:557–570. doi: 10.1023/a:1010463005396. [DOI] [PubMed] [Google Scholar]
- 47.Saphire E O, Parren P W, Pantophlet R, Zwick M B, Morris G M, Rudd P M, Dwek R A, Stanfield R L, Burton D R, Wilson I A. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 48.Finnegan C M, Berg W, Lewis G K, DeVico A L. J Virol. 2001;75:11096–11105. doi: 10.1128/JVI.75.22.11096-11105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]