Abstract
Using cDNA microarrays, we identified StarD4 as a gene whose expression decreased more than 2-fold in the livers of mice fed a high-cholesterol diet. StarD4 expression in cultured 3T3 cells was also sterol-regulated, and known sterol regulatory element binding protein (SREBP)-target genes showed coordinate regulation. The closest homologues to StarD4 were two other StAR-related lipid transfer (START) proteins named StarD5 and StarD6. StarD4, StarD5, and StarD6 are 205- to 233-aa proteins consisting almost entirely of START domains. These three constitute a subfamily among START proteins, sharing ≈30% amino acid identity with one another, ≈20% identity with the cholesterol-binding START domains of StAR and MLN64, and less than 15% identity with phosphatidylcholine transfer protein (PCTP) and other START domains. StarD4 and StarD5 were expressed in most tissues, with highest levels in liver and kidney, whereas StarD6 was expressed exclusively in the testis. In contrast to StarD4, expression of StarD5 and MLN64 was not sterol-regulated. StarD4, StarD5, and StarD6 may be involved in the intracellular transport of sterols or other lipids.
Cholesterol is an essential component of mammalian cell membranes and is the biosynthetic precursor for steroid hormones, bile acids, and vitamin D. Precursors and metabolites of cholesterol are involved in cellular signaling events (1). Animals obtain cholesterol from their diets and synthesize it de novo from acetate, but excess cellular free cholesterol is toxic, and high plasma low density lipoprotein (LDL) cholesterol is associated with atherosclerotic vascular disease. Therefore, cholesterol homeostasis is finely regulated to ensure an adequate supply, yet avoid excess. Much of this regulation is transcriptional and mediated by sterol regulatory element binding proteins (SREBPs) and liver X receptors (LXRs) (1). When cellular sterols are abundant, SREBPs are inactive in the endoplasmic reticulum membrane, whereas LXR nuclear receptors bind their oxysterol ligands and activate genes involved in reverse cholesterol transport. Upon sterol depletion, LXRs are inactive but SREBPs are cleaved by regulated proteolysis to release the mature transcription factor domain, which translocates to the nucleus. SREBPs then bind promoter sterol-regulatory elements (SREs) to activate genes involved in the biosynthesis and uptake of cholesterol and fatty acids (2).
Since cholesterol and other sterols are hydrophobic lipids, intracellular sterol transport is mediated either by vesicles or by soluble protein carriers (3). An example of the latter is the steroidogenic acute regulatory protein (StAR/StarD1), which delivers cholesterol to mitochondrial P450 side-chain-cleavage enzymes in steroidogenic cells (4). There is a family of proteins with homology to StAR, each containing a 200- 210-aa StAR-related lipid transfer (START) domain (5). The START domain of MLN64/StarD3, which is 36% identical to StAR, has also been shown in vitro to bind cholesterol (6) and stimulate steroidogenesis (7). The MLN64 START domain crystal structure shows an internal hydrophobic tunnel that could accommodate a cholesterol molecule (6). The only other START domain with a known lipid ligand is the phosphatidylcholine transfer protein (PCTP/StarD2) (8).
In this study, cDNA microarrays were used to identify cholesterol-regulated genes. As an in vivo physiological model, C57BL/6 mice were fed a high-cholesterol diet to raise liver cholesterol. StarD4 (START-domain-containing 4) was identified as a gene whose hepatic expression decreased more than 2-fold upon cholesterol feeding. StarD4 expression was coordinately regulated with known SREBP-target genes, suggesting that StarD4 is also SREBP regulated. StarD5 and StarD6 were identified on the basis of homology to StarD4, and the three form a subfamily among START domain-containing proteins. StarD4 and StarD5 were ubiquitously expressed, with highest levels in liver and kidney, whereas StarD6 expression was limited to the testis. These three proteins may function in the intracellular shuttling of sterols or other lipids.
Materials and Methods
Animals and Diets.
Six-week old C57/BL6 mice (The Jackson Laboratory) were housed in a specific pathogen free, humidity- and temperature-controlled room with a 12-h light-dark cycle. Mice were fed pelleted PicoLab Rodent Diet 20 (product code 5053), which contains 0.02% cholesterol (wt/wt), or the same diet supplemented to 0.5% cholesterol (wt/wt) (Harlan Teklad, Madison, WI). After 3 weeks, mice were fasted for 6 h, anesthetized with ketamine, and killed during the last 3 h of the light cycle. Livers were harvested, frozen in liquid nitrogen, and stored at −80°C. Liver cholesterol was assayed by gas chromatography (9).
cDNA Microarrays.
Fluorescent cDNA probes were synthesized by reverse transcribing (Invitrogen Superscript II) 100 μg of liver total RNA (prepared by using RNeasy from Qiagen, Valencia, CA) in the presence of Cy3- or Cy5-labeled dUTP (Amersham Pharmacia Biotech). cDNA microarrays with ≈9,000 mouse expressed sequence tags (ESTs) were a generous gift of Raju Kucherlapati at Albert Einstein College of Medicine (10). Array protocols are online at http://sequence.aecom.yu.edu/bioinf/microarray/protocol4.html. Scanned arrays were analyzed with ScanAlyze (by Michael Eisen, http://rana.lbl.gov/EisenSoftware.htm), and results were compiled by using Microsoft Excel and Access.
Cloning and PCR.
Molecular cloning followed standard techniques using enzymes from New England Biolabs. Except where noted, PCR reagents were Advantage cDNA polymerase (CLONTECH), primers were from Gene Link (Hawthorne, NY; sequences in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org), and thermal cycling was on a Perkin–Elmer 9700. TA cloning of PCR products was carried out with pCR-2.1-TOPO (Invitrogen). All DNA constructs were sequence-verified.
Multiple-Tissue Northern Blots.
Radiolabeled DNA probes were synthesized from 20 ng of template by random priming using the DECA prime II kit (Ambion) and [32P]dATP (Perkin–Elmer). Blots were purchased and hybridized by using Express Hyb rapid hybridization buffer according to the manufacturer's instructions (CLONTECH).
Cell Culture.
Mouse 3T3-L1 cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium (DME) with 10% FBS at 37°C in a 5% CO2/95% air atmosphere. Cells were grown as fibroblast-like cells and were not differentiated to adipocytes. Lipoprotein-depleted serum (LPDS) was prepared from FBS by ultracentrifugation. Stock solutions were made for three reagents (Sigma): cholesterol (10 mg/ml in ethanol), 25-hydroxycholesterol (0.5 mg/ml in ethanol), and mevinolin (lovastatin, 0.8 mg/ml). Subconfluent cells in 6-well plates were cultured for 20 h in one of three media: control (DME with 10% LPDS), sterols (DME with 10% LPDS, 10 μg/ml cholesterol, and 1 μg/ml 25-hydroxycholesterol), or lovastatin (DME with 10% LPDS and 1 μg/ml mevinolin).
Real-Time Quantitative Reverse Transcription–PCR (RT-PCR).
Total RNA was isolated from cultured cells or frozen liver by using RNeasy kits (Qiagen) and treated with DNase I (DNA-free from Ambion, Austin, TX). Five micrograms of RNA was reverse transcribed in 20 μl by using random hexamers and Superscript II (Invitrogen). To perform relative quantification of gene expression with a standard curve, the genes of interest and the endogenous control gene (cyclophilin) were amplified in separate tubes for each cDNA sample. Duplicate 50-μl PCRs were carried out with 1× Jumpstart PCR buffer, 1.00 unit of Jumpstart Taq DNA polymerase (Sigma), 3.5 mM MgCl2, 200 μM each dNTP, 300 nM forward primer, 300 nM reverse primer, and 100 nM 6-carboxyfluorescein (6FAM)-labeled TaqMan probe. TaqMan primer pairs and probes (sequences in Table 3) were designed with Primer Express (Applied Biosystems) to span splice junctions. The template was 10 μl of a 1:100 dilution of cDNA, and the standards were a serial dilution of cDNA.
A 7700 Sequence Detection System (Applied Biosystems) was used with the default thermal cycling profile (95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 1 min; 4°C soak). The quencher dye (TAMRA, N,N,N′,N′-tetramethyl-6-carboxyrhodamine) was the passive reference. The threshold was set at 0.05 unit of normalized fluorescence, and a threshold cycle (Ct) was measured in each well. Relative standard curves were plotted for each gene, and the mean Ct for each cDNA sample was expressed as an arbitrary value relative to standard. For each cDNA, values for genes of interest were normalized to the corresponding value for cyclophilin and expressed as a ratio. Groups were compared by a two-tailed type 2 Student's t test.
Results
Cholesterol Feeding cDNA Microarray.
The initial experiment sought to identify hepatic genes regulated by dietary cholesterol. C57BL/6 male mice were fed a chow diet low (0.02%) or high (0.5%) in cholesterol for 3 weeks. Liver cholesterol was measured for five mice on each diet, and the 0.5% cholesterol diet raised the total liver cholesterol (1.87 ± 0.23 to 4.47 ± 0.85 μg/mg of liver, P < 0.001), with an increase in both esterified (0.48 ± 0.08 to 2.64 ± 0.81 μg/mg, P < 0.001) and free (1.40 ± 0.23 to 1.83 ± 0.25 μg/mg, P < 0.05) cholesterol. cDNA microarrays were used to screen for genes whose hepatic expression changed upon cholesterol feeding. Among the six genes on the array down-regulated more than 2-fold by dietary cholesterol (Table 1), five were known SREBP-target genes involved in cholesterol or fatty acid biosynthesis (2, 11, 12). The sixth was an EST (AA239481) not previously characterized.
Table 1.
Microarray genes down-regulated by dietary cholesterol
| Mean fold decrease | GenBank accession no. | Gene product | Fold decrease on six arrays
|
|||||
|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 2a | 2b | 3a | 3b | |||
| 4.6 | AA237469 | Isopentenyl diphosphate isomerase | 3.3 | 7.6 | 2.4 | 3.6 | 4.9 | 5.9 |
| 2.8 | AA239481 | Uncharacterized EST (StarD4) | 3.6 | 2.3 | 4.8 | 1.6 | 2.8 | 1.9 |
| 2.6 | AA268608 | Squalene epoxidase | 3.2 | 2.7 | 2.8 | 1.3 | 2.9 | (44) |
| 2.5 | AA061468 | Hydroxymethylglutaryl-CoA synthase | 2.6 | 0.9 | 5.2 | 2.0 | 2.4 | 2.1 |
| 2.0 | AA116513 | Fatty acid synthase | 1.4 | 1.7 | 2.0 | 2.8 | 2.3 | 2.0 |
| 2.0 | AA500330 | Farnesyl pyrophosphate synthetase | 1.2 | 1.7 | 2.0 | 2.8 | 2.3 | 2.0 |
Experiments 1–3 each compared liver gene expression in a pair of individual mice fed different diets (0.02% versus 0.5% cholesterol). Each experiment was performed on duplicate arrays (a, b) by reversing the Cy3 and Cy5 probe labeling. Since there was marked variability within and between experiments, the following criteria were used for regulated genes: expression differed 2-fold or greater on at least four of the six arrays with the higher expression level at least 25% over background. For each of the six genes down-regulated by the high-cholesterol diet, the fold regulation on each array is shown, as well the average from all six (the outlying value in parenthesis was eliminated).
Identification of the StarD4 Gene.
To identify the novel gene, the 1,114-bp insert of EST AA239481 was sequenced. There were no long protein-coding regions in any reading frame, suggesting the sequence was 3′ untranslated region (3′ UTR). blast searches placed the EST sequence on a mouse bacterial artificial chromosome (BAC) clone (AC020796), about 3 kb downstream of a 230-bp coding sequence homologous to START genes. This 230-bp sequence (the coding part of exon 6, see below) was in multiple mouse ESTs, allowing assembly of a 675-bp ORF by in silico EST walking. The mouse gene encodes a 224-aa protein consisting almost entirely of a START domain, so it was named stard4. The human STARD4 orthologue encodes 205 amino acids 87% identical to the mouse protein.
Mouse and human StarD4 ORFs were RT-PCR amplified from liver, cloned, and sequence-verified. The sequence of mouse StarD4 from C57BL/6 disagreed with some ESTs at ORF positions 121 and 152. Since other ESTs agreed with the C57BL/6 sequence, StarD4 was cloned from a second inbred mouse strain, FVB. Sequencing confirmed that these nucleotide and amino acid positions are indeed polymorphic between strains: C57BL/6 has guanine-121, cytosine-152 (Glu-41, Ala-51), whereas FVB has adenine-121, thymine-152 (Lys-41, Val-51). Based on the StarD4 crystal structure (13), both side chains are surface exposed and do not contribute to the putative lipid-binding tunnel, so it is uncertain whether these polymorphisms have functional consequences.
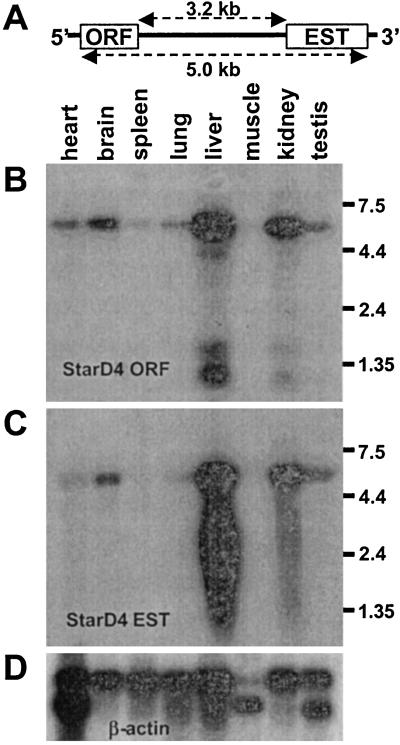
Northern blots were performed to verify that the array EST and ORF sequences were present on the same mRNA (Fig. 1A). On a mouse multiple-tissue Northern blot, a StarD4 ORF probe hybridized to a predominant mRNA at ≈5.5 kb in all eight tissues, with the highest levels in liver and kidney (Fig. 1B). The array EST insert probe also hybridized to this ≈5.5-kb mRNA with the same relative tissue levels (Fig. 1C).
Figure 1.
Multiple-tissue Northern blots of mouse StarD4. (a) Schematic of the mouse StarD4 mRNA, at least 5 kb long with 3.2 kb of 3′ UTR between the ORF and EST AA239481 sequence. A mouse multiple-tissue Northern blot was hybridized with three probes: StarD4 ORF (B), EST AA239481 insert (C), or β-actin control for RNA loading (D). Longer exposures revealed the ≈5.5-kb StarD4 band in skeletal muscle.
Sterol Regulation of StarD4 Expression in Vivo and in Vitro.
To confirm cholesterol-regulated expression of StarD4, quantitative RT-PCR primers and probes were designed. Upon feeding 0.5% cholesterol, mouse liver StarD4 mRNA decreased 2- to 3-fold (Fig. 2A), as expected from the microarray data. In addition, 3T3 cells were cultured in sterols to repress SREBP activation, and StarD4 expression decreased 4-fold compared with the control media. Conversely, when 3T3 cells were cultured in lovastatin (to inhibit cholesterol biosynthesis, deplete cellular sterols, and activate SREBPs), StarD4 expression increased 3.4-fold compared with control media and 14-fold compared with sterol-containing media (Fig. 2B). Four known SREBP-target genes (2) showed the same pattern of regulation (Fig. 2C).
Figure 2.
Sterol regulation of StarD4 expression. (A) Regulation by dietary cholesterol in mouse liver. C57BL/6 female mice were fed a semisynthetic diet with no cholesterol (0.00%, n = 5) or the same diet supplemented to 0.50% cholesterol (n = 5). StarD4 expression was measured in each liver by quantitative RT-PCR. (B and C) Regulation by sterols in cultured 3T3 cells. 3T3-L1 cells were cultured in one of three media: control, sterols, or statin (n = 4 wells each, see Materials and Methods). Expression of StarD4, hydroxymethylglutaryl-CoA reductase (HMGR) and synthase (HMGS), low density lipoprotein receptor (LDLR), and ATP:citrate lyase (ACLY) was measured in each well. All data (mean ± SD) are normalized to cyclophilin and control condition set equal to 100% for each gene. *, P < 0.001 vs. control, **, P < 0.05 vs. control.
A Subfamily of START Genes Including StarD4, StarD5, and StarD6.
blast searches against the complete human genome and EST databases identified 15 START domain-containing genes. Brown fat inducible thioesterase (BFIT) gene is alternatively spliced to encode two C termini (14), so the amino acid sequences of 16 human START domains were aligned (Fig. 3A). Two other uncharacterized START proteins, which were named StarD5 and StarD6, shared 26–32% identity with StarD4 and with one another. These three proteins formed a subfamily among START proteins, sharing only 16–21% identity to MLN64 and StAR and 14% or less with PCTP and other START domains (Fig. 3B). A phylogenetic tree divided these human START domains into six distinct subfamilies (Fig. 3C). The three novel proteins formed the StarD4 subfamily, and the nearest branch included StAR and MLN64. The other four subfamilies were proteins with PCTP-like START domains, proteins with N-terminal Rho GTPase activator protein (RhoGAP) domains, proteins with N-terminal acyl-CoA hydrolase (ACH) domains, and the hypothetical protein KIAA1300. Additional subfamilies of START proteins are present only in plants (13).
Figure 3.
StarD4, StarD5, and StarD6 are a subfamily among START domains. (A) The START domains of 16 human proteins were aligned by using ClustalW. The colored bars above the alignment indicate positions with a strong consensus. Amino acid agreements with StarD4 are boxed, while agreements with consensus are colored yellow. (B) The percent amino acid identity for each pairwise comparison in the alignment. Yellow indicates the similarity between the StarD4 subfamily (violet) and the StAR/MLN64 subfamily (blue). The remaining 9 proteins fall into four groups: PCTP-like (orange), Rho GTPase activator protein (RhoGAP) domain-containing (green), acyl-CoA hydrolase (ACH) domain-containing (pink), and other (gray). (C) A phylogenetic tree based on the alignment shows the same subfamilies. DNAstar software (Lasergene, Madison, WI) was used for this analysis.
The human and mouse sequences for StarD4, StarD5, and StarD6 were analyzed in silico by using resources available publicly through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) or commercially through Celera Genomics (http://www.celera.com/). Full-length coding sequences, UniGene clusters, and chromosomal positions were determined (Table 2). The three genes do not reside together in a cluster, but the mouse and human orthologues for each gene reside in syntenic chromosomal regions. While there is only one copy per genome of StarD4 and StarD5, Celera mouse chromosome 10 includes an intronless StarD6 sequence, perhaps a processed pseudogene. Mouse StarD6 ORF PCR primers in different exons amplified a strong ≈700-bp product from genomic DNA (data not shown), confirming this intronless gene.
Table 2.
Gene information for human (h) and mouse (m) StarD4, StarD5, and StarD6
| Gene | GenBank accession no. | UniGene cluster(s) | Celera gene | Chromosome | Genetic map position, cM | Northern mRNA length, kb | ORF length, bp | Protein length, aa |
|---|---|---|---|---|---|---|---|---|
| mStarD4 | AF480297–8 | Mm. 31508, 23344 | mCG21633 | 18 | 12 | ≈5.5 | 675 | 224 |
| hStarD4 | AF480299 | Hs.162205 | hCG37443 | 5q22 | 116–121 | ≈5.5, ≈4.5 | 618 | 205 |
| mStarD5 | AF480302 | Mm. 25702 | mCG8260 | 7 | 41 | ≈3.0, ≈1.5 | 642 | 213 |
| hStarD5 | AF480304 | Hs. 172803 | hCG27342 | 15q26 | 72–77 | ≈3.0, ≈1.5 | 642 | 213 |
| mStarD6 | AF480303 | Mm. 83623, 195613 | mCG9256 | 18 | 44 | ≈1.5 | 702 | 233 |
| hStarD6 | AF480305 | Hs. 304542, 143962 | hCG1643548 | 18q21 | 71–83 | ≈1.5 | 663 | 220 |
cM, centimorgan.
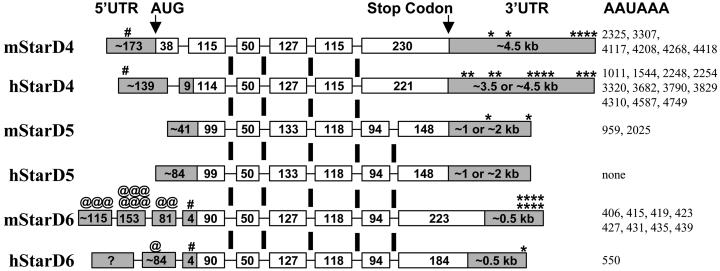
To determine the exonic organization of each gene, cDNA contigs and genomic sequences were aligned to reveal introns that followed the GT-AG rule. The StarD4 subfamily genes were remarkably similar, with most splice junctions conserved (Fig. 4, splice junction sequences in Table 4, which is published as supporting information on the PNAS web site). The exonic organization is different for the MLN64, StAR, and PCTP genes (not shown), supporting a distinct StarD4 subfamily. Mouse StarD4 has 12 aa encoded by exon 1, but this exon is noncoding in human StarD4 and there are no counterparts in StarD5 or StarD6. Upstream in-frame stop codons allow identification of initiator codons in StarD4 and StarD6, whereas StarD5 has no upstream exons in over 180 mouse and human Unigene ESTs. The mouse StarD6 mRNA has a long, multiexon 5′ UTR with multiple initiation codons, and such upstream AUGs may function as regulators of translation (15). Whereas most other START proteins have additional N-terminal domains (6), StarD4, StarD5, and StarD6 are 205–233 aa proteins consisting almost entirely of START domains.
Figure 4.
Exonic organization of StarD4, StarD5, and StarD6. Exons for the mouse (m) and human (h) genes, represented as boxes with lengths in nucleotides, were determined by alignment of cDNA with genomic DNA. Vertical bars show splice junctions that are conserved among the genes. Coding sequences are white and UTRs are gray. The 5′ UTR lengths were based on the longest 5′ sequences in each UniGene cluster, whereas the 3′ UTR lengths were estimated on the basis of Northern blot mRNA sizes (Table 2). #, First upstream in-frame stop codon in 5′ UTR; @, upstream AUG codon in 5′ UTR; *, poly(A) signal (AAUAAA) in the 3′ UTR. The distances from the stop codon for each poly(A) signal are indicated at the far right. The 5′ end of human StarD6 is uncertain because very little sequence data are available for this gene.
Expression of StarD5 and StarD6.
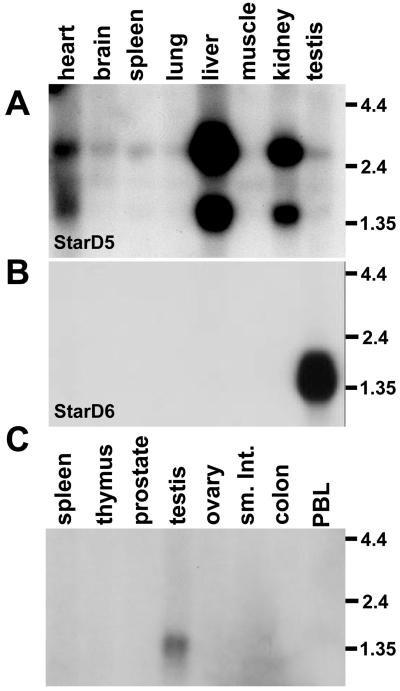
StarD5 and StarD6 ORFs were RT-PCR amplified and cloned from liver and testis, respectively, and used to probe mouse multiple-tissue Northern blots. Like StarD4, StarD5 messages showed ubiquitous expression, highest in liver and kidney (Fig. 5A). In contrast, StarD6 expression was restricted to testis (Fig. 5 B and C). The mRNA lengths revealed by Northern blots (Table 2) agree well with predictions based on the AAUAAA polyadenylation signals in the 3′ UTRs (Fig. 4), and multiple messages appear to represent alternative sites of RNA polyadenylation and cleavage.
Figure 5.
Multiple-tissue Northern blots of StarD5 and StarD6. (A) Mouse blot hybridized with a mouse StarD5 ORF probe. (B) Mouse blot hybridized with a mouse StarD6 ORF probe. (C) Human blot hybridized with a mouse StarD6 ORF probe (sm. int., small intestine; PBL, peripheral blood leukocyte).
Finally, quantitative RT-PCR was performed to determine whether the widely expressed START genes StarD5 and MLN64 are sterol-regulated like StarD4. Neither StarD5 nor MLN64 expression was regulated in mouse liver upon cholesterol feeding or in 3T3 cells upon culture in sterols (data not shown).
Discussion
This study describes StarD4, a sterol-regulated gene encoding a StAR-related lipid transfer protein. StarD4 was initially identified by using cDNA microarrays, as liver StarD4 expression decreased more than 2-fold in mice fed a high-cholesterol diet. In cultured 3T3 cells, StarD4 expression was likewise coordinately regulated by sterols with known SREBP-target genes. Two other START proteins, StarD5 and StarD6, were identified on the basis of homology to StarD4, and these three constituted a distinct subfamily of START proteins. StarD4 and StarD5 were ubiquitously expressed, with highest levels in liver and kidney, whereas StarD6 was expressed exclusively in the testis.
Three lines of evidence support the hypothesis that StarD4, StarD5, and StarD6 bind cholesterol or other sterols. First, like many other genes in cholesterol metabolism, StarD4 is regulated by sterols, and a functional sterol-regulatory element has been identified in the StarD4 promoter (R.E.S., unpublished results). Because SREBPs also regulate genes involved in the metabolism of other lipids (2), nonsterols cannot be ruled out as potential ligands. Second, these three proteins are most homologous to StAR and MLN64, which bind cholesterol, and more distantly related to PCTP and other START domains, which bind phospholipids and perhaps other lipids. Third, the crystal structure of StarD4 shows a hydrophobic tunnel with dimensions similar to those of MLN64 (13). StarD4, StarD5, and StarD6 may be cytosolic lipid carriers like PCTP (16), since all four lack additional N-terminal domains or other localization signals [based on PROSITE searches (17)].
Given their ubiquitous expression, we speculate that StarD4 and StarD5 may be involved in fundamental processes such as intracellular sterol transport or cholesterol biosynthesis. While some cholesterol transport is vesicular, cytosolic protein carriers have been proposed for other transport events, such as movement of newly synthesized cholesterol to the plasma membrane (3) and plasma membrane cholesterol to the endocytic recycling compartment (18). A START protein could also mediate sterol transport to mitochondrial sterol 27-hydroxylase (Cyp27) (19). In cholesterol biosynthesis, squalene, lanosterol, and every subsequent intermediate are hydrophobic molecules that may require protein carriers. The squalene carrier supernatant protein factor (SPF) stimulates the enzymatic activity of squalene epoxidase, transfers squalene between membranes in vitro, and stimulates cholesterol biosynthesis when overexpressed (20). StarD4 could serve an analogous function for a postsqualene sterol intermediate, as StarD4 shows coordinate regulation with cholesterol biosynthetic enzymes.
Testis-specific expression suggests a specialized role for StarD6, although a StAR-like role in steroidogenesis is unlikely because it is not expressed in the ovary. While it may also be expressed in Leydig or Sertoli cells, StarD6 appears to be present in the germ cells because several mouse ESTs have been cloned from spermatocyte or spermatid libraries. Cholesterol precursor sterols may have important roles in reproduction. Meiosis-activating sterols (MAS) stimulate resumption of meiosis (21), and testis-MAS (T-MAS, the cholesterol precursor 4,4-dimethyl-5α-cholesta-8,24-dien-3β-ol) accumulates in postpubertal testis (22). Other postlanosterol sterols are surprisingly abundant in the testis and epididymis (23), where they may play a role in sperm maturation. If StarD6 binds a sterol involved in meiosis or germ cell development, this gene may be important for fertility.
In summary, the lipid-binding protein StarD4 was identified by microarray analysis of gene expression in cholesterol-fed mouse liver. StarD5 and StarD6 were then identified on the basis of homology to StarD4, and these three genes form a subfamily within the START family. We hypothesize that the StarD4 subfamily is involved in the intracellular metabolism of cholesterol or other sterols, although it is possible that they bind other lipids instead. Since StarD4, StarD5, and StarD6 are only ≈30% identical to one another, each may bind specifically to a distinct lipid. Further studies including the generation of knockout mice should help to elucidate the physiological functions of these three proteins.
Supplementary Material
Acknowledgments
We thank Raju Kucherlapati, Joerg Heyer, and Sandra Merscher for assistance with cDNA microarray technology, Zhihua Han for help with Northern blots, Michael Sinensky and Doug Thewke for useful discussions, and Jonathan Smith, Kara Maxwell, and Leslie Castelo-Soccio for critical reading. This work was supported by National Institutes of Health Medical Scientist Training Program Grant GM07739 (R.E.S.) and by National Institutes of Health Grant HL32435 (J.L.B.).
Abbreviations
- StAR
steroidogenic acute regulatory protein
- MLN64
protein of unknown function
- PCTP
phosphatidylcholine transfer protein
- START
StAR-related lipid transfer
- StarD
START domain-containing
- SREBP
sterol regulatory element binding protein
- EST
expressed sequence tag
- RT-PCR
reverse transcription–PCR
- UTR
untranslated region
Footnotes
References
- 1.Edwards P A, Ericsson J. Annu Rev Biochem. 1999;68:157–185. doi: 10.1146/annurev.biochem.68.1.157. [DOI] [PubMed] [Google Scholar]
- 2.Edwards P A, Tabor D, Kast H R, Venkateswaran A. Biochim Biophys Acta. 2000;1529:103–113. doi: 10.1016/s1388-1981(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 3.Liscum L, Munn N J. Biochim Biophys Acta. 1999;1438:19–37. doi: 10.1016/s1388-1981(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 4.Stocco D M. Annu Rev Physiol. 2001;63:193–213. doi: 10.1146/annurev.physiol.63.1.193. [DOI] [PubMed] [Google Scholar]
- 5.Ponting C P, Aravind L. Trends Biochem Sci. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- 6.Tsujishita Y, Hurley J H. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 7.Watari H, Arakane F, Moog-Lutz C, Kallen C B, Tomasetto C, Gerton G L, Rio M C, Baker M E, Strauss J F., 3rd Proc Natl Acad Sci USA. 1997;94:8462–8467. doi: 10.1073/pnas.94.16.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Brouwer A P, Bouma B, van Tiel C M, Heerma W, Brouwers J F, Bevers L E, Westerman J, Roelofsen B, Wirtz K W. Chem Phys Lipids. 2001;112:109–119. doi: 10.1016/s0009-3084(01)00171-2. [DOI] [PubMed] [Google Scholar]
- 9.Sehayek E, Ono J G, Shefer S, Nguyen L B, Wang N, Batta A K, Salen G, Smith J D, Tall A R, Breslow J L. Proc Natl Acad Sci USA. 1998;95:10194–10199. doi: 10.1073/pnas.95.17.10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung V G, Morley M, Aguilar F, Massimi A, Kucherlapati R, Childs G. Nat Genet. 1999;21:15–19. doi: 10.1038/4439. [DOI] [PubMed] [Google Scholar]
- 11.Paton V G, Shackelford J E, Krisans S K. J Biol Chem. 1997;272:18945–18950. doi: 10.1074/jbc.272.30.18945. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura Y, Sakakibara J, Izumi T, Shibata A, Ono T. J Biol Chem. 1996;271:8053–8056. doi: 10.1074/jbc.271.14.8053. [DOI] [PubMed] [Google Scholar]
- 13.Romanowski M J, Soccio R E, Breslow J L, Burley S K. Proc Natl Acad Sci USA. 2002;99:6949–6954. doi: 10.1073/pnas.052140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams S H, Chui C, Schilbach S L, Yu X X, Goddard A D, Grimaldi J C, Lee J, Dowd P, Colman S, Lewin D A. Biochem J. 2001;360:135–142. doi: 10.1042/0264-6021:3600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris D R, Geballe A P. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirtz K W. Annu Rev Biochem. 1991;60:73–99. doi: 10.1146/annurev.bi.60.070191.000445. [DOI] [PubMed] [Google Scholar]
- 17.Falquet L, Pagni M, Bucher P, Hulo N, Sigrist C J, Hofmann K, Bairoch A. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao M, Lin S X, Karylowski O J, Wustner D, McGraw T E, Maxfield F R. J Biol Chem. 2002;277:609–617. doi: 10.1074/jbc.M108861200. [DOI] [PubMed] [Google Scholar]
- 19.Sugawara T, Lin D, Holt J A, Martin K O, Javitt N B, Miller W L, Strauss J F., 3rd Biochemistry. 1995;34:12506–12512. doi: 10.1021/bi00039a004. [DOI] [PubMed] [Google Scholar]
- 20.Shibata N, Arita M, Misaki Y, Dohmae N, Takio K, Ono T, Inoue K, Arai H. Proc Natl Acad Sci USA. 2001;98:2244–2249. doi: 10.1073/pnas.041620398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byskov A G, Baltsen M, Andersen C Y. J Mol Med. 1998;76:818–823. doi: 10.1007/s001090050286. [DOI] [PubMed] [Google Scholar]
- 22.Tacer K F, Haugen T B, Baltsen M, Debeljak N, Rozman D. J Lipid Res. 2002;43:82–89. [PubMed] [Google Scholar]
- 23.Lindenthal B, Aldaghlas T A, Kelleher J K, Henkel S M, Tolba R, Haidl G, von Bergmann K. J Lipid Res. 2001;42:1089–1095. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.