Abstract
The x-ray structure of the mouse cholesterol-regulated START protein 4 (StarD4) has been determined at 2.2-Å resolution, revealing a compact α/β structure related to the START domain present in the cytoplasmic C-terminal portion of human MLN64. The volume of the putative lipid-binding tunnel was estimated at 847 Å3, which is consistent with the binding of one cholesterol-size lipid molecule. Comparison of the tunnel-lining residues in StarD4 and MLN64-START permitted identification of possible lipid specificity determinants in both molecular tunnels. Homology modeling of related proteins, and comparison of the StarD4 and MLN64-START structures, showed that StarD4 is a member of a large START domain superfamily characterized by the helix-grip fold. Additional mechanistic and evolutionary studies should be facilitated by the availability of a second START domain structure from a distant relative of MLN64.
Cholesterol is required for synthesis of all steroid hormones, and delivery of this hydrophobic precursor to the sites of further processing is the rate-limiting step in steroidogenesis. The steroidogenic acute regulatory (StAR) protein, a 30-kDa phosphoprotein associated with mitochondria, appears to transport cholesterol in steroidogenic cells (1). Mutations in the StAR gene cause one form of congenital lipoid adrenal hyperplasia, a condition characterized by blockade of steroid hormone synthesis at the site of delivery of cholesterol to the cytochrome P450 side-chain-cleavage enzyme (P450ssc) that converts cholesterol to pregnenolone (1, 2).
The StAR protein is a compact single-domain molecule with an N-terminal extension that targets it to the mitochondria. This C-terminal 200- to 210-residue motif is known as the StAR-related lipid transfer (START) domain, and it is found in many proteins in multicellular eukaryotes (3–5) with putative functions ranging from signal transduction, transcriptional regulation, and GTPase activation to thioester hydrolysis. Sequence profile searches documented that these proteins are members of the START domain superfamily (6). Many START domain-containing proteins appear to bind lipophilic molecules. In addition to participating in lipid transport, START domain-containing proteins may target proteins to membranes (3, 7) and/or perform certain catalytic functions (6).
The three-dimensional structure of a START domain-containing protein came from the x-ray studies of the C-terminal portion of human MLN64 (PDB ID code 1EM2) (8), a late endosomal membrane protein with a cytoplasmic cholesterol-binding START domain (9, 10) that is 35% identical to the START motif of the human StAR protein (8). The crystal structure of MLN64-START (referred to hereafter as MLN64) revealed an α/β structure of the helix-grip fold. The most striking feature of the domain is an elongated hydrophobic tunnel thought to accommodate one cholesterol molecule (8).
In this paper, we report the crystal structure of the mouse cholesterol-regulated START protein 4 (StarD4), which shares a 21% sequence identity with human MLN64 (11). This second START domain structure permitted a useful comparison with the structure of MLN64 (8). In addition, automated homology modeling with both StarD4 and MLN64 templates provided additional structural information for many START domain superfamily members.
Materials and Methods
Cloning, Protein Expression, and Purification.
The full-length mouse StarD4 ORF was amplified by PCR using a forward primer containing a BamHI restriction site (GTCGTGGATCCATGGCTGACCCTGAGAGCCCG), a reverse primer containing an XhoI restriction site (GTCGTCTCGAGTCATGCCTTGCGTAGACCTTTTCG), and mouse liver cDNA as template, using standard protocols (12). The resulting fragment was cloned into the corresponding restriction sites of the pGEX-6P-1 plasmid (Promega). Glutathione S-transferase (GST)-tagged selenomethionine (Se-Met)-containing protein was expressed in Escherichia coli BL21 cells (Novagen) during overnight induction at 18°C. Fusion protein was purified by GST affinity chromatography. The N-terminal GST tag was removed by digestion with PreScission protease (Pharmacia), and StarD4 was separated from GST and uncleaved fusion protein on glutathione-agarose. After overnight dialysis in 20 mM Hepes, pH 8.4/100 mM KCl/5 mM DTT, the target protein was further purified by Q Sepharose ion-exchange chromatography. Protein for crystallization was dialyzed against 20 mM Hepes, pH 7.5/100 mM KCl/5 mM DTT, concentrated to 12 mg/ml, and passed through a 0.1-μm-pore filter. Gel filtration experiments documented that StarD4 is monomeric in solution (data not shown). The purity and homogeneity of the protein and incorporation of Se-Met were confirmed by gel electrophoresis and matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) (data not shown).
Crystallization.
Diffraction-quality Se-Met StarD4 crystals were obtained at 4°C by sitting-drop vapor diffusion against a reservoir containing 0.1 M sodium cacodylate (pH 6.5), 0.2 M magnesium acetate, 16% (wt/vol) polyethylene glycol 8000, and 22% (vol/vol) glycerol. Crystals grew as hexagonal rods with two molecules per asymmetric unit in the trigonal space group P31 (a = 70.0 Å, c = 85.2 Å). Crystals were frozen by transfer to mother liquor for 15–30 sec and rapid immersion in liquid propane.
Data Collection, Structure Determination, and Refinement.
Diffraction data were collected under standard cryogenic conditions with a MARCCD detector on Beamline X9A at the National Synchrotron Light Source, Brookhaven National Laboratory. The structure of Se-Met StarD4 was determined by means of multiwavelength anomalous dispersion (13) with data recorded at three x-ray wavelengths corresponding to the peak (λ1) and inflection point (λ2) of the selenium K absorption edge, plus a high-energy remote (λ3). Each dataset was independently integrated and scaled by using hkl (14). Positions of 10 of the possible 12 selenium atoms (6 each for two molecules in the asymmetric unit) were determined with SnB (15) and anomalous difference Fourier syntheses, and refined by using mlphare (16) (figure of merit of 0.42 at 2.2-Å resolution). Density modification yielded a good-quality experimental electron density map that was suitable for model building with o (17), and the atomic model was refined to convergence with cns (18). Residues 1–23, 223–224, and five residues from the N-terminal cloning artifact (Gly-Pro-Leu-Gly-Ser) for each monomer were not visible in the electron density map and were omitted from refinement. In the course of analyzing the electron density maps we discovered an unidentified bulky feature that appeared to be covalently attached to Cys-169. MALDI-MS analysis of the protein sample used for crystallization did not reveal evidence of a bound ligand. It is possible that this residual electron density corresponds to a product formed by a reaction between the protein and a product of water radiolysis such as a hydroxyl free radical (19). During the latter stages of refinement, this density feature was modeled as water. The final model, consisting of 398 of 448 residues and 290 water molecules, was refined at 2.2-Å resolution to an R factor of 23.4% with an Rfree value of 28.1%. procheck (20) revealed no disallowed (φ, ψ) combinations and excellent stereochemistry (overall G value = 0.2; see Table 1 for a summary of x-ray data and refinement statistics).
Table 1.
Crystallographic data and refinement statistics for PDB ID code 1JSS
| λ1 (Se peak) | λ2 (Se inflection) | λ3 (Se high remote) | |
|---|---|---|---|
| Crystal characteristics and data collection statistics | |||
| Space group | P31; two molecules per asymmetric unit | ||
| Cell constants | a = 70.0 Å, c = 85.2 Å | ||
| Wavelength, Å | 0.97910 | 0.97950 | 0.96108 |
| Resolution, Å | 30.0–2.2 | 30.0–2.2 | 30.0–2.2 |
| Number of observations | 617,842 | 517,103 | 525,017 |
| Number of reflections | 23,657 | 23,729 | 23,775 |
| Completeness, % | 99.8 | 99.8 | 100 |
| (in 2.2–2.28-Å shell) | (98.3) | (97.6) | (100) |
| Mean I/σ(I) | 27.8 | 26.7 | 29.5 |
| (in 2.2–2.28-Å shell) | (6.8) | (3.7) | (3.1) |
| Rmerge on I* | 0.054 | 0.062 | 0.062 |
| (in 2.2–2.28-Å shell) | (0.301) | (0.471) | (0.530) |
| Sigma cut-off | I < −3σ(I) | I < −3σ(I) | I < −3σ(I) |
| Figure of merit† | 0.42 (20.0–2.2-Å resolution) for 23,665 reflections | ||
| Model and refinement statistics | |||
| Data set used in structure refinement | λ1 (Se peak) | ||
| Resolution range, Å | 30–2.2 | ||
| Number of reflections | 46,556 (42,013 in working set; 4,543 in test set) | ||
| Completeness, % | 98.5 (88.9 in working set; 9.6 in test set) | ||
| Cutoff criteria | | F | > 0.0 | ||
| Number of amino acid residues | 398 | ||
| Number of water molecules | 290 | ||
| Rcryst‡ | 0.234 | ||
| Rfree | 0.281 | ||
| rmsd | |||
| Bond lengths, Å | 0.008 | ||
| Bond angles, ° | 1.60 | ||
| Luzzati error, Å | 0.30 | ||
| Ramachandran plot statistics§ | |||
| Residues in most favored regions | 303 (86.1%) | ||
| Residues in additional allowed regions | 48 (13.6%) | ||
| Residues in generously allowed regions | 1 (0.3%) | ||
| Residues in disallowed regions | 0 (0%) | ||
| Overall G factor§ | 0.2 | ||
| modpipe statistics (StarD4 template)¶ | |||
| Total number of models (model score > 0.7, model length > 100 residues) | 49 | ||
| Models with >50% sequence identity | 1 | ||
| Models with 30–50% sequence identity | 2 | ||
| Models with <30% sequence identity | 46 | ||
The x-ray source was Beamline X9A at the National Synchrotron Light Source.
Rmerge = Σhkl Σi | I(hkl)i − 〈I(hkl) 〉 |/Σhkl Σi 〈I(hkl)i〉.
Figure of merit calculated by using mlphare (16).
Rcryst = Σhkl | Fo(hkl) − Fc(hkl) |/Σhkl | Fo(hkl) |, where Fo and Fc are observed and calculated structure factors, respectively.
Computed with procheck (20).
Computed with modpipe (25). Models are publicly available from MODBASE (http://pipe.rockefeller.edu) by means of advanced search with the keyword NYSGRC_1JSS.
Multiple Sequence Alignments.
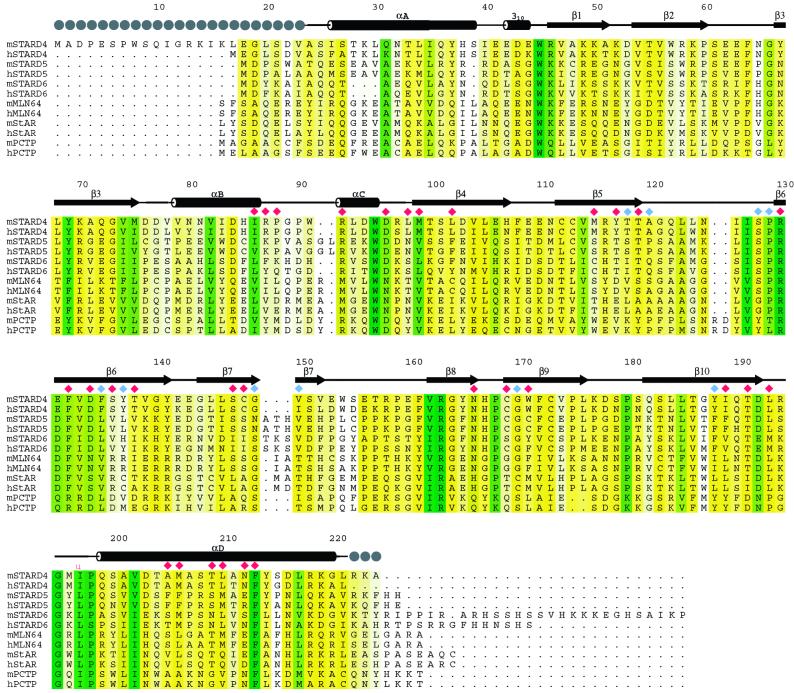
Proteins similar to mouse StarD4 and human MLN64 were identified by using psi-blast (21). Multiple sequence alignments were prepared by using ClustalW (22), and conservation was calculated with BLOSUM62 (23). Alignments of selected StarD4-related proteins are shown in Fig. 1. Identities of the sequences used in the alignment and generation of the phylogenetic tree are provided in Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org.
Figure 1.
Alignment of proteins similar to mouse StarD4 identified in an iterative psi-blast (21) search. Secondary structural elements are shown above the aligned sequences, and the locations of linear motifs are labeled. Gray spheres indicate disordered residues. Color-coding denotes sequence conservation (white-to-green ramp, 30–100% sequence identity). Residues with side-chain atoms lining the tunnel are marked with red diamonds, those with backbone atoms lining the tunnel are shown as blue diamonds. mStarD4 (GenBank AF480298); hStarD4 (GenBank AF480299); mStarD5 (GenBank AF480302); hStarD5 (GenBank AF480304); mStarD6 (GenBank AF480303); hStarD6 (GenBank AF480305); mMLN64, residues 231–446 of mouse MLN64 (GI 6225683; GI indicates the GenInfo Identifier); hMLN64, residues 230–445 of human MLN64 (GI 6225682); mStAR, residues 66–284 of mouse StAR (GI 1236243); hStAR, residues 67–285 of human StAR (GI 1351124); mPCTP, mouse phosphatidylcholine transfer protein (PCTP) (GI 6679235); hPCTP, human PCTP (GI 14771310).
Tunnel Volume Calculations and Display.
Tunnel volumes within StarD4 and MLN64 were computed with voidoo (24) using a probe radius of 1.4 Å, and were visualized with o (17).
Homology Modeling.
Automated comparative protein structure modeling (hereafter referred to as homology modeling) with modpipe (25) was carried out with the experimentally determined structures of mouse StarD4 and human MLN64 used as modeling templates. Homology models were computed with psi-blast alignments subject to the fulfillment of spatial restraints as implemented in modeller (26), and they were assessed by computing a model score that uses a statistical energy function, sequence similarity with the modeling template, and a measure of structural compactness (27). Tests with known structures have shown that models with scores from 0.7 to 1.0 have the correct fold at a 95% confidence level (27).
Results and Discussion
Overall Structure of Mouse StarD4.
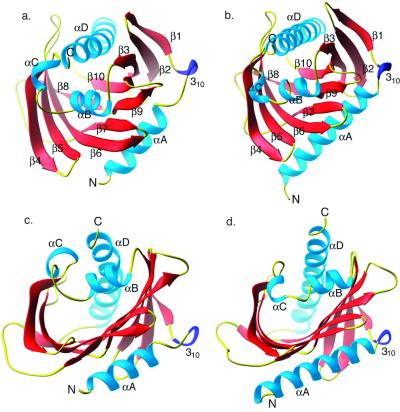
Mouse StarD4 is a single α/β domain protein (dimensions 40 Å × 46 Å × 46 Å) with a 10-stranded antiparallel (with the exception of β9, which is a continuation of β8), twisted U-shaped β-sheet flanked by two long α-helices, αA and αD. Helix αD stretches over the opening that leads into the putative lipid-binding tunnel (Fig. 2a). The strand order in StarD4 is β1-β2-β3-β10-β8 followed by β9-β7-β6-β5-β4. The linear arrangement of the secondary structural elements within the polypeptide chain is αA, 310, β1, β2, β3, αB, αC, β4, β5, β6, β7, β8, β9, β10, and αD (Fig. 1).
Figure 2.
(a and b) ribbons (32) drawing of StarD4 (a) and MLN64-START (b) with labeled N and C termini and secondary structural elements. (c and d) View into the tunnel of StarD4 (c) and MLN64 (d) with α-helices and termini labeled.
Comparison with MLN64.
Both the linear and spatial arrangements of the secondary structural elements in StarD4 (Fig. 2a) are remarkably similar to those observed in MLN64 (Fig. 2b). There are two minor differences in the assignment of the secondary structural elements in both structures. The region encompassing αA and 310 in StarD4 was interpreted as one long α-helix in MLN64. The 310 helix is present in both structures and we have decided not to merge it with the preceding αA helix. Similarly, β8 and β9 in StarD4 and MLN64 are represented as one long β-strand (β8) in the previously reported MLN64 structure (8).
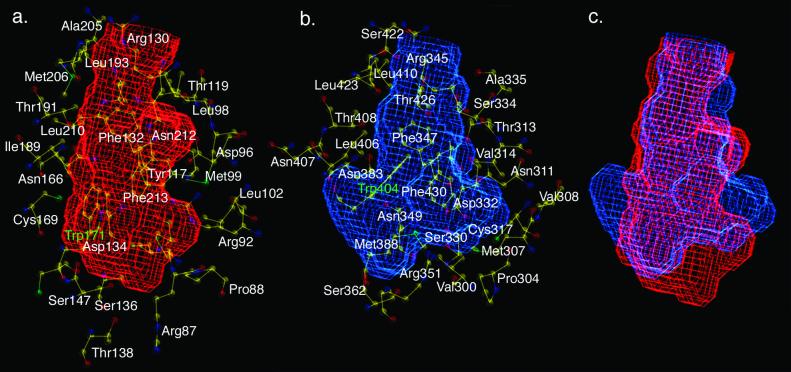
Visual inspection of a superposition of the putative lipid-binding tunnels computed with voidoo permitted identification of tunnel-lining amino acids: 41 in StarD4 and 42 in MLN64. These residues can be divided into two groups presenting either backbone (8 plus 2 Gly in StarD4, 10 plus 4 Gly in MLN64) or side-chain atoms to the tunnel interior (31 in StarD4, 28 in MLN64; Fig. 3 and Table 2, which is published as supporting information on the PNAS web site).
Figure 3.
StarD4 and MLN64 tunnel volume plots generated with voidoo (24). (a) Volume plot of the putative StarD4 lipid-binding tunnel. (B) Volume plot of the MLN64 tunnel. (c) An overlay of StarD4 (red) and MLN64 (blue) volume plots. Residues lining the tunnels are shown in a and b.
We believe that residues presenting backbone atoms are unlikely to be conserved among START domains unless they contribute to the hydrophobic core of the protein. Eight such residues found in StarD4 (Thr-118, Ala-120, Ser-128, Pro-129, Phe-135, Tyr-137, Val-150, and Tyr-188) are equivalent to MLN64 residues Val-333, Ala-335, Ser-343, Pro-344, Arg-352, Ile-365, and Ile-405, respectively. Gly-364 and Gly-387 in MLN64 are the counterparts of Gly-149 and Gly-170 in StarD4. MLN64 has two other Gly residues, Gly-384 and Gly-386, which in StarD4 are replaced by His-167 and Cys-169. MLN64 has additional nonconserved residues available for backbone interactions with tunnel-binding ligands: Ala-316, Ser-363, Ala-366, Pro-385, and Ile-419 that in StarD4 correspond to Ser-101, Cys-148, Ser-151, Pro-168, and Val-202, respectively.
Residues contributing side-chain atoms to the tunnel lining are shown in Fig. 3. We anticipate that some of these amino acids will be highly conserved if they are important for the structural integrity of the tunnel or binding of invariant parts of lipid ligands. Tunnel-lining residues that vary among START domains may determine binding specificity. In all, nine pairs of residues are conserved between the tunnel linings of StarD4 and MLN64 (StarD4 residues listed first): Pro-88/Pro-304, Arg-130/Arg-345, Phe-132/Phe-347, Ser-147/Ser-362, Asn-166/Asn-383, Thr-191/Thr-408, Leu-193/Leu-410, Thr-209/Thr-426, and Phe-213/Phe-430. Trp-171 in StarD4 and Trp-404 in MLN64 are also relevant in this analysis. Although these two aromatic side chains do not occur in the same relative position within the START domain sequence, they are located on the same plane within the tunnel and play a similar architectural role (α-carbon separation = 4.9 Å).
Previous work on MLN64 identified Met-307 and Asn-311 as likely ligand specificity determinants in START domains (8). In StarD4, Met-307 is replaced by Arg-92, and Asn-311 by Asp-96. We have identified additional pairs of residues that may be responsible for determining lipid specificity of a START domain (StarD4 residues listed first, MLN64 second): Met-99/Val-314, Leu-102/Cys-317, Asp-134/Asn-349, Ser-136/Arg-351, Cys-169/Gly-386, Trp-171/Met-388, Gln-190/Asn-407, and Leu-210/Met-427. Moreover, there are other residues in one START domain that lack structural counterparts in the other. For example, all of the side-chain atoms of Arg-87 in StarD4 are located within the tunnel. In MLN64, there is an extended loop between Val-300 and Pro-304, and the three residues within the loop are not exposed to the interior of the cavity. The Asp-332–Arg-351 salt bridge pair in MLN64, which may play a role in cholesterol binding (8), does not exist in StarD4 (the two residues are replaced by Tyr-117 and Ser-136, respectively). Instead, the Arg-92–Asp-96 pair (distance between their closest side-chain atoms = 2.9 Å) appears likely to form a salt bridge in the StarD4 tunnel. It is possible that tunnel salt bridges contribute to the binding of zwitterionic lipids instead of uncharged sterols (8). Alternatively, these salt bridges may stabilize the structure of the tunnel.
In sum, it seems likely that lipid specificity of a START domain is determined in at least two ways. One group of side chains within the tunnel could participate in direct lipid–START interactions, whereas the same and/or other side chains might impose spatial constraints on the shape and volume of the tunnel (e.g., StarD4 Trp-171 and MLN64 Trp-404). Both variations in the shape of the tunnel and local chemical environments within the tunnel probably contribute to lipid binding specificity. Verification of this hypothesis must await conclusive identification of cognate ligands that will almost certainly become cocrystallization candidates.
To gain additional insights into the mechanism of action of StarD4, we measured the volume of the putative lipid-binding tunnel by using voidoo (24) (see Fig. 2 c and d for StarD4 and MLN64 tunnel entrance views). Initial calculations indicated that the StarD4 tunnel volume is 1,424 ± 294 Å3 (a similar value of 1,820 ± 481 Å3 was observed with MLN64). These results compare favorably with the previously reported estimate of 1,900 Å3 for MLN64 (8), which is compatible with the binding of two molecules of cholesterol (28). Visual inspection of the two tunnels, however, suggests that only one cholesterol molecule can be accommodated by either protein. Visualization of the voidoo-generated volume plots using o (17) revealed that a large part of the calculated volume in both proteins fell outside the confines of the tunnel. The “spill-over” forms within the opening at the top of both tunnels surrounded by αD, the β5–β6 and αC–β4 loops. To estimate the true tunnel volumes, we blocked the opening of the tunnel with a 7-residue α-helix and repeated the calculations. The measured volume of the tunnel proper in StarD4 is 847 ± 106 Å3, and that of MLN64 = 848 ± 107 Å3. Both estimates are consistent with a START domain binding only one cholesterol molecule, reported to occupy a volume of 741 Å3 (29), or a lipid of similar size.
Sequence Analysis and Homology Modeling.
A dali (30) search of the Protein Data Bank (http://www.rcsb.org/, January 2002) with the coordinates of StarD4 identified three structurally similar proteins with high Z scores, including the START domain of human MLN64 [Z score = 26.5, root-mean-square deviation (rmsd) = 1.7 Å for 196 α-carbon pairs, PDB ID code 1EM2], birch pollen allergen (Bet v 1) from Betula verrucosa (Z score = 9.3, rmsd = 3.1 Å for 126 α-carbon pairs, PDB ID code 1BV1), and the Rattus norvegicus phosphatidylinositol transfer protein (PIPT; Z score = 6.7, rmsd = 3.4 Å for 131 α-carbon pairs, PDB ID code 1FVZ). Other structures identified in the dali search include members of the helix-grip fold family of proteins. Neither Bet v 1 nor PITP contains a recognizable START domain, and StarD4, Bet v 1, and PITP share very few amino acid identities. The putative lipid-binding tunnel of StarD4 is replaced by a deep cleft in Bet v 1. The closed lipid-binding tunnel in PITP is more than twice as large as the StarD4 tunnel (the PITP tunnel volume is 1,889 ± 259 Å3), and it appears to be compatible with the binding of diacyl phospholipids. Although there have been suggestions to the contrary (31), we favor the view that these protein structures are the result of divergent evolution.
Automated homology modeling with modpipe (25) using the StarD4 and MLN64 templates yielded models (length >100 residues, model score >0.7) for 107 proteins (January 2002): 23 with both templates, 26 with StarD4 only, and 58 with MLN64 only. In both cases, the templates yielded models for eukaryotic proteins bearing START domains and a hypothetical protein from Pseudomonas aeruginosa (PA1579; GI: 15596776). Models for the START domains of the Goodpasture antigen-binding proteins (GABP) were derived solely from the MLN64 template, which reflects higher sequence conservation between GABPs and MLN64 STARTs. Using the StarD4 template, we were able to generate models for both mouse and human StarD5 and StarD6 proteins.
Both StarD4 and MLN64 templates yielded models of each other. The StarD4 model derived from the MLN64 template versus the experimental structure of StarD4 gave an rmsd of 1.7 Å (182 α-carbon pairs, Z score = 23.2), and the MLN64 model derived from the StarD4 template versus the structure of MLN64 gave similar results (rmsd = 1.7 Å for 184 α-carbon pairs, Z score = 22.6). The agreement between our computed models and the structures of both proteins provides further evidence of the reliability of homology modeling with modpipe. In cases in which models for the same sequence were obtained from both structural templates, the average rmsd between alternative models was 1.8 Å with Z scores = 23.0 ± 3.0.
Phylogeny of the START Domain Superfamily.
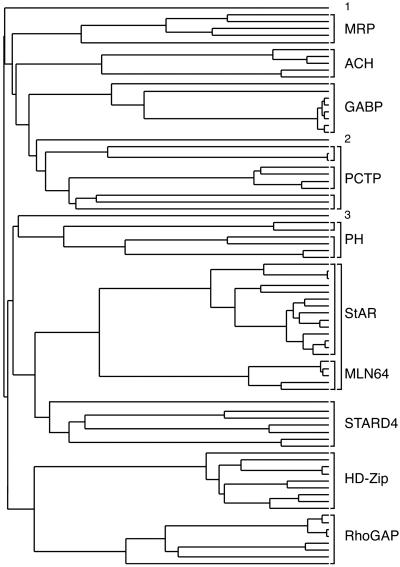
We used clustalw to align all of the START domains identified by modpipe modeling with StarD4 and MLN64, and we generated a phylogenetic tree depicted schematically in Fig. 4. START domain sequences were grouped into nine distinct subfamilies (naming reflects sequence features N-terminal to the START domain): StarD4-like proteins, MLN64/StAR, RhoGAP, HD-Zip, PCTP-like proteins divided into three subgroups, Goodpasture antigen-binding proteins (GABP), acyl-CoA thioester hydrolases (ACH), A. thaliana membrane-related proteins (MRP), and plant pleckstrin homology (PH) proteins divided into two subgroups. A subset of these subfamilies is found in the human genome (11). The StarD4 subfamily includes StarD4, StarD5, and StarD6, which are ≈30% identical to each other. In addition, we found a StarD4-like START domain in the Caenorhabditis elegans KD02D3.2 protein of unknown function (TrEMBL Q21130; residues 99–271 27% identical to mouse StarD4). This protein was tentatively included in the StarD4-like subfamily. The MLN64 and StAR START domains share an ≈35% sequence identity and have been shown to bind cholesterol. Three additional START domains are more distantly related to the other subfamilies, including Arabidopsis thaliana K21G20 (TrEMBL Q9FGQ8, outlier no. 1), Dictyostelium discoideum ChtA (TrEMBL Q9U9U5; outlier no. 2) with the closest relatives in the PH START subfamily, and a single bacterial protein from P. aeruginosa PA1579 (outlier no. 3). PA1579 shares a 15% sequence identity with StarD4 and a 16% identity with human MLN64-START over 180 residues. Homology modeling with both templates produced reliable models for this protein, with model scores of 0.79 (StarD4 template) and 0.71 (MLN64 template). A psi-blast search with PA1579 did not reveal similar eubacterial sequences containing START domains, which raises the possibility of host gene acquisition by P. aeruginosa and the gene product's involvement in pathogenicity.
Figure 4.
Phylogenetic tree representing the evolutionary relationships among the aligned START domains. Subfamily members are shown in brackets (see text). See Fig. 5 in the supporting information for identification of each protein.
In summary, the x-ray structure of the mouse StarD4 protein determined at 2.2-Å resolution revealed a putative lipid-binding tunnel comparable in volume and size to cholesterol. Comparative analysis of StarD4 and MLN64 tunnels facilitated the identification of likely lipid specificity determinants in START domain-containing proteins. Phylogenetic studies indicated the presence of multiple START subfamilies in mammals. The StarD4 and MLN64 structures, together with homology models of other START domain proteins, provide a framework for interpreting biochemical function and evolutionary relationships.
Supplementary Material
Acknowledgments
We thank Dr. K. Rajashankar for help with data collection, and Drs. J. B. Bonanno and V. Ilyin for useful discussions. This work was supported by National Institutes of Health Grants P50-GM62529 (to S.K.B.) and GM20276 (to M.J.R.). S.K.B. is an Investigator in the Howard Hughes Medical Institute. StarD4 represents target T526 from the New York Structural Genomics Research Consortium.
Abbreviations
- StAR
steroidogenic acute regulatory protein
- START
StAR-related lipid transfer
- StarD4
cholesterol-regulated START domain protein 4
- PCTP
phosphatidylcholine transfer protein
- GST
glutathione S-transferase
- Se-Met
selenomethionine
- GI
GenInfo Identifier
- MALDI-MS
matrix-assisted laser desorption ionization mass spectrometry
- rmsd
root-mean-square deviation
Footnotes
Data deposition: The atomic coordinates and structure factors for the StarD4 protein have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1JSS).
References
- 1.Stocco D M. J Endocrinol. 2000;164:247–253. doi: 10.1677/joe.0.1640247. [DOI] [PubMed] [Google Scholar]
- 2.Lin D, Sugawara T, Strauss J F, 3rd, Clark B J, Stocco D M, Saenger P, Rogol A, Miller W L. Science. 1995;267:1828–1831. doi: 10.1126/science.7892608. [DOI] [PubMed] [Google Scholar]
- 3.Ponting C P, Aravind L. Trends Biochem Sci. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- 4.Newton A C. Curr Biol. 1995;5:973–976. doi: 10.1016/s0960-9822(95)00191-6. [DOI] [PubMed] [Google Scholar]
- 5.Shaw G. BioEssays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- 6.Iyer L M, Koonin E V, Aravind L. Proteins. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 7.Hurley J H, Misra S. Annu Rev Biophys Biomol Struct. 2000;29:49–79. doi: 10.1146/annurev.biophys.29.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsujishita Y, Hurley J H. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 9.Akiyama N, Sasaki H, Ishizuka T, Kishi T, Sakamoto H, Onda M, Hirai H, Yazaki Y, Sugimura T, Terada M. Cancer Res. 1997;57:3548–3553. [PubMed] [Google Scholar]
- 10.Alpy F, Stoeckel M E, Dierich A, Escola J M, Wendling C, Chenard M P, Vanier M T, Gruenberg J, Tomasetto C, Rio M C. J Biol Chem. 2001;276:4261–4269. doi: 10.1074/jbc.M006279200. [DOI] [PubMed] [Google Scholar]
- 11.Soccio R E, Adams R M, Romanowski M J, Sehayek E, Burley S K, Breslow J L. Proc Natl Acad Sci USA. 2002;99:6943–6948. doi: 10.1073/pnas.052143799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson W A. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Weeks C M, Miller R. Acta Crystallogr D. 1999;55:492–500. doi: 10.1107/s0907444998012633. [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z. In: Isomorphous Replacement and Anomalous Scattering. Wolf W, Evans P R, Leslie A G W, editors. Warrington, U.K.: SERC Daresbury Lab.; 1991. pp. 80–85. [Google Scholar]
- 17.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Weik M, Ravelli R B, Kryger G, McSweeney S, Raves M L, Harel M, Gros P, Silman I, Kroon J, Sussman J L. Proc Natl Acad Sci USA. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 21.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins D G, Bleasby A J, Fuchs R. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 23.Henikoff S, Henikoff J G. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleywegt G J, Jones T A. Acta Crystallogr D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 25.Pieper U, Eswar N, Stuart A C, Ilyin V A, Sali A. Nucleic Acids Res. 2002;30:255–259. doi: 10.1093/nar/30.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez R, Sali A. Proc Natl Acad Sci USA. 1998;95:13597–13602. doi: 10.1073/pnas.95.23.13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrescu A D, Gallegos A M, Okamura Y, Strauss J F, 3rd, Schroeder F. J Biol Chem. 2001;276:36970–36982. doi: 10.1074/jbc.M101939200. [DOI] [PubMed] [Google Scholar]
- 29.Schroeder F, Jefferson J R, Powell D, Incerpi S, Woodford J K, Colles S M, Myers-Payne S, Emge T, Hubbell T, Moncecchi D, et al. Mol Cell Biochem. 1993;123:73–83. doi: 10.1007/BF01076477. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 31.Yoder M D, Thomas L M, Tremblay J M, Oliver R L, Yarbrough L R, Helmkamp G M., Jr J Biol Chem. 2001;276:9246–9252. doi: 10.1074/jbc.M010131200. [DOI] [PubMed] [Google Scholar]
- 32.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.