Summary
Calcium signaling is crucial for endothelial cell homeostasis. Alterations in intracellular calcium levels due to shear stress are linked to vascular dysfunction and diseases. Here, we present a protocol to perform live calcium imaging by using a live calcium indicator on human lung endothelial cells subjected to shear stress in a commercially available microfluidic device (Ibidi Luer VI). We also outline data analysis procedures. This protocol can test altered shear stress effects in the vasculature and identify potential drug treatments.
For complete information on the generation and use of this protocol, please refer to Amoakon et al.1
Subject areas: Biotechnology and bioengineering, Cell-based Assays, Tissue Engineering
Graphical abstract

Highlights
-
•
Guidelines for cultivation of human endothelial cells
-
•
Cultivation of human endothelial cells in a microfluidic device under flow
-
•
Guidelines for live-cell calcium imaging in a microfluidic device
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Calcium signaling is crucial for endothelial cell homeostasis. Alterations in intracellular calcium levels due to shear stress are linked to vascular dysfunction and diseases. Here, we present a protocol to perform live calcium imaging by using a live calcium indicator on human lung endothelial cells subjected to shear stress in a commercially available microfluidic device (Ibidi Luer VI). We also outline data analysis procedures. This protocol can test altered shear stress effects in the vasculature and identify potential drug treatments.
Before you begin
We developed a protocol to perform live calcium imaging on endothelial cells subjected to shear stress in a commercially available microfluidic device. This protocol can be especially useful for studying calcium-permeable mechanosensitive channels such as PIEZO1, which is highly expressed in endothelial cells and is the primary shear stress sensor.
The experimental steps outlined below can be used to successfully culture endothelial cells in the Ibidi Luer VI microfluidic device and utilize a live calcium indicator, Fluo-8, to examine change in intracellular calcium levels. Fluo-8 has a higher brightness than other dyes, such as Fluo-3 and Fluo-4, and is more versatile due to its temperature stability and ability to be loaded into cells at room temperature. This procedure requires a cell culture incubator dedicated to containing a peristaltic pump (Ismatec, #ISM4312). Peristaltic pumps are a more cost-effective option than syringe pumps with a lower risk of contamination. Use of a peristaltic pump allows for continuous operation of the pump, instead of limiting the user to the volume of media in the syringe. A peristaltic pump can handle larger volumes than a syringe pump, which is necessary to achieve the desired rate of shear stress for our usage. However, if precision is required and a low shear stress is all that is needed, a syringe pump would be ideal. While this protocol has been optimized for human lung microvascular endothelial cells (HMVEC-Ls), it can also be utilized with other types of endothelial cells by adjusting the cell culture medium, seeding density, and time of culture prior to imaging. Imaging can be performed using microscope systems other than the Lionheart microscope suggested here, provided they are equipped with a 20X objective and an incubation chamber. This protocol may be applicable for use with other commercially available single-channel microfluidic devices or even adapted to other chip designs. The tips mentioned in the protocol apply to the Ibidi Luer VI microfluidic device and may or may not be applicable to other devices.
Cell culture maintenance for later use in the device
Timing: 1 week before experiment
-
1.
Prepare complete medium for HMVEC-Ls by adding the supplements to the base medium (EGM-2 Bulletkit (Lonza, CC-3162)).
-
2.
Culture HMVEC-Ls to 70%–90% confluency in a T-25 or T-75 flask. Replace cell culture medium every day with fresh medium warmed at 37°C for 15–30 min.
Note: For reference, in a T-75 flask, the culture should reach around 1.5 × 106 cells when confluent.
Note: We recommend using cells between passage 3–7 for experiments.
Note: We recommend using a T-25 or T-75 flasks instead of tissue culture dishes because the flasks can be vented through the filtered cap to allow for gas exchange while minimizing medium evaporation.
Note: Confluency can be assessed quantitatively using previously established methods if desired.2
Modification and sterilization of supplies for media flow
Timing: >1 day before experiment, 1–2 h
-
3.Prepare the pump and inlet tubing:
-
a.Assemble the inlet tubing (Figure 1A):
-
i.Cut 5.5 cm of tubing X (the tubing with an inner diameter of 1/8″).
-
ii.Cut 22.5 cm of tubing Y (the tubing with an inner diameter of 1/16″).
-
iii.Connect tubing X to the end of tubing Y.
-
i.
-
b.Assemble the outlet tubing (Figure 1B):
-
i.Cut 4.5 cm of tubing XX (the tubing with an inner diameter of 1/8″).
-
ii.Cut 22 cm length of tubing YY (the tubing with an inner diameter of 1/16″).
-
iii.Connect tubing XX and tubing YY.
-
i.
-
c.Place the tubing in a sterilization pouch and seal.
-
a.
-
4.Prepare the media bottle:
-
a.Use a soldering iron to make 2 holes in the cap of a 15 mL conical. Ensure that the hole is large enough to insert the appropriate tubing pieces (Ibidi pump tubing, tubing XX, and tubing YY).
-
b.Secure the lid on the 15 mL conical tube, leaving it slightly loosened.
-
c.Place the 15 mL conical, 1 male luer lock, and the tubing from step 3 into a sterilization pouch and seal.
-
a.
-
5.
Place 4–6 of each luer lock size (3/32″ and 1/16″) in a sterilization pouch and seal.
-
6.
Autoclave the supplies using the glass cycle to ensure the tubing and media bottle remain intact.
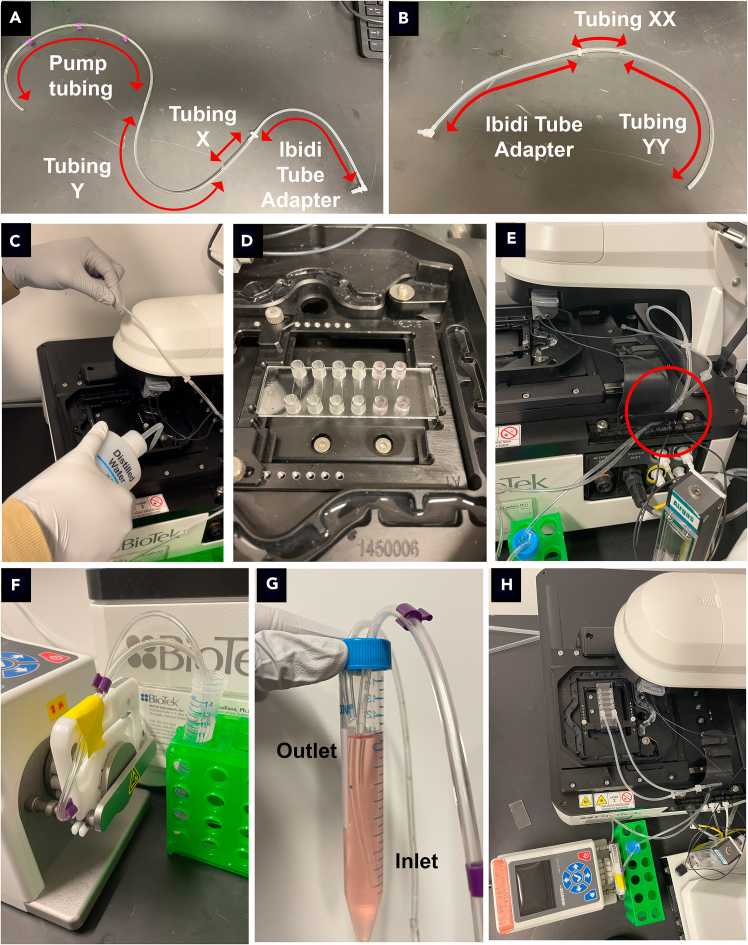
Figure 1.
Assembling the flow system for live imaging of the Ibidi Luer VI
To assemble the pump system for calcium imaging, assemble the inlet tubing (A) and the outlet tubing (B). Then, fill the moat surrounding the chip with distilled water (C), and place the chip on the microscope stage (D). Secure the inlet and outlet tubing in the clip on the side of the microscope (circled in red) (E). Place the inlet tubing into the 15 mL conical containing medium, securing the tubing to the cassette (F), and then place the outlet tubing into the 15 mL conical (G). The assembled system is shown in (H).
Equipment setup: Establishing the plate type for imaging chips on the Lionheart microscope
Timing: >1 day before experiment, 1 h
-
7.
Turn on the Lionheart microscope and open the “Gen5” software.
-
8.
Select Protocols --> Create New.
-
9.To make the appropriate plate type:
-
a.In Menu, go to System --> Plate Types.
-
b.Highlight “Chamber Slide 6x2 HTC” and click “Copy.”
-
c.Make the selections shown in Figure 2, and then click “OK”.
-
d.Select the box next to the plate type, and then click “Close.”
-
a.
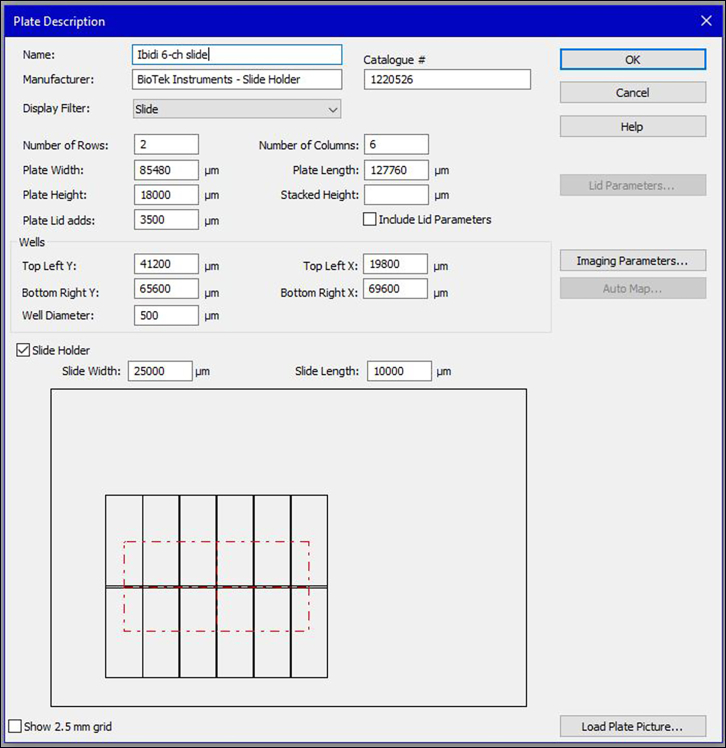
Figure 2.
The settings for the plate type in the Gen5 software for imaging acquisition
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| EGM-2 Bulletkit culture medium with supplements | Lonza | CC-3162 |
| Penicillin-Streptomycin | Gibco | 15-140-163 |
| Plasmocin prophylactic | InvivoGen | ANT-MPP |
| Phosphate buffer solution | Invitrogen | 10010 |
| TrypLE Express without phenol red | Life Technologies | 12604013 |
| Trypsin neutralizing solution (TNS) | Lonza | CC-5002 |
| Dimethyl sulfoxide | Sigma-Aldrich | D2650-100 mL |
| Corning Hank’s balanced salt solutions | Fisher Scientific | MT21023CV |
| Pluronic F-127 | MilliporeSigma | P2443 |
| Critical commercial assays | ||
| Fluo-8 Calcium Flux Assay Kit - No Wash | Abcam | ab112129 |
| Experimental models: Cell lines | ||
| Human Lung Microvascular Endothelial Cells (HMVEC-L) | Lonza | CC-2527 |
| Software and algorithms | ||
| LC Pro Plugin for Fiji | ImageJ | https://imagej.nih.gov/ij/plugins/lc-pro/index.html |
| Other | ||
| Instant sealing sterilization pouch | Fisher Scientific | 01-812-54 |
| PYREX Media/storage bottles, narrow mouth, round, with screw cap, Corning, 50 mL | Avantor | 16157–068 |
| Acrodisc 25 mm syringe filter | Pall Corporation | 4612 |
| μ-Slide VI 0.4 IbiTreat (surface modification) | ibidi | 80606 |
| Ismatec REGLO ICC Digital peristaltic pump; 3-channel, 12 roller | Cole-Parmer | ISM4312 |
| E Tygon tubing, 2.06 mm ID, purple/purple/purple | Ismatec | MFLX96461-42 |
| PVC tubing, inner diameter 1/16″, outer diameter 1/8″, thickness 1/32″ | Fisher Scientific | 14-387-345 |
| PVC tubing, inner diameter 1/8″, outer diameter 1/4″, thickness 1/16″ | Fisher Scientific | 14-387-338 |
| Fisherbrand Male Luer with 1/16 in. ID Barb - polypropylene | Fisher Scientific | 01-000-116 |
| Fisherbrand Male Luer with 3/32 in. ID Barb - polypropylene | Fisher Scientific | 01-000-124 |
| BioTek Lionheart FX automated microscope | Agilent | N/A |
| 10 mL plastic syringe | Fisher Scientific | 14955459 |
| Allegra X-14R centrifuge | Beckman Coulter | A99465 |
| Cellometer Auto T4 cell counter | Nexcelom Bioscience | AutoT4-302-1136 |
| Forma Series II Water Jacket CO2 incubator | Thermo Scientific | 3110 |
| Kimberly-Clark Professional Kimtech Science Kimwipes Delicate Task Wipers, 1-Ply | Fisher Scientific | 06–666 |
| ANBES soldering iron | Amazon | 00612677965225 |
Materials and equipment
EGM-2 Bulletkit culture medium
| Reagent | Final concentration | Amount |
|---|---|---|
| EBM-2 Basal Medium | N/A | 500 mL |
| Fetal Bovine Serum | 2% | 10.00 mL |
| Hydrocortisone | 0.20 μg/mL | 0.20 mL |
| hFGF-B | 4 ng/mL | 2 mL |
| VEGF | 2 ng/mL | 0.50 mL |
| R3-IGF-1, 0.50 mL | 5 ng/mL | 0.50 mL |
| Ascorbic Acid | 75.00 μg/mL | 0.50 mL |
| hEGF | 10 ng/mL | 0.50 mL |
| GA-1000 | Gentamicin: 30 μg/mL Amphotericin: 15 ng/mL |
0.50 mL |
| Heparin | 1 ng/mL | 0.50 mL |
| Penicillin-Streptomycin | 1% | 5 mL |
| Plasmocin Prophylactic | 0.2% | 1 mL |
| Total | N/A | 521.20 mL |
Note: Store at –4°C for up to 1 month. All supplies are contained in the BulletKit culture medium.
Step-by-step method details
Cell seeding onto the microfluidic device
Timing: 1 day
-
1.
Warm the EGM-2 medium, phosphate buffer solution (PBS), and TrypLE at 37°C for 15–30 min.
Note: Make sure TNS is completely thawed and warmed up. This may take longer than 30 min.
-
2.
Wash cells cultured on tissue culture flask with PBS (use 2 mL for a T-25, 6 mL for a T-75).
Note: Cells should have a cobblestone-like appearance.
-
3.
Remove PBS and add TrypLE (2 mL for T-25, 6 mL for T-75). Immediately move the flask into a cell culture incubator (set at 37°C) for 6–7 min.
CRITICAL: After 6–7 min, the cells should have lifted off the base of the flask and look like light, round circles floating in the flask.
-
4.
Inactivate reaction by adding media directly to cells in TrypLE (6 mL for T-25, 15 mL for T-75).
Note: HMVEC-Ls attach strongly to flask, and 6 min incubation is typically not enough to detach all the cells. However, longer incubation time presents the risk or cytotoxicity to the cells. Therefore, after adding additional media, use a 10 mL pipette to vigorously pipet medium up and down onto the bottom of the flask to manually detach as many cells as possible.
Note: If using trypsin to detach cells, we recommend using TNS instead of EGM-2 culture medium to inactivate the reaction as EGM-2 contains only 2% fetal bovine serum (FBS), which is not sufficient to quickly inactivate trypsin. Typical medium contains 10% FBS.
-
5.
Pipet solution into 15 mL conical tubes. If you used a T-25, then one tube will be sufficient, but for a T-75, split the solution equally into two 15 mL conical tubes (10.5 mL per tube).
-
6.
Centrifuge at 1200 RPM for 5 min at 4°C.
-
7.
Take the conical tubes back into the cell culture hood.
-
8.
Remove supernatant and resuspend in EGM-2 medium in 1 mL.
Note: Ideally 1 mL of EGM-2 should be used, but as little as 200 μL could be used to achieve higher concentration.
-
9.
Count cells using the Cellometer Auto T4 Cell Counter.
Note: If an automated cell counter is not available, cell counting can be done manually using previously published protocols.3
-
10.
Dilute cells to the desired concentration (25,000–30,000 cells in 30 μL [833–1,000 cells/μL])
-
11.
Seed 25,000–30,000 endothelial cells in 30 μL (maximum volume one channel can hold) of EGM-2 medium per channel of the chip.
CRITICAL: Return chip to 37°C incubator for 10 min to allow cells to attach. Without this step, the subsequent addition of medium to the inlet and outlet will push cells out of the channel.
Note: Chips come pre-sterilized, and no additional sterilization steps are necessary before use.
-
12.
Add 60 μL of EGM-2 medium to the inlet and outlet.
-
13.
Place the chip in a 150 mm petri dish and place a wadded KimWipe soaked in sterile water next to the chip to prevent medium from evaporating overnight (Figure 3).
Note: As an alternative, sterile water can be added directly into a 35 mm Petri dish that is placed in the 150 mm Petri dish containing the chip.
Note: Adding sterile water directly to the 150 mm Petri dish is not recommended, as it may inadvertently enter the chip.
-
14.
Cover the dish and leave the chip in the incubator overnight for cells to attach and proliferate.
-
15.
If the chip is not used for live imaging the following day, change medium daily if it is not connected to the flow system.
Note: Culturing under flow is recommended, as this maintains a higher percentage of cell attachment.
Figure 3.
Set up for overnight incubation of the Ibidi Luer VI microfluidic device
The microfluidic chip is to be incubated overnight at 37°C inside a 100-mm Petri dish with a wet, wadded KimWipe soaked in sterile water.
Preparation and culture of the microfluidic device under flow
Timing: 1–2 h
-
16.
Warm the EGM-2 medium at 37°C for 15–30 min.
-
17.
Place the sterilized pouches of tubing in the incubator to warm during this time, as well.
-
18.
Place the pump in the incubator. Turn on the pump and set the tube ID to the appropriate tube inner diameter (2.06 mm) and the flow rate to 11.83 mL/min.
Note: This corresponds to a shear stress of 15 dynes/cm2, which is the physiologically relevant amount of shear stress in pulmonary arteries.
-
19.
In the cell culture hood, fill the chip inlet and outlet to the brim with medium (Figure 4A).
-
20.
Flush the pump tubing with 70% ethanol twice and sterile phosphate-buffered saline once, then bring into the cell culture hood.
-
21.Assemble the pump tubing in the hood:
-
a.Assemble the inlet tubing (Figure 1A):
-
i.Connect tubing Y into the end of the pump tubing that is closest to a purple stop.
-
ii.Ensure that tubing X is connected to the end of tubing Y.
-
iii.Connect the straight connector on one of the Ibidi tube adapters to the end of tubing X.Note: It can be easier to insert the connector into the tubing by placing one side of the connector in the pump tubing while squeezing the tubing about 5 mm away from the end and then focus on inserting the other side of the connector into the tubing. Do not touch the end of the tubing, as this may cause contamination.
-
i.
-
b.Assemble the outlet tubing (Figure 1B):
-
i.Ensure tubing XX and tubing YY are connected.
-
ii.Connect the straight connector on one of the Ibidi tube adapters to the end of tubing XX.
-
i.
-
a.
-
22.
Fill the autoclaved 15 mL conical with at least 10 mL of media.
-
23.
In the cell culture hood, attach a 10 mL syringe to the 3/32″ luer lock.
-
24.
Fill the syringe with media.
-
25.
Connect the syringe to the pump tubing (Figure 1A).
-
26.
Fill the inlet with media until the tip of the Ibidi tube adapter is overflowing with media.
-
27.
Connect to chip inlet plug (Figure 1B).
Note: Insert the inlet fully into the chip to prevent the formation of bubbles.
CRITICAL: Ensure there are no bubbles in the inlet tubing before connecting it to the chip. If bubbles enter the chip, the cells may become detached.
-
28.
Attach a 10 mL syringe to the 1/16″ luer lock.
-
29.
Fill the syringe with media.
-
30.
Connect the syringe to tubing YY of the outlet tubing.
-
31.
Fill the outlet with media until the tip of the Ibidi tube adapter is overflowing with media.
-
32.
Connect to chip outlet plug.
Note: Insert the outlet fully into the chip to prevent the formation of bubbles.
CRITICAL: Ensure there are no bubbles in the outlet tubing before connecting it to the chip. If bubbles enter the chip, the cells may become detached.
-
33.
Place a binder clip about 5 cm from the end of the outlet tubing (tubing YY).
CRITICAL: Addition of the binder clip ensures media will not leave the tubing. If media leaves the tubing, air bubbles will form in the tubing.
-
34.
Detach the syringe.
-
35.
Without touching the end of the outlet tubing, insert the outlet tubing into the media bottle.
Note: The outlet tubing should remain above the surface of the media.
-
36.
Remove the syringe connected to the pump tubing, and carefully place the pump tubing into the bottom of the media bottle, below the surface of the media.
-
37.
Move the pump/tubing system into the incubator.
-
38.
Place the 3-stop pump tubing into the cassette, using the two stops furthest away from what would be the inlet side of the tubing.
Note: Using these two stops ensures that there is enough tubing length at the inlet.
-
39.
Remove the binder clip from the outlet tubing.
-
40.
Begin flow.
Note: If media droplets are leaving the end of the outlet tubing, media is successfully flowing through the chip.
-
41.
Change medium every 2 days by aspirating media from the 15 mL conical, leaving a small drop at the end of the inlet to prevent the introduction of air bubbles.
Figure 4.
Design of the Ibidi Luer VI microfluidic device
The Ibidi Luer VI contains 6 channels where the inlet and outlet flank the cell culture surface (A). The inlet and outlet are female Luer adapters, suitable to attach the male Luer tubing adapters (B).
Live calcium imaging in the microfluidic device
Timing: 2–3 h
Timing: the total experiment involves 10 min of imaging (for step 65)
This step utilizes Fluo-8 live cell calcium tracker to track calcium transients through the endothelial cells in the microfluidic device.
Note: Before beginning, confirm that the cells on the chip are confluent and have a cobblestone shape. Additionally, using an incubator while conducting imaging will ensure the system remains at 37°C, which will increase reproducibility of results.
-
42.
Warm 45 mL of EGM-2 medium per chip used at 37°C for 15–20 min.
Note: All 45 mL of medium will not be used, but it is recommended to have extra in case there are air bubbles in the tubing system that need to be removed.
-
43.
Make the appropriate amount of loading dye solution by combining the following amounts of the components.
Note: X is the number of channels to be incubated with Fluo-8.
Note: For example, to fill 6 channels, mix 562.5 μL HHBS, 62.502 μL Pluronic F-127, and 1.248 μL Fluo-8 Dye.
Loading dye solution
| Reagent | Amount |
|---|---|
| Hanks’ Buffer w/ 20 mM HEPES (HHBS) | 93.75∗X μL |
| Pluronic F-127 | 10.417∗X μL |
| Fluo-8 Dye | 0.208∗X μL |
| Total | 104.375∗X μL |
Note: The Fluo-8 Calcium Flux Assay Kit protocol is designed for use with a 96-well plate. Here, we have adapted it for use with the Ibidi Luer VI chip by dividing the given volumes for a full 96-well plate (9000 μL HHBS, 1000 μL Pluronic F-127, and 20 μL Fluo-8 Dye) by 96.
-
44.
Mix the loading dye made in the previous step with an equal amount of HHBS.
Note: For example, to fill 6 channels total and have 200 μL per channel, mix 600 μL of loading dye with 600 μL HHBS.
Note: Each channel can only hold 30 μL; however, approximately 200 μL of loading dye per channel is needed to ensure that previous medium is completely exchanged with loading dye by replacing several times.
-
45.If the chip is connected for flow, remove the flow lines from the chip:
-
a.Place a binder clip on the outlet side of the chip, approximately 6 cm from the chip.
-
b.Cut the tubing on the outlet side at least 7 cm away from the chip.
-
c.Slowly remove the inlet connector from the chip, giving liquid time to fill the inlet.
-
d.After ensuring the inlet is overflowing, slowly remove the outlet connector from the chip, filling the inlet with more media to prevent air from entering the chip.
-
a.
CRITICAL: Remove the connectors slowly and ensure the inlet and outlet remain filled with liquid to prevent air from entering the chip. If air enters the chip, the cells will detach.
-
46.For each channel, remove medium in the following order to completely exchange medium.
 CRITICAL: Remove medium by pipetting with a P-200 tip that is against the wall away from the outlet channel opening (Figure 5). Some media will still remain in the channel. Additionally, if there are air bubbles in the inlet or outlet, remove using the pipette. If there is a bubble in the pipette tip, change tips to prevent the bubble from entering the channel.
CRITICAL: Remove medium by pipetting with a P-200 tip that is against the wall away from the outlet channel opening (Figure 5). Some media will still remain in the channel. Additionally, if there are air bubbles in the inlet or outlet, remove using the pipette. If there is a bubble in the pipette tip, change tips to prevent the bubble from entering the channel.-
a.Pipet 60 μL of loading mixture (loading dye+HHBS) into the inlet channel.
-
b.Remove 60 μL of old medium from the outlet.
-
c.Repeat steps a & b.
-
d.Add 60 μL of loading mixture into the inlet.
-
a.
-
47.
Incubate chip for 30 min at 37°C in the cell culture incubator.
-
48.Incubate chip for 1 hour at room temperature in the dark.Note: To incubate the chip in the dark, place the chip in a 100-mm Petri dish, and add Kimwipes soaked in PBS in a 35 mm uncovered Petri dish. (Instead of a Kimwipe soaked in PBS, PBS can also just be added to the 35 mm Petri dish.) Cover the 100-mm Petri dish with aluminum foil.
-
a.During this incubation period:
-
i.Turn on the Lionheart microscope and open the “Gen5” software. Begin warming the incubation chamber to 37°C by going to Task Manager → Instrument Control → Incubate. Set “Requested” to 37°C and check the “on” box.Note: Preparing the microscope before will reduce the amount of time the chip is outside at room temperature, which could impact cell function, reducing the quality of the experiment.
-
i.
-
a.
-
49.While the chip is incubating, prepare the pump and inlet tubing.
-
a.Turn on the pump and set the tube ID to the appropriate tube inner diameter (2.06 mm) and the flow rate to 11.83 mL/min.Note: Do not start flow at this time.
-
b.Assemble the inlet tubing (Figure 1A):
-
i.Flush the pump tubing with 70% ethanol twice and sterile Phosphate-buffered saline once.
-
ii.Connect tubing Y into the end of the pump tubing that is closest to a purple stop.
-
iii.Place the 3-stop pump tubing into the cassette, using the two stops furthest away from what would be the inlet side of the tubing. This ensures that there is enough tubing length at the inlet.
-
iv.Connect the straight connector on one of the Ibidi tube adapters to the end of tubing X.Note: It can be easier to insert the connector into the tubing by placing one side of the connector in the pump tubing while squeezing the tubing about 5 mm away from the end and then focus on inserting the other side of the connector into the tubing.
-
i.
-
c.Place the cassette on the pump.
-
d.Assemble the outlet tubing (Figure 1B):
-
i.Connect the straight connector on one of the Ibidi tube adapters to the end of tubing XX.
-
i.
-
a.
-
50.
Add distilled water to the hydration tray of the microscope (Figure 1C).
-
51.
Prepare 15 mL conical tubes with the desired compounds to test (for example, 20 μM CFTRinh-172). Make at least 10 mL total of the medium/compound solution.
-
52.When the chip is done incubating, exchange the loading mixture completely with new medium using the following steps:Note: This step will minimize background during imaging.
-
a.Pipette 60 μL of medium into the inlet.
-
b.Remove 60 μL of old loading mixture from the outlet.
-
c.Repeat steps a & b.
-
d.Add 60 μL of medium into the inlet.
-
a.
-
53.
Fill the chip inlet and outlet to the brim with medium, and then place the chip on the microscope (Figure 1D).
-
54.
Secure the inlet and outlet tubing in the clip on the side of the microscope (as shown in Figure 1E).
-
55.Connect the 15 mL conical tubes with the desired compounds to the chip.
-
a.In the Task Manager menu, select Instrument Control and click “Plate In.”
 CRITICAL: If this step is not done prior to connecting the tubing, this can unintentionally disconnect the tubing from the chip.
CRITICAL: If this step is not done prior to connecting the tubing, this can unintentionally disconnect the tubing from the chip. -
b.Place the end of the inlet tubing into the 15 mL conical containing medium and secure the inlet tubing to the cassette using tape (as shown in Figure 1F). Be sure to place the tubing as far into the conical as possible without touching the bottom of the tube.
-
c.Start flow on the pump and watch the inlet tubing fill with medium until a small amount of medium drips out of the end of the inlet tubing. If you see bubbles in the tubing, run the flow until the bubble is removed.Note: Paper towels or a Petri dish can be used to catch excess medium that flows from the tubing.
-
d.Once there is a convex meniscus at the end of the inlet tubing connector, attach it to the chip inlet. The medium will slightly overflow, but this is important to prevent air bubbles.
-
e.Connect a second Ibidi tube adapter to the chip outlet and place in the 15 mL conical with medium. This tubing should not be as far down in the conical as the pump tubing. (Figure 1G).Note: Maintaining the differential height of the pump tubing and outlet tubing will prevent introduction of air bubbles into the chip.
-
a.
-
56.
Next, start a new experiment by going to Task Manager → Imager Manual Mode → Capture now.
-
57.
In the “Imager Manual Mode – Load Vessel” menu, select the plate created above under “Vessel type.”
-
58.
Determine the focus to be imaged by ensuring the appropriate 4X lens and “GFP 469, 525” are selected. Also, adjust the focus by clicking “autofocus” or by manually scrolling.
-
59.
To determine the area to be imaged, click on the value next to “well” to ensure the desired well is being imaged. Ensure that the microscope is focused on a confluent area in the middle of the channel using the arrows.
-
60.
Determine the optimum exposure for your sample under the “Exposure” heading. Using the autoexposure option may be helpful.
-
61.Under the “Imaging Mode” heading, ensure that Kinetic Imaging is checked. Click “Edit Imaging Step.”
-
a.Under imaging step, ensure that “Use current focus height” and “slow kinetics” is selected.
-
b.Click on “Kinetic step” and set the run time to 10 min and interval to 2 Seconds. (Reads will adjust to 301).
-
c.Click on “Read GFP 469,525”, and ensure that channel 1 is selected.
-
d.Ensure that the Imaging Procedure box matches the selections in Figure 6, and then click OK to exit.
-
a.
-
62.
Select the camera icon to begin recording.
-
63.
Begin flow on pump immediately after first image is taken during recording. This first image can serve as the baseline image before flow is applied.
Note: Because change of intracellular calcium in response to shear stress occurs very rapidly, it is important not to start the pump until recording is underway.
-
64.
Stop the pump after 1 min of reading has elapsed, and let the microscope continue reading for 9 more min to visualize sustained changes in intracellular calcium levels.
-
65.
To save images manually, select File → Create Zip File. Images can be moved to the desired location for quantification and analysis.
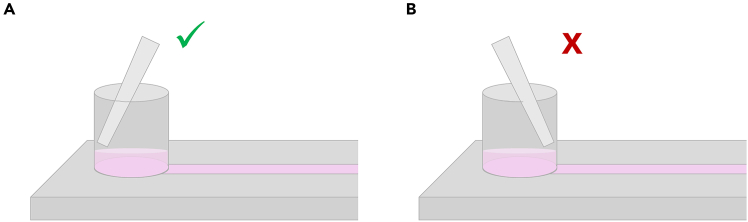
Figure 5.
Medium exchange in the Ibidi Luer VI chip
How to properly remove medium from the microfluidic device. Ensure the pipette tip is against the wall away from the channel opening (A), and not directly against the channel opening (B) to avoid aspirating cells and prevent the introduction of air bubbles.
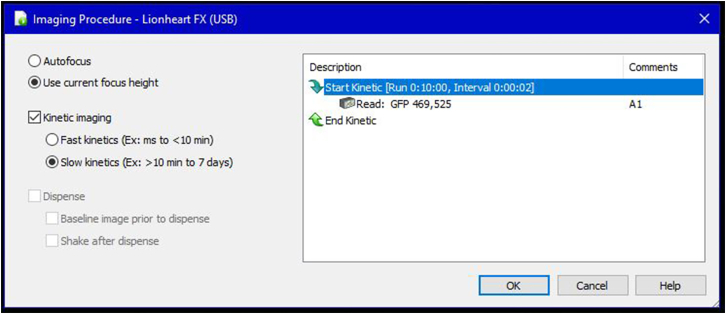
Figure 6.
The settings for the imaging procedure in the Gen5 software
Quantification and statistical analysis of change in intracellular calcium concentrations
Timing: 30 min to 1 h
This step utilizes ImageJ to quantify images of calcium transients taken in the previous section. LC Pro, an ImageJ plugin, performs automated regions of interest (ROI) analysis on grayscale image sequences. After the user specifies parameters such as ROI diameter and the frame rate of the imaging sequence, LC Pro will algorithmically detect local maxima or changes in intensity to identify regions where increases in fluorescence intensity occur. For each ROI, the fluorescence intensity over time is measured, and time traces of intensity versus time for each ROI are generated. The plugin provides output files including the magnitude, duration, and spatial spread of each detected calcium event.
The spatial and temporal complexity of calcium imaging data can be challenging to analyze manually. LC Pro streamlines analysis of calcium imaging data by automating ROI selection and intensity quantification, increasing the efficiency and reproducibility of calcium image analysis. The format of outputs is easily translatable into graphical outputs for publication. Additionally, LC Pro is a freely available, open-source tool that is accessible to users without high programming expertise. This facilitates broader implementation of calcium imaging analysis across the scientific community.
Other options for analysis of calcium transients include CardiAP,4 which relies on manual selection of ROIs, and Suite2P,5 which utilizes frequency-based approaches for transient identification and ROI selection.
Note: LC Pro must be downloaded prior to analysis. Instructions are available at: https://imagej.net/ij/plugins/lc-pro/.
-
66.Use ImageJ to open raw images folder from the experiment:
-
a.Select File → Import → Image Sequence and choose the first image of the stack of images.
-
a.
-
67.
In the “Sequence Options” box, select “Convert to 8-bit Greyscale,” “Sort names numerically,” and “Use virtual stack.” Save the image stack by selecting to File → Save As → Tiff.
Note: Save this stack in a folder containing only this stack.
-
68.Select and analyze calcium transients of ROIs:
-
a.Open LC Pro by going to Plugins → LC Pro.
-
b.In the “LC_Pro Settings” box, select “15” for the ROI diameter and “1” for frame rate with a p-value of 0.05. Click “OK”.
-
c.Choose folder containing Tiff image stack created in Step 1.
-
d.Open “ROI normalized.txt” from LC Pro’s output folder inside the folder containing the Tiff image. This file contains the intensity versus time values for the ROI.
-
e.Copy and paste values from the .txt file into Excel.
-
a.
-
69.
In Excel, normalize all values by the initial fluorescence value.
-
70.
Make graph and run statistics with the desired program (for example, GraphPad Prism) using the grouped table option with the number of replicate values being equal to the sample with the least number of ROIs (Figure 7).
Figure 7.
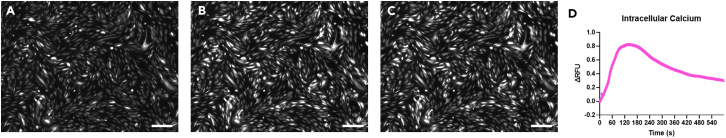
Images and analysis from live imaging of calcium transients in the Ibidi Luer VI chip
(A and B) Representative raw images of live recording of calcium transients of HMVEC-Ls under shear stress at time = 0 (A), at time = 1 min (B) and at time = 2 min (max intensity) (C) (15 dyne/cm2) used to quantify data.
(D) Time course from raw images. Change in intracellular calcium levels of HMVEC-Ls subjected to shear stress (15 dyne/cm2) for only 1 min out of the 10-min total recording time (n = 93 cells; data are mean ± SE). Scale bar = 200 μm.
Expected outcomes
This protocol provides a platform to reliably record and impartially analyze changes in intracellular calcium concentration in endothelial cells in response to shear stress on a commercial microfluidic device. This method has the advantage of increased throughput through the use of a commercially available chip and utilized a microscope with a stage setup that is compatible with this device. If the system is kept closed to maintain sterility, the automated microscope could be used for more long-term studies, including measuring multiple time points on the same chip overnight. Additionally, it also reduces user bias by performing an automated analysis of the calcium signaling. This protocol can be used to examine the effects of shear stress on calcium signaling on other cell types as well as the effects of pharmaceutical drugs and possible biomarkers of disease on calcium signaling, furthering our understanding of diseases such as atherosclerosis, hypertension, and stroke as well as neurovascular disorders.
Limitations
Culturing of cells on the chip can be challenging due to small volumes of medium, difficulties achieving optimal confluency, and the constant need to maintain stable flow and temperature conditions while preventing air bubbles and leaks. Cell number plays a role in calcium signaling, and performing a cell count of the conditions or cell types used for experiments can ensure similar numbers of cells are on each chip. While cell counts can be performed on these chips, the use of a fluorescent dye prevents other forms of analysis of these chips. The quality of images taken will affect the results obtained by analysis.
Troubleshooting
Problem 1
After step 40 or 63, cells are washed away during shear flow application.
Potential solutions
There are multiple solutions to this problem:
-
•
Reduce the flow rate.
-
•
Ensure the cells are not overconfluent – this can impact the cell viability and the ability to withstand high levels of shear stress.
-
•
Ensure the cells are a low passage number – this can impact cell viability and their ability to withstand high stress.
-
•
Use the Ibitreat-coated Ibidi chips. If Ibidi chips are not being used, always coat microfluidic channels with an appropriate coating (examples are poly-amino acids, basement membrane matrix, and extracellular matrix proteins such as fibronectin, gelatin, and collagen). Additionally, if coated chips are used, ensure that the chips are not expired.
Problem 2
After step 40 or 63, microfluidic system is leaking.
Potential solution
Inspect the tubing and connectors to determine where the leak originates:
-
•
If the leakage is at the pump tubing/inlet tubing junction, make sure the tubing is the correct size to fit securely over the connector for the inlet tubing. Also confirm that there is not a tear in the tubing.
-
•
If the leakage is at the inlet tubing/chip inlet junction, ensure the connector from the inlet tubing is firmly embedded in the chip inlet.
-
•
If the leakage is at the chip outlet/outlet tubing junction, ensure the connector from the outlet tubing is firmly embedded in the chip outlet.
Problem 3
After steps beginning flow in the device (step 40 or 63) or conducting medium changes in the device (step 13, 46, or 52), there are air bubbles in the microfluidic device.
Potential solution
Air bubbles may be introduced during medium changes or during preparation of the tubing before beginning flow.
-
•During medium changes:
-
○When adding and removing medium, reduce the amount of medium removed from the outlet to not completely drain the inlet.
-
○Make sure to place the cell culture tip against the wall away from the channel when removing medium from the outlet.
-
○When pipetting into the inlet, do not go past the first stop on the pipet.
-
○Ensure medium is fully warmed, as temperature differences between medium and incubator could create small bubbles.
-
○
-
•During preparation of tubing before beginning flow:
-
○Check the filled inlet tubing for bubbles before connecting to the chip inlet. If there are bubbles, then run medium through the inlet tubing until the bubble exits, and then connect the inlet tubing to the chip inlet.
-
○Confirm that the inlet tubing is at the bottom of the medium container, while the outlet tubing is closer to the top. If this is indeed the case, then bubbles in the outlet tubing should be released and go to the surface of the medium without risking the introduction of bubbles into the inlet tubing.
-
○Before connecting tubing to the chip, fill the inlet and outlet so high the medium slightly domes above the inlet/outlet. Also fill the tubing with enough medium to produce a similar dome at the connector.
-
○
-
•
If there is a bubble in the channel, then place the pipette tip next to the inlet and forcefully expel medium from the tip, flushing the channel with enough flow to dislodge the bubble. On some occasions, tilting the chip can be enough to move an air bubble to either the inlet or outlet, especially when the bubble is small enough to easily pass through.
-
•
A bubble trap can be added between the inlet tubing and the chip inlet to prevent bubbles that originate in the tubing. Bubble traps are commercial or custom-fabricated devices that capture and remove bubbles from liquid prior to flow into microchannels, preventing damage to cells. Common types of bubble traps include hydrodynamic traps, membrane-based traps, and coalescing filters. These utilize fluid flow patterns, selective permeability, and density, respectively, to divert and trap bubbles. To add a bubble trap to the system, first select a trap compatible with the system requirements (fluid type, flow rate, and connection type). Install the bubble trap upstream of your microfluidic device, but downstream of the pump tubing. (Anywhere between the pump tubing and the Ibidi tube adapter is appropriate.) If a PTFE membrane-based trap is used, ensure the trap is mounted with fluid ports at the lowest point to allow bubbles to naturally rise to the top to be expelled through the membrane.
Problem 4
Poor fluorescent signal of the Fluo-8 calcium indicator (step 60).
Potential solution
Exposing the indicator to light can reduce fluorescent signal. If the indicator was not exposed to light, adjust the microscope settings (exposure time, power of the excitation source, and gain) to improve visualization. Also, confirm that the indicator is not expired.
Problem 5
Unsure if medium exchange is occurring properly in step 13, 46, or 52.
Potential solution
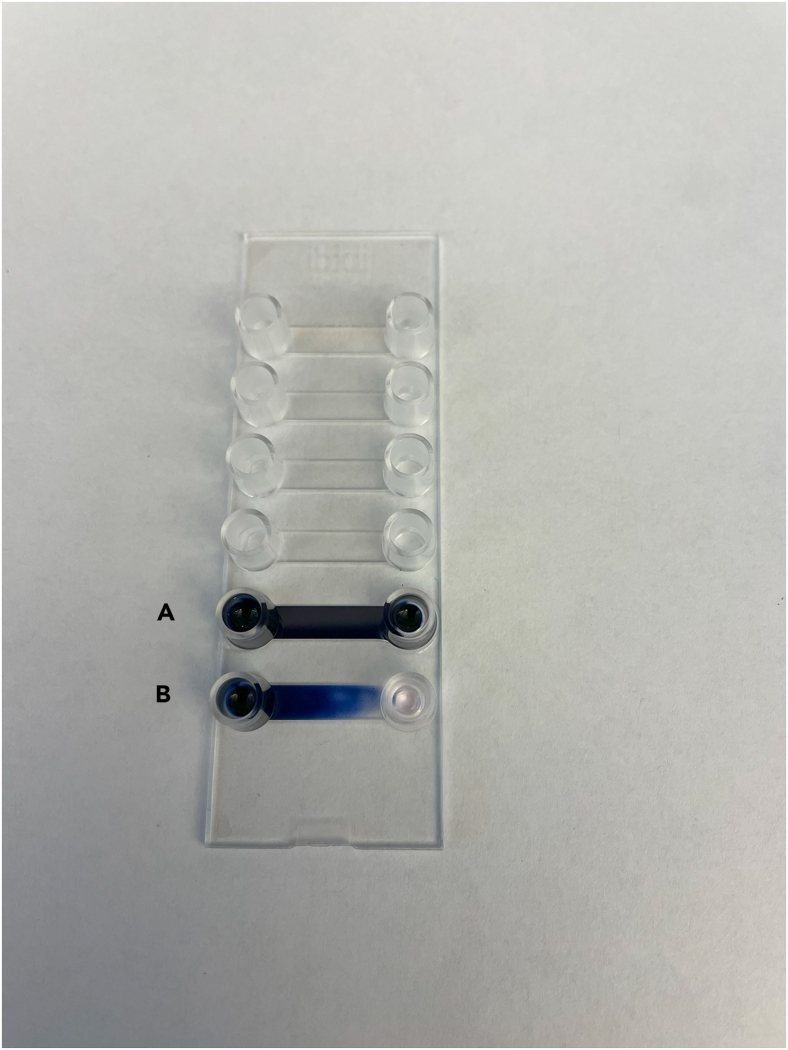
If unsure whether medium exchange is occurring as expected, practice by filling the chip with medium and then replacing the medium with a dye/water solution (Figure 8).
Figure 8.
Troubleshooting medium exchange in the Ibidi Luer VI chip
Channels A and B of this chip were filled with medium, and then the medium exchange protocol was completed using a blue dye solution. Channel A demonstrates a successful medium exchange, and Channel B demonstrates an unsuccessful medium exchange, where medium still remains in the channel.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nethika Ariyasinghe (nethika.ariyasinghe@cshs.org).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by either technical contact, Nethika Ariyasinghe (nethika.ariyasinghe@cshs.org) or Jean-Pierre Amoakon (amoakoja@mail.uc.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
These data do not report original code.
-
•
Data are available from the lead contact upon request.
Acknowledgments
This protocol was supported by the American Heart Association Pre-Doctoral Fellowship (J.-P.A., AHA award 903099), the NIH grants (A.P.N., NHLBI HL147351; NIDDK P30-DK117467), and the Cystic Fibrosis Foundation (A.P.N., AMIN19A0). We thank Makayla Roberts for her technical assistance with troubleshooting the Lionheart microscope.
Author contributions
N.A. and J.-P.A. contributed to the writing and editing of this protocol, and A.P.N. contributed to the review, editing, and generation of funding.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Nethika Ariyasinghe, Email: nethika.ariyasinghe@cshs.org.
Jean-Pierre Amoakon, Email: amoakoja@mail.uc.edu.
References
- 1.Amoakon J.P., Lee J., Liyanage P., Arora K., Karlstaedt A., Mylavarapu G., Amin R., Naren A.P. Defective CFTR modulates mechanosensitive channels TRPV4 and PIEZO1 and drives endothelial barrier failure. iScience. 2024;27 doi: 10.1016/j.isci.2024.110703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busschots S., O’Toole S., O’Leary J.J., Stordal B. Non-invasive and non-destructive measurements of confluence in cultured adherent cell lines. MethodsX. 2015;2:8–13. doi: 10.1016/j.mex.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phelan M.C., Lawler G. Cell Counting. Curr. Protoc. Cytom. 2001;Appendix 3 doi: 10.1002/0471142956.cya03as00. Appendix 3A. [DOI] [PubMed] [Google Scholar]
- 4.Velez Rueda A.J., Gonano L.A., Smith A.G., Parisi G., Fornasari M.S., Sommese L.M. CARDIAP: calcium transients confocal image analysis tool. Front. Bioinform. 2023;3 doi: 10.3389/fbinf.2023.1137815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang A., Zhao C., Sheffield M.E.J. A Preprocessing Toolbox for 2-Photon Subcellular Calcium Imaging. eNeuro. 2025;12 doi: 10.1523/ENEURO.0565-24.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
These data do not report original code.
-
•
Data are available from the lead contact upon request.