Abstract
Somatic hypermutation generates variants of antibody genes and underpins the affinity maturation of antibodies. It is restricted to the V-gene segments, and although it decays exponentially toward the 3′end, it includes recognizable hot spots. Although the detailed mechanism of hypermutation remains elusive, the process may take place in two separate stages, preferentially targeting G/Cs in the first and A/Ts in the second stage. It seems that MSH2 is involved in the second stage, and that activation induced deaminase (AID) is implicated in the control of hypermutation. The constitutively hypermutating cell line Ramos expresses AID, and we have prepared transfectants that express a chimeric AID-green fluorescent protein. The fluorescence is strongly detected in the cytoplasm but not in the nucleus. Yet, the chimeric protein increases the hypermutation rate either directly or, more likely, indirectly, by favoring the transport of AID into the nucleus. Thus, in Ramos, AID seems to be rate limiting. Unexpectedly, the proportion of deletions also is increased. The increase in mutation rate detected by a fast cytofluorimetric method based on the accumulation of sIgM-loss mutants correlates with the increase measured by mutations defined by sequence analysis. The higher mutation rate is largely explained by the higher proportion of mutated clones, indicating that AID controls the number of cells that undergo hypermutation but not the number of mutations that are incorporated in each mutation round.
Two somatic events are characteristic of the maturation of antibodies. The first is the class switch, whereby the same V-gene fragment, initially expressed as IgM, is transposed to other positions in the same heavy chain gene complex to be expressed as one of the other heavy chain classes/subclasses. During the second event, the V-segment is subjected to a very high level of point mutations (hypermutation) that provides variants that are subsequently selected by their increased affinity for antigen (1). Common features point to a molecular connection in the initiation of both events. For example, both class switch recombination and hypermutation are focused on the motif AGCT (2). Furthermore activation induced deaminase (AID), a factor initially recognized as essential to class switch recombination, also affects hypermutation (3, 4). Thus, AID is proposed to be a common early element required for class switch recombination, gene conversion in chicken cells, and hypermutation (5, 6).
In this paper, we describe results obtained by transfecting Ramos with a construct encoding a chimeric protein AID attached to the green fluorescent protein (GFP). Ramos is a lymphoblastoid cell line that expresses AID (7) and hypermutates in vitro with the characteristics of in vivo hypermutation; namely, it is restricted to both light and heavy chains of antibody genes and, more specifically, targets the variable gene fragment while the constant segment is excluded (8). However, there is an interesting difference. In vivo, there are two mutational hot spots that revolve around the consensus motif GAGCT or TA when read in the coding strand (9–11). There is evidence that in vivo mutations occur in both strands and, therefore, the complementary sequences of those motifs are also (albeit to a lesser extent) mutational hot spots (12, 13). There is, however, a general bias toward mutations of A in the coding strand, suggesting a superimposed polarity (13, 14). A candidate responsible for this polarity is the error prone polymerase η (15, 16). In the case of Ramos, the TA hot spots and A bias are absent, and the same is found in vivo in MSH2−/− mice (8, 17, 18). This finding led to the proposal that the mutation events occur in two stages (18). The first introduces GAGCT hot spots and the second, dependent on the first and indeed on the expression of the MSH2 repair-related gene, is strongly biased toward TA sequences. Therefore by working with Ramos we are restricting our observations to the putative first stage.
Below we describe our attempt to localize the intracellular site of action of AID. By confocal imaging we find the AID-GFP fluorescence only in the cytoplasm. We further demonstrate that despite this unexpected localization the chimeric protein displays a biological activity, because it is able to increase the number of cells that undergo hypermutation. Indeed the mutation rate of transfected lines reach values of almost 10−4, which is still somewhat lower than the one derived from in vivo studies (19). The difference may be accounted for by the additional mutations introduced in vivo during the MSH2-dependent phase.
Material and Methods
Vector Construction.
Human AID cDNA was cloned by reverse transcription (RT)-PCR from total RNA from the Ramos cell line by using the primers 5′-ATGGACAGCCTCTTGATGAACCGGAGG-3′ and 5′-TCAAAGTCCCAAAGTACGAAATGCGTC-3′ and cloned into pCR2.1 TOPO vector (Invitrogen). After checking the correct clone by restriction analysis and sequencing, AID was subcloned into pEGFP-N3 (CLONTECH) as an N terminus fusion to enhanced GFP by the addition of an EcoRI site upstream of the ATG of AID, the removal of the termination codon, and the addition of a Sal I site at the 3′ end of the cDNA using the following primers: 5′-CGGGAATTCATGGACAGCCTCTTGATGAACCGGAGG-3′ and 5′-GCGGCGTCGACAAGTCCCAAAGTACGAAATGCGTC-3′. The final clone, hA2pEGFP-N3, was confirmed to be in frame by sequencing with the primer 5′-GTCAGATCCGCTAGCGCTAC-3′.
Cell Cultures and Transfections.
A random clone of Ramos was grown for a period of more than a month, and cells were taken for subsequent experiments. Ramos (107 cells) was transfected with 20 μg of linear hA2pEGFP-N3 or pEGFP-N3 DNA and then distributed in 24-culture wells. After 24 h, G418 was added to a concentration of 4 mg/ml. Cells were grown in RPMI medium 1640 (GIBCO) supplemented with 10% (vol/vol) FCS (HyClone) and 50 μM β-mercaptoethanol. Clones were derived by fluorescence activated cell sorter (FACS; MoFlo, Cytomation, Fort Collins, CO) delivering single cells on 96-well plates (Corning) containing 0.1 ml of medium. It was noticed that when the resistant clones were grown in medium containing G418 (at 4.0 mg/ml), the cell division time was considerably slower than either the wild-type or the transfected cells grown in ordinary medium. Thus, to measure the mutation frequency of IgM to nonexpression and the accumulation of mutants by sequence analysis, the transfected clones were grown in ordinary medium.
Sequence Analysis.
The rearranged VH segment of Ramos was amplified from genomic DNA as described (8) and cloned into M13mp18. Sequences were done on an ABI377 (Perkin–Elmer) with BigDye Terminator chemistry and M13(-21) primer. Sequences were aligned and analyzed with PREGAP4 and GAP4 software (20).
Fluorescent Staining and Confocal Microscopy.
About a million cells were pelleted and washed with PBS/1% (vol/vol) BSA. The pellet was resuspended in 250 μl of PBS/1% BSA containing 1:100 phycoerythrin-coupled goat anti-human IgM (Serotec, UK). After 1 h in ice, an excess of the PBS/BSA was added and the pelleted cells were resuspended in 0.5 ml of PBS/BSA containing 10 μg of propidium iodide.
Cells were attached to poly-l-lysine-coated slides and fixed in 4% (vol/vol) paraformaldehyde/PBS. The localization of the AID-GFP fusion protein was imaged by confocal microscopy with a Nikon E800 microscope and a Bio-Rad Radience-Plus scanning system.
Mutation Rates.
To measure mutation rates, cells were expanded in the absence of G418. Two methods were used. In the first method, we followed the accumulation of mutants of a phenotypic selected population (21). We took advantage of the fact that the emergence of IgM− Ramos cells was largely because of several nonsense mutations (8). Thus, live cells that strongly stained with the anti-IgM were purified by FACS. Cells then were grown, and the accumulation of IgM− cells was followed over time by using a Facscalibur (Becton Dickinson). There is a linear increase over a period of several days, and the slope is the rate of accumulation of mutants over time. This slope is not the mutation rate of a single base but the sum of the accumulation of a number of nonsense mutations. Therefore, it gives a good comparison of the hypermutation rates in closely related populations.
The second method was based on sequence analysis. Individual cells were grown continuously and exponentially for 28 or 29 days (cell viability was well over 95% in all cases) starting with a single cell in 100 μl of medium to a final volume of 1 liter. All variations from the consensus sequence were taken as mutations. The mutation frequency (m) was calculated as m = N/bp/gen, where N is the total number of mutations, bp is the total number of bases sequenced, and gen is the estimated number of generations elapsed based on a division time of 20 h.
Results
AID-GFP Is Not Detected in the Nucleus.
Ramos was transfected with an artificial gene driven by a cytomegalovirus promoter and encoding a chimeric protein AID joined to the N terminus of the GFP. Only 14 of 24 cultures grew, suggesting that most, and probably all, were single clones. Cytofluorimetric analysis revealed that only a small fraction (up to 12%) of the cells were strongly fluorescent. Two of those cultures were selected as representative for further studies. The GFP+/IgM+ fraction of each of them was sorted, expanded (B1), and then subjected to a new round of sorting that after expansion (B2) was subjected to a third round of sorting (B3). Although there was a clear enrichment of GFP+ cells in the first round of selection, no obvious improvement was achieved by further rounds (Fig. 1). The persistent negative population arose because the reversion to the GFP− phenotype was fast. This reversion was observed with cultures kept in G418 suggesting that the loss was largely phenotypic rather than the result of loss of vector (see below).
Figure 1.
Expression of AID-GFP and sIgM in transfectants. (Upper Left) Clone RhA2/12 was not stained for sIgM. The 3.5% GFP+ fraction was sorted and grown for 25 days to derive the B1 population shown (Lower Left) after 25 days in culture. The bright IgM and GFP fraction was sorted and grown to derive B2 that is shown after 14 days of growth (Upper Right). The bright sIgM and GFP+ fraction of panel B2 was sorted and analyzed after 4 days of growth (Lower Right).
Fluorescence microscopy revealed a strong cytoplasmic stain with no evidence of nuclear fluorescence (Fig. 2). Western blots of cytoplasmic extracts did not reveal any cleaved GFP in the transfectants (not shown). Although we could not exclude the possibility that there was a minuscule amount of fluorescent protein undetectable by our methods that penetrated the nucleus, the control transfectants expressing GFP alone disclosed indiscriminate fluorescence subcellular localization (not shown). This result would suggest that cleavage of the chimeric protein was not likely responsible for the phenotype of the transfectants. The apparently exclusive cytoplasmic location of the AID-GFP chimeric protein does not prove that AID itself does not penetrate the nucleus. The chimeric protein could have been totally inactive and/or unable to penetrate the nuclear membrane. Therefore, it was necessary to establish whether the chimeric protein was biologically active.
Figure 2.
Confocal microscopy of AID-GFP-transfected cells. Three serial confocal sections (top is the most distal) of the same field are shown on the left column. The spacing between sections was 1.5 μm. (Bar = 5 μm.) Differential interference transmission images of the sections are shown to the right. The cytoplasmic indentations can be clearly seen in the transmission images.
Overexpression of AID-GFP Increases the Rate of sIgM Loss.
We first analyzed the rate at which sIgM− cells arose. Such cells would largely derive by nonsense mutations at any of the triplets that differ by a single base from a stop codon (8). Examination of the DNA sequence of wild-type Ramos showed that about 1 in 20 of the possible point mutations would lead to a nonsense codon. In addition, some of the mutations were likely to be deletions leading to loss of sIgM. Therefore, the rate of mutation to sIgM− was expected to be above 20-fold the average V-region mutation rate per bp, making this type of analysis a very convenient approach. Thus, the chosen clonal populations were further selected to eliminate all sIgM−/GFP− and the sIgM+/GFP+ cells were grown for several days. At different time points, we measured the relative number of cells expressing a mutated phenotype. The results were used to calculate the apparent mutation rate at which the sIgM− cells arose. Although on average at least 95% of the selected cells were initially GFP+, after 8 days of growth, only ≈80% of them remained, and these were excluded from the calculation. The slope of the linear graphs shown in Fig. 3 was the rate of accumulation of sIGM− cells per day. Controls of Ramos transfectants expressing GFP alone were used for comparison. Table 1 summarizes the experiments performed with the cultures analyzed. The apparent mutation rate of sIgM+ to sIgM− within the fluorescent AID-GFP cells was more than two-fold higher than that of the control. The difference was highly significant (P < 0.005).
Figure 3.
Rate of loss of sIgM determined by cytofluorimetry. sIgM+ and GFP+ cells were sorted (day 0), and the emergence of sIgM− cells was plotted against time. The slope of the regression line represents the rate at which mutant cells arise per day.
Table 1.
Rate of mutation to sIgM negative
| RhA2 (AID-GFP) | Controls | ||
|---|---|---|---|
| 11 B1 | 7.5 | RGFP2 | 3.2 |
| 11 B2 | 7.5 | RGFP2 | 2.4 |
| 11 B3 | 9.6 | RGFP8 | 4.4 |
| 12 B2 | 12.3 | ||
| 12 B3 | 8.9 | ||
| Average* | 9.2 | 3.3 | |
103 cells per day.
Significance of t test, P < 0.005.
AID-GFP Increases the Probability That a Cell Initiates Hypermutation and the Frequency of Deletions.
To ascertain that overexpression of AID led to an increase in hypermutability, we analyzed the rate of accumulation of mutants by sequence analysis. For this purpose, we grew several clones of Ramos and of transfectants in parallel. We started with single cells, and the cultures were slowly diluted to keep them in logarithmic growth until they contained about 109 cells in 1 liter of medium. This result was achieved in about 28 and 29 days in Ramos and the transfectants, respectively, confirming that the generation time of both was very similar. DNA from each culture was then prepared and the VH segment was amplified and cloned. The sequences of the isolated clones were then compared. Apart from a higher prevalence of deletions (see below), the distribution and nature of the mutants generated by transfected clones (Fig. 4) was not significantly different from the ones generated by Ramos either in this work (not shown) or those from previous experiments (8).
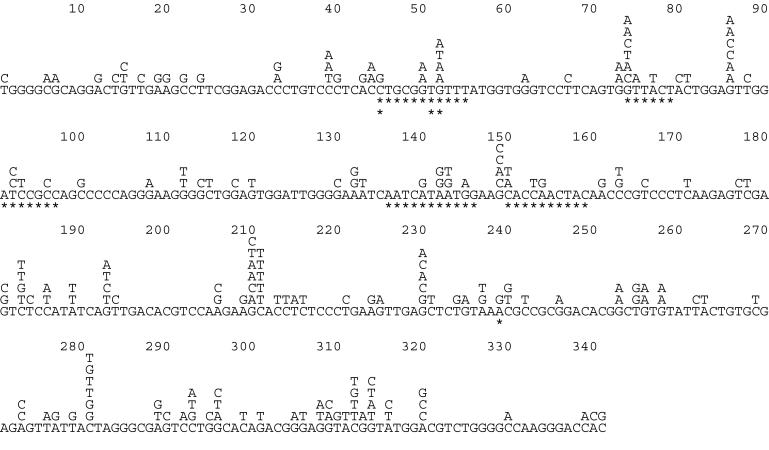
Figure 4.
Mutations in the VH segment of AID-transfected Ramos. All mutations that could have a clonal origin have been excluded from the figure. * under the consensus sequence marks deletions. Differences in the consensus sequences were observed in positions 96 (C to T) and 181 (G to A) in clone A46 and in positions 293 (C to A) and 310 (T to A) in clone A93.
The results supported the conclusions derived by the fluorescence method (Fig. 5 pie graphs and Table 2 with mutations). The increase in mutation rates caused by AID over-expression was significant, although not as marked as the one derived by the fluorescence method. The difference between both sets of data (1.7 vs. 2.8) could be because of the increased proportion of frame-shift mutations observed among the transfected cells. Indeed, whereas only about 1 of 20 point mutations leads to a stop codon, at least 2/3 of deletions will give rise to IgM loss. In the analysis of Ramos clones, no deletions were found, whereas the transfectants accumulated 11 deletions, at least 7 of which were not clonally related (shown in Fig. 4).
Figure 5.
Pie graphs of mutation analysis. The wedges represent the fraction of clones with the indicated number of mutations in the VH segments. The number of clones analyzed is shown in the center. Deletions are included as a single mutation event.
Table 2.
Sequence defined mutations
| Clone | Sequences
|
Total bases | Deletions | Point mutation | Mutation rate* | ||
|---|---|---|---|---|---|---|---|
| Mutated | Not mutated | Ratio | |||||
| AID Transfected RHA2/12B3 Subclones (35 generations) | |||||||
| A06 | 21 | 12 | 1.75 | 11253 | 3 | 31 | 8.1 |
| A21 | 17 | 13 | 1.31 | 10230 | 0 | 21 | 5.9 |
| A46 | 15 | 12 | 1.25 | 9207 | 1 | 26 | 8.4 |
| A63 | 19 | 21 | 0.90 | 13640 | 0 | 34 | 7.1 |
| A88 | 38 | 29 | 1.30 | 22847 | 4 | 52 | 7.0 |
| A93 | 46 | 32 | 1.44 | 26598 | 3 | 67 | 7.5 |
| Total | 156 | 119 | 1.31 | 93775 | 11 | 231 | 7.4† |
| RAMOS Subclones (34 generations) | |||||||
| R17 | 17 | 23 | 0.74 | 13640 | 0 | 23 | 5.0 |
| R38 | 10 | 27 | 0.37 | 12617 | 0 | 10 | 2.3 |
| R87 | 19 | 24 | 0.79 | 14663 | 0 | 28 | 5.6 |
| Total | 46 | 74 | 0.62 | 40920 | 0 | 61 | 4.3† |
10−5 × bp/gen.
Significance of t test P < 0.01.
The proportion of unmutated cells was significantly lower in the AID-transfected clones than in Ramos (Fig. 5). The fact that there were occasional cells among transfectants that accumulated four or five mutations was not likely to be pure chance. On the contrary, they were best explained by the statistically increased probability of cells repeatedly entering the mutation cycle. Indeed, a closer examination of the data reveals that the ratio of mutated vs. unmutated cells increased by a factor of 2.1, which parallels the 1.7 fold increase in mutation rate (Table 2). Thus the increased mutation frequency can be fully explained by an increase in the proportion of cells that mutated.
Discussion
In this paper, we test a fast and simple method to estimate changes in mutation rates of cell lines that is based on the phenotypic change from IgM+ to IgM−. This method gave an average value of 9.2 × 10−3 per cell per day for the AID-GFP transfectants, significantly higher than the 3.3 × 10−3 per cell per day for controls. These results were confirmed by direct sequence analysis, which also gave a clear indication that over-expression of AID-GFP increased the rate of hypermutation. Indeed, after 1 month of continuous growth, all transfectant clones accumulated many more mutations than Ramos clones, although the increase in the rate of mutation was somewhat smaller than the one obtained by the fluorescence method. Interestingly, the difference could be explained by the increase in the proportion of frame-shift mutants (Table 2). This increase is because at least two of three deletions give rise to premature chain terminations whereas only about 1 in 20 point mutations gives rise to stop codons. Thus, the results demonstrate the reliability of the much simpler fluorescence approach to reveal alterations in mutation rates.
We cannot offer a convincing explanation of why the expression of the chimeric protein increases the proportion of deletions. Deletions have been shown to be an integral part of hypermutation (22), and, therefore, we believe that far from being artifactual, there is a mechanistic basis for the increase. It is possible that the increase reflects an alteration in the balance of the different steps of the establishment and fixation of mismatches. For instance, one could imagine that as the rate-limiting activity of AID is overcome, the enzymes involved in normal hypermutation become limiting and other repair processes are put in operation. This interpretation could help to explain the variations in the reported frequency of deletions in cell lines and in vivo (23–25).
The results of this paper provide further evidence for the involvement of AID in hypermutation. Such a role was strongly suggested by the observation that in AID−/− mice and humans affected by hyperIgM syndrome with a defective AID, there was a significant decrease in hypermutation frequency in vivo (3, 4). In addition, our results suggest that although the constitutive hypermutating cell Ramos expresses AID, the amount present in the wild-type cell is a rate-limiting step of its hypermutation phenotype. While this manuscript was being completed, new evidence for the involvement of AID in hypermutation, V-gene conversion, and class switch recombination was published (5, 26–28). Particularly relevant to our results is that Martin et al. (26) showed that the hypermutation rate of Ramos could be substantially increased in AID transfectants to a maximum mutation rate of about 10−5 bp per generation. This rate is almost 10-fold lower than that of our transfectants, perhaps because they started with a hypermutation-defective Ramos clone. Although we have been able to increase the constitutive hypermutation rate of a lymphoblastoid cell line to an estimated value of almost 10−4 bp per generation, this rate remains somewhat lower than the in vivo 3–5 × 10−4 bp per generation derived from the rate of accumulation of silent mutations (29).
Ramos was transfected with the chimeric protein AID joined to GFP in an attempt to localize the site of action of AID. Initially, the majority of the transfected cells that contained the plasmid (resistant to G418) failed to express AID-GFP. This finding was true for a variety of independently transfected clones, perhaps because cells expressing the chimeric protein have a growth disadvantage. Sorting and expanding the small fraction of fluorescent transfectants solved this problem. When the cells were analyzed by confocal microscopy, the fluorescence was found in the cytoplasm, and we could not detect nuclear expression. Taken at face value, our results suggest that the site of action of AID-GFP is in the cytoplasm. Indeed, it has been proposed that AID may be an editing enzyme that modifies the mRNA of an unknown protein (30, 31). Specific modifications of mRNA sequences by deamination of A or C is one of the posttranscription editing functions that regulates protein expression (32). In general, RNA editing is a nuclear event, but the possibility remains that at least in the case of AID, it could be primarily cytoplasmic (33). Of course, although we see no trace of nuclear fluorescence in transfected cells, we cannot exclude that an undetectable quantity of AID-GFP is sufficient for its action.
However we prefer an alternative explanation for the cytoplasmic action of AID-GFP because the absence of nuclear fluorescence does not necessarily mean that AID itself is not in the nucleus. Indeed, AID (like GFP that has a similar molecular weight) but not AID-GFP may freely enter the nucleus. In such a case, the phenotype of the transfectants can be explained if the free AID concentration is controlled by interaction with a cytoplasmic protein and if AID-GFP competes with this interaction, thus increasing the concentration of AID. For example, the nuclear concentration of β-catenin depends on binding to other proteins controlling subcellular localization and targeted degradation in the cytoplasm (34).
Based on the analyses of the base substitution biases in normal mice and of the hypermutation pattern of MSH2−/− mice, we suggest that hypermutation involves two distinct stages, and that each stage is characterized by biases in the mutated bases (13, 18). In the case of cell lines, the constitutive nature of the hypermutation seems to be restricted to only one stage. Each of those stages must include a number of steps initiated by specific events, and AID seems to be a very early one (5, 6, 27). However, AID activity could have two consequences for hypermutation. It may signal a cell to initiate the process without affecting the number of mutations introduced before cell division or it may affect both. Our results show that the increase in the mutation rate is explained by the increase in the number of cells that have initiated hypermutation. Indeed, the ratio of mutated vs. unmutated clones increased by a factor of 2.4, compared with a 1.7-fold increase in mutation rates (Table 2). Control of initiation but not of intrinsic hypermutability is an essential requirement of our kinetic model of hypermutation decay (35).
Therefore, it seems that two proteins directly implicated in hypermutation, namely AID and MSH2, act in a complementary fashion. AID is involved in the initiation step that introduces the G/C bias but does not control the number of mutants introduced in each cell cycle; the MSH2-dependent phase only acts on the already AID-targeted cells and introduces extra mutations biased toward A residues in the coding strand.
Acknowledgments
César will be sorely missed. We thank Richard Grenfell for cell sorting, Facundo Batista and Stefi Reichel for help with confocal microscopy, and Kevin Hiom for helpful discussions. This work was supported in part by the Association for International Cancer Research.
Abbreviations
- AID
activation induced deaminase
- GFP
green fluorescent protein
References
- 1.Griffiths G M, Berek C, Kaartinen M, Milstein C. Nature (London) 1984;312:271–275. doi: 10.1038/312271a0. [DOI] [PubMed] [Google Scholar]
- 2.Ehrenstein M R, Neuberger M S. EMBO J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 4.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 5. Harris, R. S., Sale, J. E., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Curr. Biol. 12. [DOI] [PubMed]
- 6.Petersen S, Casellas R, Reina-San-Martin B, Chen H T, Difilippantonio M J, Wilson P C, Hanitsch L, Celeste A, Muramatsu M, Pilch D R, et al. Nature (London) 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Bardwell P D, Woo C J, Poltoratsky V, Scharff M D, Martin A. Int Immunol. 2001;13:1175–1184. doi: 10.1093/intimm/13.9.1175. [DOI] [PubMed] [Google Scholar]
- 8.Sale J E, Neuberger M S. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- 9.Betz A G, Rada C, Pannell R, Milstein C, Neuberger M S. Proc Natl Acad Sci USA. 1993;90:2385–2388. doi: 10.1073/pnas.90.6.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goyenechea B, Milstein C. Proc Natl Acad Sci USA. 1996;93:13979–13984. doi: 10.1073/pnas.93.24.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogozin I B, Kolchanov N A. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 12.Dorner T, Foster S J, Farner N L, Lipsky P E. Eur J Immunol. 1998;28:3384–3396. doi: 10.1002/(SICI)1521-4141(199810)28:10<3384::AID-IMMU3384>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 13.Milstein C, Neuberger M S, Staden R. Proc Natl Acad Sci USA. 1998;95:8791–8794. doi: 10.1073/pnas.95.15.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebecque S G, Gearhart P J. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogozin I B, Pavlov Y I, Bebenek K, Matsuda T, Kunkel T A. Nat Immun. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 16.Zeng X, Winter D B, Kasmer C, Kraemer K H, Lehmann A R, Gearhart P J. Nat Immun. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 17.Frey S, Bertocci B, Delbos F, Quint L, Weill J C, Reynaud C A. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- 18.Rada C, Ehrenstein M R, Neuberger M S, Milstein C. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 19.Berek C, Milstein C. Immunol Rev. 1987;96:23–41. doi: 10.1111/j.1600-065x.1987.tb00507.x. [DOI] [PubMed] [Google Scholar]
- 20.Bonfield J K, Smith K, Staden R. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachl J, Dessing M, Olsson C, von Borstel R C, Steinberg C. Proc Natl Acad Sci USA. 1999;96:6847–6849. doi: 10.1073/pnas.96.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs H, Rajewsky K, Fukita Y, Bross L. Philos Trans R Soc London B. 2001;356:119–125. doi: 10.1098/rstb.2000.0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goossens T, Klein U, Kuppers R. Proc Natl Acad Sci USA. 1998;95:2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson P C, de Bouteiller O, Liu Y J, Potter K, Banchereau J, Capra J D, Pascual V. J Exp Med. 1998;187:59–70. doi: 10.1084/jem.187.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H Y, Kaartinen M. Scand J Immunol. 1995;42:52–59. doi: 10.1111/j.1365-3083.1995.tb03625.x. [DOI] [PubMed] [Google Scholar]
- 26.Martin A, Bardwell P D, Woo C J, Fan M, Shulman M J, Scharff M D. Nature (London) 2002;415:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- 27.Nagaoka H, Muramatsu M, Yamamura N, Kinoshita K, Honjo T. J Exp Med. 2002;195:529–534. doi: 10.1084/jem.20012144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okazaki I-M, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. Nature (London) 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 29.Berek C, Milstein C. Immunol Rev. 1988;105:5–26. doi: 10.1111/j.1600-065x.1988.tb00763.x. [DOI] [PubMed] [Google Scholar]
- 30.Kinoshita K, Honjo T. Nat Rev Mol Cell Biol. 2001;2:493–503. doi: 10.1038/35080033. [DOI] [PubMed] [Google Scholar]
- 31.Muramatsu M, Sankaranand V S, Anant S, Sugai M, Kinoshita K, Davidson N O, Honjo T. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 32.Smith H C, Sowden M P. Trends Genet. 1996;12:418–424. doi: 10.1016/0168-9525(96)10042-1. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Sowden M P, Smith H C. J Biol Chem. 2000;275:22663–22669. doi: 10.1074/jbc.M910406199. [DOI] [PubMed] [Google Scholar]
- 34.Rosin-Arbesfeld R, Townsley F, Bienz M. Nature (London) 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- 35.Rada C, Milstein C. EMBO J. 2001;20:4570–4576. doi: 10.1093/emboj/20.16.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]