Abstract
We have identified a subset of CD56+CD3− human natural killer (NK) cells that express CD4 and the HIV coreceptors CCR5 and CXCR4. These cells can be productively infected in vitro by both CCR5- and CXCR4-using molecular clones of HIV-1 in a CD4-dependent manner. Analysis of HIV-infected persons showed that viral DNA is present in purified NK cells, and virus could be rescued from these cells after in vitro cultivation. Longitudinal analysis of the HIV-1 DNA levels in NK cells from patients after 1–2 years of highly active antiretroviral therapy indicated that NK cells remain persistently infected and account for a substantial amount of the viral DNA in peripheral blood mononuclear cells. These results demonstrate that a subset of non-T cells with NK markers are persistently infected and suggest that HIV infection of NK cells is important for virus persistence. The properties of the virus reservoir in these cells should be considered in attempts to further optimize antiretroviral therapies.
Highly active antiretroviral therapy (HAART) is very effective in controlling HIV-1 as reflected by the dramatic decrease (100- to 1,000-fold) in plasma viral load that follows the initiation of treatment in most HIV-1 patients (1–5). Despite this effectiveness, virus is never eradicated. The presence of a pool of latently infected cells (6, 7) that appears to be established early during primary HIV-1 infection (8, 9) provides a mechanism for HIV-1 persistence even in patients receiving HAART. It has been established that quiescent CD4+ T lymphocytes harbor replication-competent HIV-1 in a latent form (6–9). This pool of long-living, latently infected CD4+ T cells represents a major obstacle for the eradication of HIV-infected cells in patients receiving HAART. Some reports have indicated a lack of correlation between the estimated frequency of resting CD4+ T cells harboring infectious virus and the kinetics of viral rebound upon cessation of antiretroviral therapy (10). In addition, lack of genetic identity between the rapidly rebounding virus after therapy interruption and the virus present in the T cell latent reservoir was noticed in several patients (11, 12). This finding suggested that the origin of the rebounding virus may not be only the infected resting T lymphocytes, at least in some patients, and supported the existence of unidentified, long-term HIV-1 reservoir(s) in patients receiving HAART. Toward a better control of HIV infection, it is important to characterize all cell types that contribute to long-term HIV persistence.
Materials and Methods
Clinical Samples and Natural Killer (NK) Cell Purification.
Blood samples were collected in ACD tubes under approved protocols for human subjects research. Peripheral blood mononuclear cells (PBMC) were obtained by gradient centrifugation over Histopaque gradients. Monocytes were depleted by magnetic separation with anti-CD14-coated magnetic beads. T lymphocytes were positively selected by using anti-CD3-coated beads. NK cells were purified from the CD14- and CD3-depleted PBMC by a two-step magnetic separation. First, the cells were labeled with a mouse anti-human CD56 mAb for 30 min at 4°C, and, after washing out the excess of free antibody, the NK cells were purified with beads coated with an anti-mouse IgG antibody. The purity of the selected NK preparations was typically higher than 98%; the low frequency of contaminating cells were mostly CD56(−) and CD3(−) as judged by flow cytometric analysis of the samples. Contaminating T cells were typically less than 0.5% (Fig. 1b).
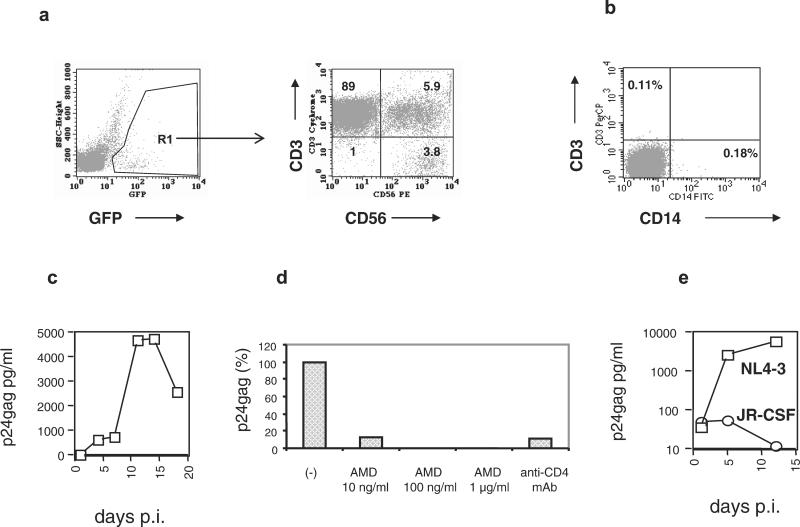
Figure 1.
In vitro infection of primary NK cells. (a) Primary PBMC were infected with GFP-tagged HIV-1 molecular clones and stained with anti-CD3 and anti-CD56 mAbs. Infected cells were gated according to the expression of GFP (Left). The phenotype of 10,000 green cells is shown (Right). Numbers within the quadrants represent the percentage of positive cells. The NK cells (CD3−CD56+) in the lower right quadrant represent 3.8% of the infected cells. The presented results were obtained at day 14 after infection. (b) Purified NK cells are free of T cells and monocyte/macrophages. After depleting monocytes and T lymphocytes from PBMC, CD56+ cells were positively selected by immunomagnetic beads and analyzed by FACS. (c) HIV-1 infection of purified human NK cells with the R5 HIV-1 strain JR-CSF at a multiplicity of infection of 0.1. Similar results were obtained in an additional experiment. (d) Anti-CD4 mAbs and the CXCR4-specific antiviral compound AMD3100 inhibit HIV-1 entry into NK cells. Purified primary NK cells were incubated with 5 μg of an anti-CD4 HIV-neutralizing antibody or different concentrations of AMD3100 for 30 min before exposure to the X4 HIV-1 strain NL4–3. After 1 h, the cells were washed and cultured in the absence of anti-CD4 antibodies. AMD3100 was present in the culture medium throughout the entire experiment. (e) NK cells from a blood donor homozygous for the Δ32 CCR5 mutation were purified and exposed to NL4–3 (X4) and JR-CSF (R5) HIV-1 strains. Viral replication was monitored by p24gag determinations in culture supernatants.
Flow Cytometric Analysis.
PBMC were washed twice with PBS containing 0.5% heat-inactivated human AB serum (Sigma). The cells were incubated for 20 min at 4°C in the dark with directly conjugated mAbs (PharMingen). Cells were washed twice and analyzed either in a FACScan or LSR. The data were analyzed with cellquest software. The following directly conjugated antibodies (PharMingen) were used in these studies: CD4, CD3, CD56, CCR5, CXCR4, α/β TCR (T cell antigen receptor), and γ/δ TCR.
Viral Infection.
The following HIV-1 molecular clones were used for in vitro infection experiments: pNL4–3, pJR-CSF, and pNL4–3GFP (13). Infectious stocks were generated by transfection in 293 cells as described (13). AMD 3100 (a gift from Julie Strizki, Schering–Plough Research Institute, Kenilworth, NJ) was used in inhibition experiments. Viral replication was monitored by measuring HIV-1 p24gag accumulation in culture supernatant with a commercial ELISA kit (Zeptometrix, Buffalo, NY).
Real-Time Quantitative PCR.
A taqman probe and gag primers specific for a wide range of clade B HIV-1 clinical isolates were used. The forward and reverse primer sequences were 5′-AGCCCAGAAGTAATACCCATGTTT3-′ and 5′-CCCCCCACTGTGTTTAGC-3′, respectively, and the fluorogenic probe was 5′-FAM-CAGCATTATTCAGAAGGACCACCCCA-TAMRA-3′. The reaction mixture (50 μl) contained 25 μl DNA, 5 μl 10× TaqMan buffer, 4 μl deoxynucleotide triphosphates (10 mM each of dATP, dCTP, and dGTP and 20 mM dUTP), 7 μl MgCl2 (25 mM), 1 μl forward primer (45 mM), 1 μl reverse primer (2.5 mM), 1 μl fluorogenic probe (12.5 μM), 0.5 μl AmplErase (1 unit/μl), and 0.5 μl AmpliTaq Gold DNA polymerase (5 units/μl; Perkin–Elmer). Real-time PCR was performed in an ABI PRISM 7700 Sequence Detector (Applied Biosystems). Activation of UNG (2 min at 52°C) and AmpliTaq (10 min at 95°C) was followed by 45 cycles (15 sec at 95°C and 1 min at 60°C). Each DNA sample was analyzed in duplicate. As control for cross-contamination, a sample not containing DNA also was amplified in duplicate. To quantify the cell equivalents in the input genomic DNA, we used real-time PCR assays measuring either porphobilinogen deaminase (PBGD) or CCR5 DNA, as described (14, 15). To measure HIV mRNA levels by reverse transcription–PCR, the forward primer was 5′-GCACGGCAAGAGGCGA-3′, located in exon 1; the reverse primer was 5′-GAGGTGGGTTGCTTTGATAGAG-3′, located in the boundary between exons 5 and 7 (second and third exons of nef). The TaqMan probe sequence was 5′-FAM-CGGCGACTGGAAGAAGCGGAGA-TAMRA-3′, located in the boundary of exons 1 and 5. This primer–probe combination is specific for the detection of the HIV-1 nef mRNA 1.5.7. For a subset of samples, real-time PCR measurements of cell-associated HIV-1 viral load were performed independently in three different laboratories by using different HIV-1 gag primers and specific probes.
Results
NK Cells Are Targets for HIV-1 Infection in Vitro.
To identify other targets for HIV-1 infection, we used PBMC cultures to characterize the cell types infected by HIV-1 clones tagged with green fluorescent protein (GFP) (13, 16). This allowed the detection and quantification of the infected cells by monitoring intracellular GFP produced as a Nef-GFP fusion by the integrated provirus. Two molecular clones with different coreceptor specificities were used: a CCR5-tropic clone (R5) derived from JR-CSF and a CXCR4-tropic (X4) clone derived from pNL4–3 (13). This technique combined with the use of different mAbs permits the immunophenotyping of HIV-1-infected cells. Flow cytometric analysis of human PBMC infected with GFP-tagged HIV-1 in vitro (Fig. 1a) demonstrated that a fraction (3.8%) of the infected cells displayed the CD3−CD56+ phenotype characteristic of human NK cells, whereas 5.9% of infected cells had both T and NK cell markers (CD3+CD56+), characteristic of NK/T cells. HIV-1 infection of NK cells in vitro has been observed previously, but the subject remained somewhat controversial (17–19) and no evidence of in vivo infection has been reported. The reported in vitro infection was found to be CD4-independent (18) or nonproductive (17). We studied in vitro infection by using isolated NK cells purified by a combination of negative and positive selection steps (see Materials and Methods). The purity of the isolated NK cells was assessed by FACS analysis. It was found that the preparations were highly pure (>99%) and contained less than 0.4% and 0.2% of T cells and monocytes, respectively (Fig. 1b). Exposure to HIV-1 (JR-CSF) at a multiplicity of infection of 0.1 showed productive viral infection, as detected by the presence of increasing amounts of HIV-1 gag in culture supernatants (Fig. 1c). These results demonstrated that NK cells were infected productively by HIV-1 in the absence of any other cell types. Additional infection experiments were performed to test whether CD4 and HIV coreceptors were required for NK infection. Incubation of purified NK cells with an anti-CD4 mAb before exposure to HIV resulted in strong inhibition of viral production, indicating that HIV-1 infection of NK cells is CD4-dependent (Fig. 1d). To demonstrate involvement of CXCR4 in HIV-1 entry, we used the specific entry inhibitor AMD3100 (20). We showed that this compound was able to inhibit production of the X4-tropic HIV-1 molecular clone NL4–3 (Fig. 1d). To analyze whether CCR5 is required for HIV-1 entry, we infected purified NK cells from a blood donor homozygous for the Δ32 CCR5 mutation with two molecular clones having defined coreceptor specificity. The R5-tropic HIV-1JR-CSF failed to infect these cells, as judged by the absence of gag production (Fig. 1e). In contrast, infection of NK cells from the same individual with the X4-tropic NL4–3 resulted in productive infection with accumulation of increasing amounts of extracellular gag protein. The same viral stocks were able to infect NK cells from non-Δ32 CCR5 individuals.
CD4, CCR5, and CXCR4 Are Expressed on a Subset of Primary NK Cells.
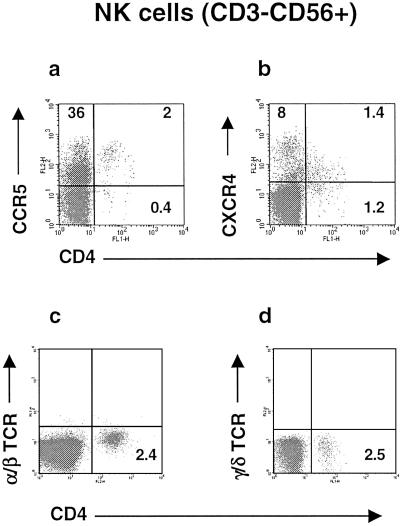
Taken together, the results from the previous experiments indicated the existence of NK cells expressing both CD4 and HIV coreceptors. Indeed, analysis of NK cells from many healthy donors by four-color flow cytometry demonstrated the presence of a NK subset characterized by low expression of CD4 and expression of CCR5 or CXCR4 (Fig. 2 a and b) and thus are potential targets for HIV-1 infection. These cells are present in all healthy donors and range between 0.3% and 6.5% of the total NK (CD3−CD56+) cells. These cells also exist in HIV patients; in fact, many HIV patients have a high proportion of CD4+ cells within the NK population (data not shown). Based on the absence of either α/β or γ/δ TCR on their surface (Fig. 2 c and d), as well as the lack of CD3, the identity of these cells is clearly different from the CD56+ subset of T lymphocytes (NK/T lymphocytes). Additional immunophenotyping showed that these cells are CD16+, CD2+, CD14−, CD19−, CD80−, and CD86− and express killer cell Ig-like receptors upon in vitro culture. Therefore, these cells cannot be classified as T cells or monocytes, because they do not have any of the characteristic lineage markers for these cells, whereas they have all of the characteristic markers of NKs.
Figure 2.
Identification of a subset of CD4+ cells with NK markers. Four-color flow cytometry was performed with PBMC obtained from healthy blood donors. NK cells were gated according to the pattern of staining with CD3 and CD56 mAbs. Dot plots show expression of CD4 and CCR5 (a) or CD4 and CXCR4 (b) by the CD3−CD56+ NK population. Numbers in the quadrants express the percentage of positive cells within the gate. Similar results have been obtained consistently with PBMC from more than 10 healthy blood donors. (c and d) Four-color flow cytometric analysis demonstrated the lack of α/β TCR and γ/δ TCR expression in both CD4− and CD4+ primary NK cells.
It has been shown previously that HHV-6 infection of NK clones induces CD4 expression and renders the NK clones susceptible to HIV-1 infection in vitro (19). We examined the hypothesis that in vivo infection by HHV-6, which is highly prevalent, may be the reason for the expression of CD4 in NK cells. Using PCR primers specific for HHV-6 (21), we detected the presence of infected T lymphocytes in both HIV-1-infected individuals and healthy controls. In contrast, HHV-6 infection of purified NK cells from the same donors was detected only in two of 15 individuals (one of eight HIV-1 patients and one of seven uninfected controls; data not shown). Therefore, CD4 expression on NK cells in vivo does not correlate with HHV-6 infection of these cells.
NK Cells Harbor Infectious HIV-1 in Vivo.
To analyze whether NK cells are infected by HIV-1 in vivo, T lymphocytes and NK cells were purified from HIV-1-infected individuals and the genomic DNA was analyzed by real-time PCR for the presence of HIV-1 DNA. The samples included in this study were obtained from HAART-treated patients with a wide range of CD4 counts (25–1,096, mean = 372 CD4 cells per μl). Cross-sectional analysis of samples from 19 patients was performed; 11 patients had undetectable viral load (<50 copies of viral RNA per ml of plasma), whereas the remaining eight patients had detectable plasma virus (range, 92–106,326 RNA copies/ml of plasma). Real-time PCR analysis demonstrated the presence of HIV-1 DNA in NK cells from all of the patients. The range for NK cells was 180–160,000 HIV-1 DNA copies per 106 cells. The range for T cells was 320–270,000 HIV-1 DNA copies per 106 cells, which is in agreement with previous estimations (22). Assuming that T cells and NK cells represent 65% and 7.5% of PBMC, respectively, we can estimate that the ranges of DNA copies in T and NK compartments were 208 to 175,500 and 13.5 to 12,000, respectively. The high levels of HIV-1 DNA in the purified NK cell samples rule out any possibility of contamination by other HIV-infected cell types.
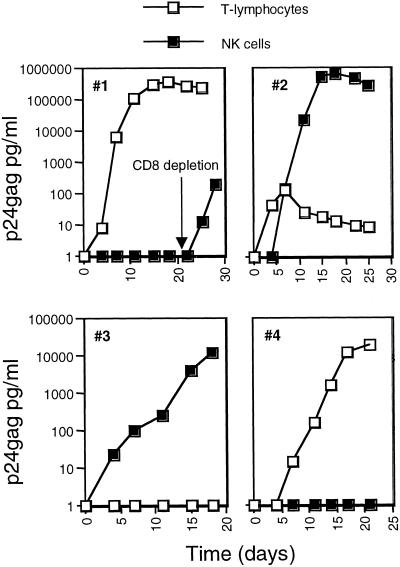
Most of the cells harboring HIV-1 DNA in peripheral blood are unable to produce infectious virus (6, 22–24). To examine whether HIV could be rescued from the NK cells, we performed viral isolation experiments from CD3−CD56+ cells purified as described above from patients under HAART and having low or undetectable virus loads (range = <50–9,300 copies per ml plasma). CD8-depleted PBMC pooled from two healthy donors were used as targets for viral recovery. Infectious HIV-1 was isolated from three of four NK samples (Fig. 3). In one case, HIV-1 could be recovered from the NK cells but not from the T lymphocytes. These viral isolation experiments were limited by the size of the available sample, typically 5 × 106 PBMC. It is expected that the efficiency of virus isolation will increase by increasing the number of cells included in the assay. These results indicate the importance of the NK cells as a source of infectious virus in HAART-treated patients with low or undetectable plasma viremia.
Figure 3.
HIV-1 isolation from highly purified (higher than 95%) NK cells. CD3+ and CD3−CD56+ cells were purified from HIV-1-infected individuals. For HIV-1 isolation, cocultures of the purified cells with CD8-depleted PBMC from healthy blood donors were established and culture supernatants were harvested at the indicated time points for the detection of p24gag production. Each of the four panels represents the viral isolation results from the T lymphocytes (□) and NK cells (■) of a single patient. For patient 1, CD8 depletion was performed after initiation of the culture as indicated.
To examine whether HIV is expressed in circulating NK cells, we measured the levels of spliced viral mRNA in NK and T cells from the same samples ex vivo. RNA samples from eight patients were analyzed by PCR by using a set of oligonucleotides specific for multiply spliced HIV-1 mRNA (15) and a TaqMan probe specific for the splice junction sequence. These conditions allow the amplification of only spliced HIV-1 mRNA and exclude the detection of full-length genomic viral RNA. No spliced viral mRNA was found in any of the eight NK samples analyzed. Similarly, no viral RNA could be detected in seven of eight T cell mRNA preparations from the same blood samples. These results show that patients receiving HAART may harbor a significant number of latently infected NK cells in the periphery, which also is the case with T cells.
HIV-1-Infected NK Cells Constitute a Long-Living Viral Reservoir in Patients Receiving HAART.
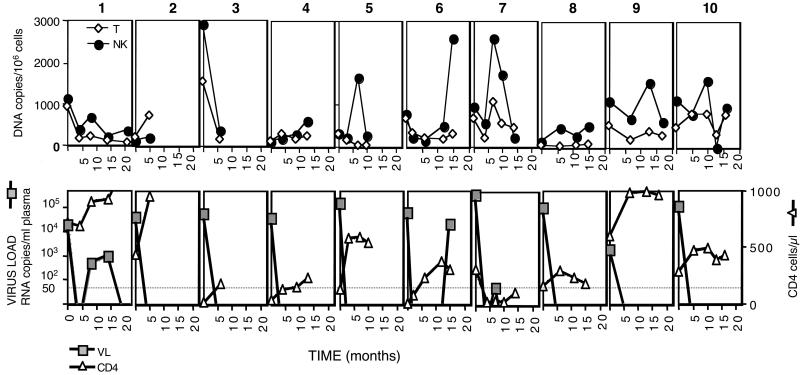
The presence of HIV-1 DNA in NK cells obtained from patients who had undetectable plasma viremia while receiving HAART showed that long-term treatment did not eliminate the virus from the NK cells. To study the impact of HAART on HIV-1-infected NK cells, we performed a longitudinal study (up to 2 years for some patients) of sequential PBMC samples from 10 HIV-1-infected individuals with no previous history of antiretroviral treatment. All of the patients started a similar therapeutic regimen, including two nucleoside analogue reverse transcriptase inhibitors (zidovudine and lamivudine for nine patients, or stavudine and didanosine for patient 3), plus one protease inhibitor (eight patients) or nevirapine (patients 2 and 10). The first sample from each individual was obtained before initiation of HAART during a phase of active viral replication as demonstrated by the presence of high plasma viral load. For patients 2 and 3, only two samples, collected 5–6 months apart, were available (before and after HAART). Purified genomic DNA was used in real-time PCR experiments to measure the frequency of HIV-1 DNA in the T and NK compartments. Results from these experiments demonstrated similar levels of viral DNA in both cell types (Fig. 4). Initiation of HAART resulted in efficient control of viral replication as evidenced by the rapid decline in HIV-1 plasma RNA (Fig. 4 Lower). In general, the drop in plasma RNA was associated with a modest decrease of HIV-1 DNA copies in both cell compartments. Patients 1, 6, and 7 had rebounds of viral expression during the time frame of the study. In these three patients, the rebound in plasma viral RNA was paralleled by an increase in the number of HIV-1 DNA copies in T lymphocytes and NK cells. Analysis of longitudinal samples from the 10 patients demonstrated that HAART failed to eliminate HIV-1 from either T lymphocytes or NK cells. The DNA copies per 106 cells found in the NK and T cell compartments were compared for the 10 patients. The HIV DNA decay slopes in T and NK cells were estimated as −14.68 HIV DNA copies per 106 cells per month and +4.82 HIV DNA copies per 106 cells per month, respectively, based on a linear random-effects model. The DNA decay slope in T cells differed significantly from zero (P = 0.047), whereas the decay slope in NK did not (P = 0.82). There was a significant difference between the two slopes (P = 0.0041), indicating that NK cells contain an increasing proportion of HIV DNA measured in PBMC during HAART. This difference may reflect a more stable reservoir in NK cells compared with total T cells. Alternatively, it may reflect changes in the kinetics of the different cell populations during treatment, because T cells increase upon HAART, whereas our measurements indicate that the NK numbers are similar in infected and uninfected individuals. Further experiments are required to characterize the mechanisms explaining our observations. Nevertheless, the presence of substantial amounts of proviral DNA shows that HAART treatment for up to 2 years does not eliminate the pool of HIV-1-infected NK cells and emphasizes the importance of NK cells as a long-term reservoir in HIV-1-infected individuals with undetectable plasma viremia.
Figure 4.
HIV-1 DNA load in the T and NK cells during HAART. T lymphocytes (CD3+) and NK cells (CD3−CD56+) were purified from sequential PBMC samples from 10 HIV-1 patients before (time 0) and at the indicated times after initiation of HAART. DNA was isolated from the two cell populations and used in real-time PCR experiments. (Upper) The kinetics of cell-associated HIV-1 DNA copies per 106 T (◊) or NK cells (●) from individual patients, numbered 1–10. (Lower) HIV-1 RNA plasma viral load (■) and CD4 counts (▵) for the individual patients. The limit of detection for the virus load determinations (50 copies), performed by the b-DNA 3.0 assay, is indicated by a dotted line.
Discussion
In this work, we have identified a subset of CD56+CD3−CD16+CD4+ cells not characterized previously. Based on immunophenotyping, these cells were classified as NK cells, because they do not have any of the characteristic markers of T cells or monocytes. These non-T cells express HIV coreceptors and are infected by HIV-1 in vitro. Infection also was detected in NK cells in vivo, based on the levels of HIV DNA found in purified NK cells. Because infection of NK cells was prevented after blocking CD4, it is reasonable to conclude that the CD4+ subset of NK cells also was infected in vivo. Because purified NK cells from HIV-positive individuals can transmit virus (Fig. 3), we concluded that infectious virus can persist in these cells even during HAART. Indeed, follow-up of 10 patients under HAART for a period of up to 2 years revealed that they continue to carry substantial levels of proviral DNA in the NK cells. Although the decay slope of proviral DNA in NK cells is less than the T cells, this may not reflect a more stable reservoir in NK cells but may be the result of differences in population kinetics, compartmentalization, or other reasons. The continuous presence of substantial levels of proviral DNA for long periods of time (Fig. 4), together with virus rescue from the NK compartment (Fig. 3), demonstrates the permanent nature of the reservoir within the NK compartment. Although the overall size of the reservoir in NK cells is smaller than in T cells, the continuous presence of virus within the NK compartment in all patients under HAART argues for the importance and relevance of the NK reservoir.
The continuous presence of HIV-1-infected NK cells in treated patients may be the result of the long half-life of these cells. At present, there is no definitive information about the half-life of NK cells, and, in any case, the life span of different subsets may be different, as is the case for the T cells. Alternatively, NK cells may have a short life span and infected NK cells may be replenished continuously by ongoing low-level replication. This second scenario implies suboptimal pharmacological effects of the anti-HIV drugs in the NK compartment. In fact, low levels of HIV-1 replication in patients receiving HAART with undetectable plasma viremia has been reported by several groups (24–29). One interesting possibility is that protease inhibitors may be less effective in NK cells, because they have high levels of p-glycoprotein activity (30). It has been shown that PIs are substrates for the p-glycoprotein pump (31, 32). We now have found that PIs block p-glycoprotein-directed efflux in NK cells less efficiently compared with T cells (unpublished data). This is similar in both normal volunteers and HIV-infected subjects. These results suggest that virus suppression could be less efficient in cells expressing high p-glycoprotein levels, such as NK cells.
NK cells are pivotal in the innate immunity against viruses and tumors. Our data demonstrate that NK cells are infected in HIV-1 patients and that currently available antiretroviral therapies fail to eliminate the virus from NK cells. Early defects in innate immunity present in HIV-1-infected individuals (33–36) thus might be a direct consequence of HIV-1 infection. Future attempts to eradicate HIV-1 from the body must take into consideration the presence of this viral reservoir. Protocols aiming to activate viral expression from latently infected T cells may fail in NK cells. To eradicate HIV-1, new interventions addressing infection of the NK pool may need to be established in the context of improved antiretroviral therapies.
Acknowledgments
We thank Vana Sypsa, School of Medicine, University of Athens, for expert assistance with data analysis; F. Merced-Galindez and the other nurses and physicians of the HIV and AIDS Malignancy Branch (HAMB), National Cancer Institute; K. Noer, R. Matthai, M. Baseler, and R. Stevens of Science Applications International Corporation, Frederick, MD; G. Alvord and D. Powell of Data Management Services, Frederick, MD; and Jeff Lifson and Mike Piatak of the AIDS Vaccine Program for their valuable assistance. L.G.K. was supported by grants from the National Institutes of Health and the Elizabeth Glazer Pediatric AIDS Foundation. A.H. and M.L. are supported by a grant from the Hellenic Centre for Infectious Diseases Control.
Abbreviations
- HAART
highly active antiretroviral therapy
- NK
natural killer
- PBMC
peripheral blood mononuclear cells
- GFP
green fluorescent protein
- TCR
T cell antigen receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Nature (London) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 2.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Nature (London) 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 3.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 4.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Nature (London) 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 5.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, et al. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 6.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finzi D, Blankson J, Siliciano J D, Margolick J B, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 8.Chun T W, Engel D, Berrey M M, Shea T, Corey L, Fauci A S. Proc Natl Acad Sci USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 10.Davey R T, Jr, Bhat N, Yoder C, Chun T W, Metcalf J A, Dewar R, Natarajan V, Lempicki R A, Adelsberger J W, Miller K D, et al. Proc Natl Acad Sci USA. 1999;96:15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun T W, Davey R T, Jr, Ostrowski M, Shawn Justement J, Engel D, Mullins J I, Fauci A S. Nat Med. 2000;6:757–761. doi: 10.1038/77481. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Chung C, Hu B S, He T, Guo Y, Kim A J, Skulsky E, Jin X, Hurley A, Ramratnam B, et al. J Clin Invest. 2000;106:839–845. doi: 10.1172/JCI10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valentin A, Lu W, Rosati M, Schneider R, Albert J, Karlsson A, Pavlakis G N. Proc Natl Acad Sci USA. 1998;95:8886–8891. doi: 10.1073/pnas.95.15.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valentin A, Trivedi H, Lu W, Kostrikis L G, Pavlakis G N. Virology. 2000;269:294–304. doi: 10.1006/viro.1999.0136. [DOI] [PubMed] [Google Scholar]
- 15.Saltarelli M J, Hadziyannis E, Hart C E, Harrison J V, Felber B K, Spira T J, Pavlakis G N. AIDS Res Hum Retroviruses. 1996;12:1443–1456. doi: 10.1089/aid.1996.12.1443. [DOI] [PubMed] [Google Scholar]
- 16.Stauber R H, Horie K, Carney P, Hudson E A, Tarasova N I, Gaitanaris G A, Pavlakis G N. BioTechniques. 1998;24:462–471. doi: 10.2144/98243rr01. [DOI] [PubMed] [Google Scholar]
- 17.Scott-Algara D, Vuillier F, Cayota A, Rame V, Guetard D, Moncany M L, Marasescu M, Dauguet C, Dighiero G. J Gen Virol. 1993;74:725–731. doi: 10.1099/0022-1317-74-4-725. [DOI] [PubMed] [Google Scholar]
- 18.Chehimi J, Bandyopadhyay S, Prakash K, Perussia B, Hassan N F, Kawashima H, Campbell D, Kornbluth J, Starr S E. J Virol. 1991;65:1812–1822. doi: 10.1128/jvi.65.4.1812-1822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lusso P, Malnati M S, Garzino-Demo A, Crowley R W, Long E O, Gallo R C. Nature (London) 1993;362:458–462. doi: 10.1038/362458a0. [DOI] [PubMed] [Google Scholar]
- 20.Schols D, Struyf S, Van Damme J, Este J A, Henson G, De Clercq E. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cone R W, Huang M L, Ashley R, Corey L. J Clin Microbiol. 1993;31:1262–1267. doi: 10.1128/jcm.31.5.1262-1267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chun T W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, et al. Nature (London) 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 23.Bukrinsky M I, Stanwick T L, Dempsey M P, Stevenson M. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharkey M E, Teo I, Greenough T, Sharova N, Luzuriaga K, Sullivan J L, Bucy R P, Kostrikis L G, Haase A, Veryard C, et al. Nat Med. 2000;6:76–81. doi: 10.1038/71569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furtado M R, Callaway D S, Phair J P, Kunstman K J, Stanton J L, Macken C A, Perelson A S, Wolinsky S M. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 26.Natarajan V, Bosche M, Metcalf J A, Ward D J, Lane H C, Kovacs J A. Lancet. 1999;353:119–120. doi: 10.1016/s0140-6736(05)76156-0. [DOI] [PubMed] [Google Scholar]
- 27.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L, Ingerman M J, Witek J, Kedanis R J, Natkin J, DeSimone J, et al. J Am Med Assoc. 1999;282:1627–1632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 28.Grossman Z, Polis M, Feinberg M B, Levi I, Jankelevich S, Yarchoan R, Boon J, de Wolf F, Lange J M, Goudsmit J, et al. Nat Med. 1999;5:1099–1104. doi: 10.1038/13410. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson A S, Korber B T, et al. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- 30.Chaudhary P M, Mechetner E B, Roninson I B. Blood. 1992;80:2735–2739. [PubMed] [Google Scholar]
- 31.Srinivas R V, Middlemas D, Flynn P, Fridland A. Antimicrob Agents Chemother. 1998;42:3157–3162. doi: 10.1128/aac.42.12.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee C G, Gottesman M M, Cardarelli C O, Ramachandra M, Jeang K T, Ambudkar S V, Pastan I, Dey S. Biochemistry. 1998;37:3594–3601. doi: 10.1021/bi972709x. [DOI] [PubMed] [Google Scholar]
- 33.Brenner B G, Gryllis C, Gornitsky M, Wainberg M A. Clin Exp Immunol. 1993;93:142–148. doi: 10.1111/j.1365-2249.1993.tb07956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ullum H, Gotzsche P C, Victor J, Dickmeiss E, Skinhoj P, Pedersen B K. J Exp Med. 1995;182:789–799. doi: 10.1084/jem.182.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirianni M C, Tagliaferri F, Aiuti F. Immunol Today. 1990;11:81–82. doi: 10.1016/0167-5699(90)90032-5. [DOI] [PubMed] [Google Scholar]
- 36.Lucia M B, Froio N, Tacconelli E, Tumbarello M, Rutella S, Rumi C, Cauda R. Eur J Histochem. 1997;41:197–198. [PubMed] [Google Scholar]