Abstract
We report a DNA microarray-based method for genome-wide monitoring of competitively grown transformants to identify genes whose overexpression confers a specific cellular phenotype. Whereas transcriptional profiling identifies differentially expressed genes that are correlated with particular aspects of the cellular phenotype, this functional genomics approach determines genes that result in a specific physiology. This parallel gene-trait mapping method consists of transforming a strain with a genomic library, enriching the cell population in transformants containing the trait conferring gene(s), and finally using DNA microarrays to simultaneously isolate and identify the enriched gene inserts. Various methods of enrichment can be used; here, genes conferring low-level antibiotic resistance were identified by growth in selective media. We demonstrated the method by transforming Escherichia coli cells with a genomic E. coli library and selecting for transformants exhibiting a growth advantage in the presence of the anti-microbial agent Pine-Sol. Genes conferring Pine-Sol tolerance (19 genes) or sensitivity (27 genes) were identified by hybridizing, on DNA microarrays containing 1,160 E. coli gene probes, extra-chromosomal DNA isolated from transformed cells grown in the presence of various levels of Pine-Sol. Results were further validated by plating and sequencing of individual colonies, and also by assessing the Pine-Sol resistance of cells transformed with enriched plasmid library or individual resistance genes identified by the microarrays. Applications of this method beyond antibiotic resistance include identification of genes resulting in resistance to chemotherapeutic agents, genes yielding resistance to toxic products (recombinant proteins, chemical feedstocks) in industrial fermentations, genes providing enhanced growth in cell culture or high cell density fermentations, genes facilitating growth on unconventional substrates, and others.
A central goal of functional genomics is to identify genes whose expression results in a specific cellular phenotype. Common strategies employ transformation, gene deletion and complementation, and identify genes by plating to enrich and separate individual transformants or deletion mutants, followed by colony amplification and sequencing of extra-chromosomal DNA (1). More recently, gene functions have been inferred by measuring changes in gene expression profiles across different physiological classes (2–5). Several techniques have been used to create deletion libraries marked with specific identifiable sequences that replace individual genes, and therefore can be monitored on specially designed oligonucleotide microarrays or standard PCR-based spotted microarrays (6–8). Alternatively, overexpression libraries have been studied by using standard plating methodologies where DNA from positive selectants were amplified by PCR and identified by hybridization to oligonucleotide arrays (9). In most of these methods, linking a set of genes with a particular cellular phenotype requires lengthy experimental plating and sequencing, does not easily lend itself to parallel scale up for high throughput applications, or, because of incorporation of PCR amplification, steps are not quantitative. Additionally, many of the previous methods rely on gene deletions, and are thus incapable of identifying physiological effects resulting from the introduction of new genes or increased gene copy numbers. In this paper we propose a parallel gene-trait mapping (PGTM) method for the direct identification of genes responsible for a particular phenotype that is amenable to parallel scale up for thousands of genes through the use of microarrays. Our motivation was to identify the effects of positive genetic modifications including not only enhanced transformant growth but also impaired survival under selective conditions.
To demonstrate the PGTM approach we sought to address two key questions in this study: (i) can we identify the accumulation of extra-chromosomal DNA fragments relative to a control by using a DNA microarray? (ii) Are the microarray results reflective of the relative levels of extra-chromosomal DNA in a heterogeneous culture of transformed host cells? To do so, we transformed Escherichia coli with an E. coli plasmid-based genomic library and selected for cells through growth in normal and anti-microbial culture conditions. We then monitored the population dynamics of these cultures by using an E. coli DNA microarray containing approximately 25% of the E. coli genome. Although we demonstrate this approach in the context of antibiotic tolerance and susceptibility, we envision broad-ranging future applications in metabolic engineering and drug discovery, among others. For example, in metabolic engineering approaches, it is of interest to determine genes that provide higher productivity because of better product formation, stress-resistance, increased or altered substrate utilization, or decreased product-associated toxicity. The approach described herein is applicable in all of these cases, and through combination with other functional genomic approaches makes for a powerful class of methods for accurately linking phenotype to gene expression.
Materials and Methods
Preparation of E. coli Genomic Library.
E. coli W3110 genomic DNA was prepared by using Qiagen Genomic Tips. Purified DNA was fragmented by using sonication for 30 s. Fragmented DNA was size separated through a 1% agarose gel and fragments between 0.5 and 3.5 kbp were extracted (Qiagen). Extracted DNA was repaired by using T4 DNA polymerase and Klenow fragment, then it was dephosphorlyated by using Calf-Intestinal Phosphatase. dATP was added to the 3′ ends of repaired DNA by using TaqDNA polymerase, and ligation was performed by using the TOPO-TA pBAD cloning vector from Invitrogen. Ligation products were electroporated into electroporation competent DH5α cells (Invitrogen) and plated on LB + Ampicillin (LBA). Transformants were grown overnight at 37°C and harvested (≈12,000 colonies) by adding LBA directly to the plates and by amplification for 3.5 h at 37°C in 300 ml of LBA at 250 rpm in a New Brunswick Scientific shaker. Plasmids were harvested by using a Qiagen Maxi kit. At an average insert size of 1.6 kbp, 12,000 colonies corresponds to a 99% probability that all sequences were included in the original library.
Growth Conditions.
Electrocompetent E. coli DH5α (Invitrogen) was transformed with the genomic library, incubated in SOC for 1 h at 37°C, enriched in LBA to an OD600 of 0.1–0.2 (≈5 h), innoculated (1% vol/vol) into LBA containing 0–2.5% Pine-Sol, and grown at 37°C. OD600 was used to monitor growth.
Plasmid Purification, Labeling, and Hybridization.
Plasmids were purified by Qiagen Mini Prep kits. Purified plasmids were sonicated for 35 s by using a 1-s pulse cycle. Fragmented plasmids were checked by 1% agarose gel to confirm a size distribution of 0.5–1.5 kbp relative to an unfragmented plasmid size distribution of 4.5–7 kbp. Fragmented plasmids were labeled by using the general random primed labeling methodology. Two micrograms of plasmid DNA was mixed with 25 nM dATP/25 nM dCTP/25 nM dGTP/10 nM dTTP/40 nM Cy3-dUTP/50 units of Klenow Enzyme (3′-5′−)/1× Klenow Buffer, and incubated for 1 h at 37°C. Labeled plasmids were mixed with an equal amount of identically and concurrently with Cy5-labeled genomic DNA (2 μg). The labeled plasmid and genomic DNA mixture was purified from unincorporated nucleotides through a DyeEx spin column (Qiagen), and was ethanol precipitated at −20°C for 1 h. Labeled pellets were resuspended in 18 μl of hybridization solution (CLONTECH), denatured for 10 min at 95°C, and applied to the arrays. Hybridization was performed overnight at 50°C (in accordance with hybridization solution) in Corning hybridization chambers in a water bath. Microarrays were washed in 100 ml of 1× CLONTECH wash buffer for 5 min, 100 ml of 0.2× SSC for 5 min, 100 ml of 0.1× SSC for 5 min, and 600 ml of 0.1× SSC for 10 min before scanning (Arrayworx charge-coupled device scanner, Issaquah, WA).
Microarray Data Quantification and Analysis.
Microarrays were analyzed by using the GenePix software program (Axon Instruments, Foster City, CA). Quantified images were exported in text file format for further analysis. Signal-to-noise ratios were routinely greater than 10 from both channels. To evaluate the similarity and reproducibility among the quantified microarray images, clustering and principal component analysis (PCA) were performed by using matlab. In the clustering analysis, similarity was based on the magnitude of the correlation coefficient, and the average linkage technique was used. Before analysis, one sample from 0.0%, one sample from 0.1%, two samples from 0.25%, and one sample from 0.4% Pine-Sol cultures were eliminated as outliers because of experimental concerns, a lack of correlation with any of the other samples, and low loadings in the PCA projections. The remaining samples were run through the clustering and PCA algorithms to reveal two general classes; the first class (class 1) contained only the sample treated with 0.4% Pine-Sol, whereas the second class (class 2) comprised samples treated with 0.25% (1 sample), 0.1% (1 sample), or 0.0% Pine-Sol (3 samples). Furthermore, an additional subdivision was observed within class 2 with the samples treated with Pine-Sol differentiated from those exposed to Pine-Sol-free media. A similar result was obtained in the PCA projection, where the samples from 0.4% Pine-Sol were separated from the others by a positive value of the second principal component (PC2).
Resistance or tolerance genes were identified on the basis of the magnitude of the Cy3 intensity values for each microarray. The resistance rank of each gene was determined for each of the three samples obtained for growth in 0.4% Pine-Sol media. These three ranks were averaged, and the mean was used to rank the gene inserts. This final rank value provides a measure of the degree of enrichment of a particular gene insert during growth in 0.4% Pine-Sol. The terms resistance and tolerance are used interchangeably and refer to enriched genes rather than clinical resistance.
Construction of Overexpression Strains.
PCR was used to amplify individual DNA constructs containing the putative tolerance genes ygcA, lpxD, kdsB, trpE, livJ, pheP, and livG, and the null genes leuC and hybC. These two null genes did not show any significant enrichment in the Pine-Sol selective conditions when compared with Pine-Sol-free conditions. Primers were designed so that the final construct contained the structural gene as well as the regions 500 bp upstream and 500 bp downstream of the gene; this was done to simulate the construct contained within the randomly fragmented genomic library. Fragments were purified and ligated into the identical pBAD-TOPOTA expression vector used in the creation of the genomic library. Insert size was confirmed by restriction analysis.
Determination of Minimum Inhibitory Concentration (MIC).
MIC was determined by applying standard 96-well dilution methodology (10). 96-well plates were set up with LBA containing a dilution series from 0–1% Pine-Sol at intervals of 0.1%. Individual wells were inoculated with 10,000 cells taken from mid-exponential phase shake flask cultures, and inoculated plates were incubated without shaking at 37°C for 18 h. MIC's are reported as the minimum concentration at which no cell growth was observed and are the result of 4–37 replicates. A pUC plasmid control was included in every 96-well plate to enable comparisons between plates. All data, therefore, is reported as the ratio of the MIC from the transformed strains over the MIC for the pUC control plasmid strains from all plates run on the same day.
Sequencing of Enriched Library Inserts.
E. coli cells harboring the genomic library were identically enriched in 0.4% Pine-Sol as described under growth conditions. Plasmid DNA was purified from the enriched culture (labeled the enriched genomic library), and this library was used to transform E. coli, which was followed by plating onto LBA + agar and overnight incubation at 37°C. A total of 192 individual colonies were picked, plasmids were purified, and restriction digestion was performed. Restriction digestion products were analyzed on a 1% agarose gel. Forty plasmids containing inserts of unique size were selected for sequencing analysis. Sequenced inserts were compared by blast to the E. coli K12 genome to identify insert genes. Identified genes were compared with PGTM results for validation of PGTM.
Results and Discussion
Parallel Gene-Trait Mapping.
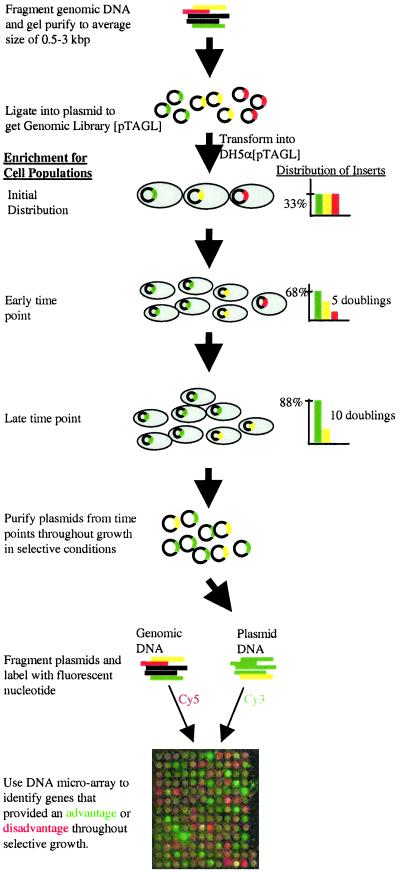
In this approach a host strain is transformed with a genomic library from the organism of interest, the transformed host population is enriched in cell subpopulation(s) exhibiting enhanced growth in selective conditions, and DNA microarrays are used to characterize the genotype of the population(s) that survived the adverse growth conditions (Fig. 1). Plasmid-based genomic libraries are commonly used to screen for genes conferring specific traits to host cells (1). Hence, in this study we created a plasmid-based genomic library by transforming E. coli DH5α with a genomic DNA E. coli library to yield E. coli strain DH5α[pTAGL]. The anti-microbial compound Pine-Sol was used as selective agent because it is known to select for mutants resistant to more traditional antibiotics such as tetracycline, chloramphenicol, and nalidixic acid. Also, the indiscriminate use of household anti-microbial agents is suggested to contribute to general expansion of antibiotic resistance (11, 12). To determine an appropriately selective Pine-Sol concentration, E. coli DH5α and DH5α[pTAGL] were grown in LB media containing various Pine-Sol levels (0–2.5% vol/vol) and the lag time, growth rate, and final cell density were monitored.
Figure 1.
PGTM. E. coli MG1655 genomic DNA was fragmented, size selected, repaired, and ligated into TOPO-TA cloning vector. E. coli DH5α were transformed with the ligation reaction, successful transformants were harvested, and their plasmids were purified. This library was used as the starting pool for all subsequent transformations. To perform the dynamic screen, chemically competent DH5α were transformed with the library and grown immediately in LB + Ampicillin (LBA) until an OD600 of 0.1–0.2 (≈5 h), at which point they were inoculated (at 1% vol/vol) into selective (0.1–2.5% vol/vol Pine-Sol) and nonselective LBA. Plasmid samples were obtained throughout growth, fragmented by sonication, labeled with Cy3, and mixed with genomic E. coli DNA labeled with Cy5. The mixture was hybridized to an E. coli DNA microarray containing 1,160 gene probes. Differences among Cy3 intensities reflect the enrichment for specific transformant subpopulations out of the heterogeneous transformant library. Transformants that contain inserts that provide a growth advantage (green) will become the majority members of the total plasmid population at the expense of transformants that contain inserts that either do not provide a growth advantage (yellow) or that provide a growth disadvantage (red). In the example bar charts, the doubling times corresponding to each transformant type were 45 min (green), 60 min (yellow), and 85 min (red). Starting from an equal distribution of transformants (33% each), transformants with green inserts will make-up close to 90% of the total cell populations after 10 doublings, whereas those with red inserts will be diluted to ≈1% of the total number of cells.
At Pine-Sol concentrations less than 0.1%, no observable effect on DH5α growth was observed. Increasing Pine-Sol concentrations did retard the growth of wild-type DH5α as reflected in a decreased final cell density as well as an increased lag time in the wild-type cells. To determine what genes of the genomic library were enriched when grown under selective conditions, we isolated plasmids at various time points from E. coli DH5α[pTAGL] cells grown at 0.0%, 0.1%, 0.25%, and 0.4% (vol/vol) Pine-Sol in LBA. We would expect that under exponential growth, cells bearing plasmids with genomic inserts that provide a growth advantage in selective media would become predominant members of the culture, whereas cells with retarded growth as result of the transformation would be lost from the culture.
DNA microarrays were used to, in essence, simultaneously perform the isolation (in lieu of plating) and sequencing (in lieu of plasmid amplification and actual DNA sequencing) steps required for insert identification. Samples from selective (LB + Pine-Sol) and nonselective (LB) conditions were obtained throughout exponential and stationary phases, and plasmids were purified, fragmented by sonication, and labeled with Cy3-dUTP. In parallel, the original fragmented MG1655 genomic DNA was labeled with Cy5-dUTP to be used as a control. These two labeled pools of extra-chromosomal and genomic DNA were mixed, purified from unincorporated nucleotides, and hybridized to an E. coli DNA microarray containing 1,160 E. coli MG1655 gene probes. We then identified genes that provided a growth advantage or disadvantage (termed antibiotic resistance and susceptibility, respectively) by monitoring plasmid population dynamics throughout growth in selective and nonselective conditions. Overall, a total of 14 plasmid samples were analyzed.
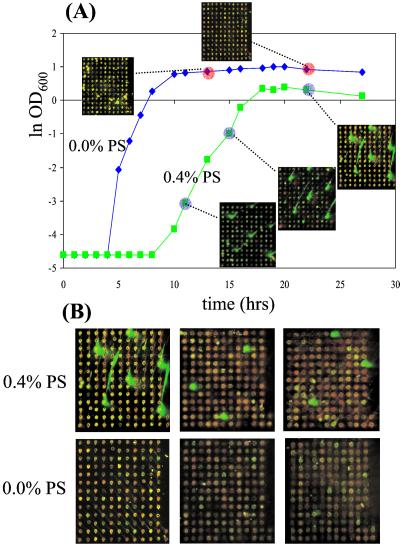
The microarray results allowed us to visualize the progression of plasmid-bearing cell populations throughout growth under selective and nonselective conditions (Fig. 2). In Fig. 2A, several bright green spots are visible at consistent locations in the 0.4% Pine-Sol samples but are not present in the 0.0% samples. These spots represent plasmids that contained genes selected for in the presence of Pine-Sol but not selected for in the absence of Pine-Sol under identical growth conditions. Most importantly, the pattern of genes enriched under selective conditions is consistent across multiple time points and is substantially different from the pattern of genes obtained under nonselective growth conditions. In Fig. 2B, the results for 435 gene probes are displayed for both the selected and nonselected cultures. Again, several very intense Cy3 signals were detected that were not observed in the antibiotic-free cultures.
Figure 2.
(A) DNA microarray images from plasmid populations throughout growth in selective and nonselective media (Pine-Sol; PS). The green signals correspond to plasmid samples labeled with Cy3-dUTP, and the red signals correspond to the control genomic DNA labeled with Cy5-dUTP and added in equal proportions to each array. Plasmids with gene inserts, identified by the large green spots on the array, were reproducibly observed in antibiotic-containing cultures and were not enriched in the antibiotic-free cultures. (B) The subarrays shown are representative blocks from the full eight-block (≈1,160 gene probes) array. The patterns of high intensity Cy3 spots in the 0.4% arrays reveal the presence of a selected subpopulation of clones not apparent in the 0.0% samples. Similar patterns of selection were observed in the remaining five blocks. The samples shown are from the last two time points indicated in A.
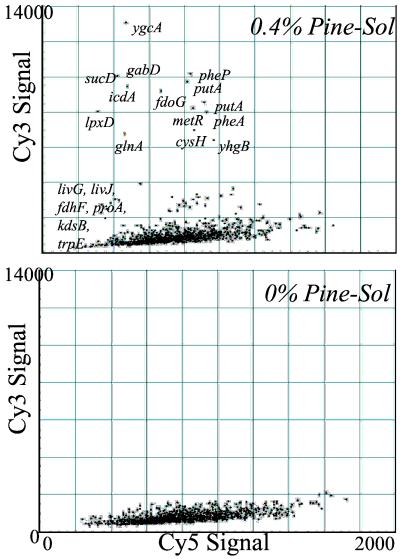
Cy3 and Cy5 fluorescence intensity values are displayed in Fig. 3 for the 0.4% and 0.0% Pine-Sol stationary phase cells. Because the Cy5 signals on all arrays originated from the same genomic DNA preparation, the distribution of these signals provides an internal control against experimental errors. On the other hand, Cy3 signals depend on the variable plasmid content and reflect, as such, the differential gene enrichment of these plasmid pools. On the same scale, the 0.4% Pine-Sol sample contained a skewed distribution of Cy3 signals when compared with the 0.0% samples, whereas the distribution of Cy5 signals from both samples were similar. More specifically, the mean and SD of the Cy3 and Cy5 intensity distributions provide quantitative evidence of the selection for a subset of cells. The SD was 125% of the mean Cy3 intensity in the enriched (0.4% Pine-Sol) cultures, and only 62% of the mean in nonselective conditions. In contrast, the SD was 38% and 41% of the mean Cy5 intensity for the selective and nonselective conditions, respectively.
Figure 3.
Quantified intensity values for the stationary phase arrays displayed in Fig. 2B. Cy3 intensity values (green Fig. 2) are located along the y axis and the Cy5 intensity values (red of Fig. 2) are represented along the x axis. The scales were set to be equal in both figures to emphasize the similar distribution of Cy5 signals (control genomic DNA) and the dramatically different distribution of Cy3 intensities. The genes with the largest Cy3 intensity correspond to genes that were selected for in the presence of Pine-Sol but were clearly not selected for in identical growth conditions without the presence of Pine-Sol. These genes were designated Pine-Sol resistance genes.
It is thus seen that transformed cells grown in a Pine-Sol-free medium do not show any significant change in the distribution of individual clones, suggesting that library amplification alone does not cause enrichment in a subpopulation of cells in the absence of a strong selective driving force. On the other hand, in selective media, the shifting in the distribution of plasmid-based signals (Cy3) reflects the strong selective pressure applied by Pine-Sol for a small subset of genes. The products of these genes provide a relative growth advantage in the presence of Pine-Sol that is not present in the absence of Pine-Sol. These genes were therefore designated as Pine-Sol resistance genes.
Mapping Cell Traits to Specific Genes.
We identified 19 genes that conferred growth advantage to their host cells when grown in the presence of Pine-Sol (Table 1). The functions of these genes are biochemically consistent with known resistance mechanisms. For example, two genes (lpxD, kdsB) are involved in lipid A biosynthesis, which is known to play a major role in antibiotic resistance and susceptibility (13, 14). Three other genes are involved in the transport of branched chain or aromatic amino acids and six are dehydrogenases. Furthermore, 8 of the 19 genes have been previously associated with some form of resistance when wild-type expression levels are altered (putA, metR, pheA, cysH, livG, livJ, trpE, proA; ref. 15).
Table 1.
PGTM identified resistance genes
| Gene | Function |
|---|---|
| ygcA | Unknown |
| gabD | Succinate-semialdehyde dehydrogenase |
| pheP | Phenylalanine specific permease |
| sucD | Succinyl–CoA synthetase |
| putA | Proline dehydrogenase |
| icdA | Isocitrate dehydrogenase |
| fdoG | Formate dehydrogenase |
| metR | Positive regulator for methionine genes |
| lpxD | Glucosamine-N-acyltransferase (lipid A) |
| pheA | Phenylalanine dehydrogenase |
| cysH | Adenylsulfate reductase |
| glnA | Glutamine synthetase |
| yhgB | Unknown |
| livG | Membrane component of high affinity branched chain amino acid transporter |
| livJ | Periplasmic binding protein of high affinity branched chain amino acid transporter |
| fdhF | Formate dehydrogenase |
| trpE | Anthranilate synthase component (tryptophan) |
| kdsB | CMP-3-deoxy-d-manno-octulosonate cytidylytransferase (lipid A) |
| proA | Glutamyl P reductase |
The major component of Pine-Sol (Pine-Oil) is α-terpineol (65%), and most of the active components of Pine-Oil are uncharged, nonaromatic cyclic hydrocarbons (11). Our results suggest that the mechanism of resistance to Pine-Sol action includes amino acid metabolism or transport, as well as altered dehydrogenase activity. Given the structural similarities of α-terpineol and many metabolites involved in aromatic amino acid biosynthesis and metabolism, this result is sensible. Also, antibiotic resistance most often occurs from alterations in the ability of the antibiotic to be transported into, stay, and remain active within the cell, all functions correlated with the identified genes. As such, these results provide a strong starting point in further investigating the specific mechanism(s) of action of antibiotic resistance-conferring compounds (5).
To identify genes that conferred a growth disadvantage in selective media, a more rigorous test of plasmid population differences was applied. It was of interest to discover those genes that conferred loss of viability when present in cells grown in the presence of Pine-Sol but did not affect growth in its absence. These susceptibility genes are very difficult to detect by conventional plating techniques, but can be identified by using the technique described in this report. For this purpose, we compared, for each gene, the mean Cy3 fluorescence intensity over four time-course samples (obtained at 1.5-h intervals) from cells grown in the presence of 0.4% Pine-Sol to the mean fluorescence intensity over three time-course samples from cells grown in the absence of Pine-Sol (0.0% samples). Genes were defined as significantly different when their means were lower (with 95% confidence) in the Pine-Sol-containing media than in the Pine-Sol-free media by using a mean hypothesis test (P < 0.05).
Many of the identified genes are consistent with a framework of antibiotic susceptibility. Specifically, rfaC and rfaD are part of a class of so called rough genes whose altered function is well known to be associated with hypersusceptibility (16). An additional set of six genes have products involved in membrane based transport activities (oppF, ptsI, ptsN, malX, cydA, cysU) (13). Furthermore, nine other genes (atpG, metK, upp, ptsI, codA, nadB, serA, cydA, pfkA) are known as selectable markers when their levels or structures are altered compared with wild type (15). A final note concerns the well-known susceptibility genes ompF and ompC. These two genes encode for outer membrane porins whose altered function produces hypersusceptibility (17). For both of these genes, the average intensity value in the 0.4% Pine-Sol samples was considerably lower than in the unselected population. For ompC and ompF, confidence could be assigned at approximately 85% (P = 0.146) and 70% (P = 0.316), respectively, indicating that overexpression of each of these two porins did lead to a reduced growth rate in the presence of Pine-Sol when compared with cells grown in the absence of Pine-Sol. Notably, identifying susceptibility genes from the experiment described herein was more challenging than for resistance genes.
Confirmation of Low-Level Pine-Sol Resistance Genes Identified by PGTM.
We confirmed the microarray results in two sets of independent experiments. In the first experiment, we sought to establish that increased copy numbers of some of the identified resistance genes do indeed confer a growth advantage in selective media. To this end we compared the values of the minimum inhibitory concentrations for individual strains transformed with a parent pUC plasmid containing no insert gene to those strains obtained with cells transformed with the same plasmid backbone but containing the original genomic library, an enriched genomic library (see below), seven of the identified resistance genes, and several “null” genes, i.e., genes that do not contribute to the enrichment or dilution of their hosts. The goal of the second set of experiments was to independently validate the identity of the genes enriched during growth in selective media. This experiment was done by usual plating and insert sequencing and comparing the identity of thus identified inserts to those of the genes identified by PGTM.
The enriched library was created by transforming cells with the original library, growing the transformants in LBA + 0.4% Pine-Sol until stationary phase, and isolating the plasmids from these cells. These experiments aimed to confirm that the enhanced growth phenotype was not caused by a chromosomal mutation or some unanticipated plasmid backbone effect that was beneficial for Pine-Sol tolerance. The MIC for strains transformed with the library increased significantly over the pUC control (see Table 2). A further increase was noted for strains transformed with the enriched library (a 20–30% increase in MIC). The insertion of a null gene into the pUC parental plasmid did not significantly alter the MIC. However, three of the genes identified by PGTM, trpE, livJ, and pheP did confer low-level resistance on overexpression. In evaluating the results of Table 2, it is important to note that the range of applicable MIC values is rather narrow because there is hardly any effect on cell growth at Pine-Sol concentrations below 0.25%, and cells grow poorly at concentrations greater than 0.6%. As shown in Fig. 2, there is a significant growth impairment of wild-type cells at Pine-Sol concentration of 0.4%. Therefore, restoring the normal growth phenotype at this Pine-Sol concentration after transforming cells with the enriched library or the genes of Table 2 is significant. These results confirm that the increased resistance to Pine-Sol is plasmid-borne rather than the result of a chromosomal mutation. It is of note that four additional resistance genes showed moderate increases in MIC over the pUC control (data not shown). Importantly, the observed ≈15% increase in MIC is sufficient to allow substantial enrichment when cells are grown over several generations (see Fig. 1). The close agreement of MIC values for cells transformed with individual genes and with the library provide further evidence of the ability of PGTM to identify enriched genetic selectants.
Table 2.
Confirmation of resistance genes
| Transformant | Trait | MIC ratio* | CV† | P value‡ | N§ |
|---|---|---|---|---|---|
| pUC19 control | Control | 1.0¶ | 0 | NA | 29 |
| Library | Res | 1.20 | 0.000 | 0‖ | 4 |
| Enriched library | Res | 1.40 | 0.165 | 0.020 | 4 |
| pBAD-hybC | Null | 1.06 | 0.065 | 0.135 | 4 |
| pBAD-leuC | Null | 1.06 | 0.065 | 0.135 | 4 |
| pBAD-trpE | Res | 1.14 | 0.000 | 0.029 | 4 |
| pBAD-livJ | Res | 1.19 | 0.080 | 0.015 | 4 |
| pBAD-pheP | Res | 1.18 | 0.112 | 0.040 | 9 |
Ratio of MIC of the corresponding strain by the MIC value of the PUC19 control.
Coefficient of variation = SD/mean.
P value is the one-tailed probability that the mean MIC between the transformant and the pUC19 control were equal using a Student's means t test for two samples of unequal variances.
Number of separate MIC assays.
MIC for pUC control was 0.6% ± 0.063% (vol/vol) Pine-Sol in LBA.
Variance in library MIC assays was zero.
In the second set of confirmation experiments, we transformed the enriched library into E. coli and determined the insert identities by plating and insert sequencing to compare insert identities to our PGTM results. Transformants were plated on LBA plates and incubated overnight, and 192 individual colonies were picked, followed by plasmid purification and restriction digestion. The digested products were analyzed by agarose gel electrophoresis, and 40 plasmids were partially sequenced. Three of the identified genes (yhgB, livM, trpC) were themselves or were directly adjacent to one of the 19 putative resistance genes identified by PGTM. For example, trpC is positioned directly adjacent to two of the top ranked resistance genes trpE and trpD. These three genes are a total of 4.1 kbp in length. The insert that contained the identified trp sequences was ≈4 kbp. Therefore, it is likely that the identified fragment contained portions of each of these genes. We have confirmed that trpE does provide low-level Pine-Sol resistance to E. coli as described above. Three additional genes carB, fumC, and glnK also were either ranked highly or had direct chromosomal neighbors (fumA, amtB) ranked among the top ≈10% of enriched clones identified by PGTM. Thus, more than 50% of the independently determined inserts were also included on the list of PGTM-identified resistance genes, with the remaining being apparently neutral to the growth phenotype.
A Tool for Functional Genomics and Target Discovery.
Genome sequencing projects are progressing at an accelerating pace, generating sequence information that must be complemented with efficient methods for gene function assignment. The approach described in this paper provides a parallel gene-trait mapping technique that is complementary, but substantially different, from transcriptional profiling (2–5). In transcriptional profiling, gene expression is only correlated to a specific cellular physiology, whereas in the trait-mapping approach, genes are identified whose presence or overexpression results in a specific physiological phenotype. We have demonstrated the ability to map specific cellular traits to all responsible genes in a parallel genome-wide screen using DNA microarrays.
PGTM differs from previous screening and microarray techniques in a number of ways. First, the majority of screening techniques rely on time-consuming sequencing of clones. In PGTM, the sequencing portion of the screen is performed by using the DNA microarray, a feature that saves considerable time and expense. Second, an advantage of PGTM is the ability to identify susceptibility genes in clones that do not survive the condition of interest (9). In the search for drug targets, it is often desirable to know which genes, when overexpressed, confer an increased susceptibility to the drug of interest. Susceptibility genes that enhance the effect of traditional antibiotics could be valuable targets for the design of novel anti-microbial strategies. Third, current microarray-based screening techniques (molecular barcoding, insertional mutant analysis) can evaluate the effect of gene deletion on overall cell physiology (7, 8). In contrast, PGTM can be used to evaluate the effect of gene overexpression on cell physiology and, similar to the molecular barcoding approach, it allows screening of an entire genome library in a single heterogeneous growth culture. Finally, most screening technologies employ selective conditions in a static manner (9), whereas PGTM allows dynamic evaluation of individual plasmid populations during competitive growth (dynamic screening).
Here we reported E. coli genes most responsible for increased Pine-Sol resistance or susceptibility. Antibiotic resistant strains of Staphylococcus aureus, Mycobacterium tuberculosis, and Streptococcus pneumonia, among others, have all been identified (18). These microbes are among the leading causes of bacterially based human disease and mortality (www.niaid.nih.gov). Each of these bacteria has recently been fully sequenced, and functional analysis of these genomes should be the next step in the discovery of new drug targets against such strains (www.tigr.org). Furthermore, low-level antibiotic resistance is now recognized as contributing to the emergence of clinically resistant microbes and it is desired to understand the mechanisms allowing these strains to evolve (12). Our confirmation studies suggested that PGTM can contribute to the discovery of low-level resistance genes and visualizing fitness landscapes in the evolution of antibiotic resistance (19).
One can envision other applications of this method as a platform technology. PGTM is applicable to any screen examining genes that confer differential growth characteristics to their hosts and, as a result, are enriched or diluted in a particular growth environment. For example, it can be applied to the screening of libraries of pathogenic bacteria within nonpathogenic hosts for genes conferring resistance or susceptibility. Similarly, one can envision screening mammalian cell libraries for genes that are able to help their respective hosts survive the presence of a cell-death-inducing factor. That is, it is of interest to screen for genes that make cancer cells more susceptible to certain chemotherapeutic agents or alternatively confer an increased lifespan for cells grown in culture (prevention of apoptosis). Identifying such genes will help to further elucidate the mechanisms of antibiotic-resistance, antibiotic-susceptibility, cancer, and programmed cell-death, and may lead to the identification of drugs and/or drug-targets. Finally, screens for overexpression strains with higher productivity because of better product formation, stress-resistance, increased or altered substrate utilization, or decreased product-associated toxicity are envisioned. The integration of this technique with other functional genomic, proteomic, and bioinformatic approaches provides a powerful portfolio of technologies for the systematic evaluation of genome function and its relation to cell physiology.
Acknowledgments
We thank Professor George Church for the E. coli microarrays used in this study. We would also like to thank William Schmitt for his help in applying Clustering, PCA, and MHT algorithms in the analysis of data, and Harry Wischnewski for excellent technical support throughout the sequencing confirmation studies. Support by the Engineering Research Program of the Office of Basic Energy Science at the Department of Energy, Grants DE-FG02-94ER-14487 and DE-FG02-99ER-15015 and National Institutes of Health Grant DK58533 is gratefully acknowledged.
Abbreviations
- PGTM
parallel gene-trait mapping
- PCA
principal component analysis
- LBA
LB + Ampicillin
- MIC
minimum inhibitory concentration
References
- 1.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 2.DeRisi J, Iyer V, Brown P. Science. 1997;278:680–685. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 3.Roberts C, Nelson B, Marton M, Stoughton R, Meyer M, Bennett H, He Y, Dai H, Walker W, Hughes T, et al. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 4.Schena M, Shalon D, Davis R, Brown P. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 5.Marton M, DeRisi J, Bennett H, Iyer V, Meyer M, Roberts C, Stoughton R, Burchard J, Slade D, Dai H, et al. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 6.Winzeler E, Shoemaker D, Davis R. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 7.Shoemaker D, Lashkari D, Morris D, Mittmann M, Davis R. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 8.Badarinarayana V, Estep P, Shendure J, Edwards J, Tavazoie S, Lam F, Church G. Nat Biotechnol. 2001;19:1060–1064. doi: 10.1038/nbt1101-1060. [DOI] [PubMed] [Google Scholar]
- 9.Cho R, Fromont-Racine M, Wodicka L, Feierbach B, Stearns T, Legrain P, Lockhart D, Davis R. Proc Natl Acad Sci USA. 1998;95:3752–3757. doi: 10.1073/pnas.95.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlowsky J, Saunders M, Harding G, Hoban D, Zhanel G. Antimicrob Agents Chemother. 1996;40:1387–1393. doi: 10.1128/aac.40.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moken M, McMurry L, Levy S. Antimicrob Agents Chemother. 1997;41:2770–2772. doi: 10.1128/aac.41.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baquero F. Drug Resist Updates. 2001;4:93–105. doi: 10.1054/drup.2001.0196. [DOI] [PubMed] [Google Scholar]
- 13.Berlyn M. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1753–1805. [Google Scholar]
- 14.Vaara M, Nurminen M. Antimicrob Agents Chemother. 1999;43:1459–1462. doi: 10.1128/aac.43.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaRossa R. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2527–2587. [Google Scholar]
- 16.Vaara M. Antimicrob Agents Chemother. 1993;37:2255–2260. doi: 10.1128/aac.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido H. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 29–47. [Google Scholar]
- 18.Henry C. Chem Eng News. 2000;78:41–58. [Google Scholar]
- 19.Kauffman S. The Origins of Order. Oxford: Oxford Univ. Press; 1994. [Google Scholar]