Abstract
A rapid approach to the 16S rRNA gene (16S rDNA)-based bacterial identification has been developed that combines uracil-DNA-glycosylase (UDG)-mediated base-specific fragmentation of PCR products with matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). 16S rDNA signature sequences were PCR-amplified from both cultured and as-yet-uncultured bacteria in the presence of dUTP instead of dTTP. These PCR products then were immobilized onto a streptavidin-coated solid support to selectively generate either sense or antisense templates. Single-stranded amplicons were subsequently treated with uracil-DNA-glycosylase to generate T-specific abasic sites and fragmented by alkaline treatment. The resulting fragment patterns were analyzed by MALDI-TOF MS. Mass signals of 16S rDNA fragments were compared with patterns calculated from published 16S rDNA sequences. MS of base-specific fragments of amplified 16S rDNA allows reliable discrimination of sequences differing by only one nucleotide. This approach is fast and has the potential for high-throughput identification as required in clinical, pharmaceutical, or environmental microbiology. In contrast to identification by MS of intact whole bacterial cells, this technique allows for the characterization of both cultured and as-yet-uncultured bacteria.
Emerging antibiotic-resistant pathogens, the actual threat of bioterrorist attacks, and bioremediation or bioprospecting efforts all require fast and accurate identification of the bacteria involved. Conventional diagnoses rely on the characterization of phenotypic traits of pure cultures obtained from specimens after cultivation and isolation of bacteria on appropriate laboratory media. These culture-based methods are time-consuming especially for slow-growing pathogens such as mycobacteria and generally are restricted to yet-culturable bacteria, which constitute only a small fraction (1–10%) of the global bacterial diversity (1). In contrast, genotypic analyses such as comparative sequencing of PCR-amplified 16S rRNA genes (rDNAs) allow for the identification of both cultured and as-yet-uncultured bacteria (1). Currently, 16S rDNA sequences constitute the largest gene-specific data set, and the number of entries in generally accessible databases is increasing continually (currently ≈30,000), making 16S rDNA-based identification of unknown bacteria isolates more and more likely. Conventional 16S rDNA sequencing is expensive and time-consuming because of the enzymatic reactions and electrophoresis or chromatography steps involved. Hence, it still is not suited for massive parallel testing as required in clinical, pharmaceutical, or environmental microbiology. Compared with standard 16S rDNA sequencing, alternative nonelectrophoretic techniques such as sequencing by MS (2) or pyrosequencing (3) are faster but generate only short read lengths, limiting their utility in 16S rDNA-based bacterial identification.
Here we introduce the concept of base-specific fragmentation of PCR-amplified DNA followed by MS analysis of the resulting fragment pattern as a method for rapid 16S rRNA-based bacterial identification. To exemplify the concept, uracil-DNA-glycosylase-based generation of T-specific fragment patterns was combined with analysis by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) MS for identification of environmental pathogenic and nonpathogenic bacteria strains. The inherent accuracy of mass spectrometric analysis and speed of signal acquisition make this an attractive candidate for high-throughput microbial identification.
Materials and Methods
Bordetella Species and As-Yet-Uncultured Bacteria.
The following Bordetella species were used in this study: Bordetella trematum DSM 11334T, Bordetella avium DSM 11332T, obtained from the Deutsche Sammlung von Mikroorgansimen und Zellkulturen (Braunschweig, Germany), and Bordetella petrii DSM 12804T, which was recently isolated in our laboratory (4). Cultivation of strains and PCR amplification of nearly full-length 16S rDNA (≈1,500 bp) was described previously (4). Yet-uncultured bacteria of anaerobic, organochlorine-reducing microbial consortia were represented by 16S rDNA sequence regions (≈350 or ≈1,500 bp) directly PCR-amplified and cloned from dechlorinating bioreactors (16S rDNA amplicons B and SHA; refs. 5 and 6).
MS on Base-Specific Fragmentation Patterns of Amplified 16S rDNA.
PCR.
Aliquots (1-μl) of 16S rDNA amplicons obtained from standard PCR in previous studies (5, 6) served as templates for PCR in the presence of dUTP instead of dTTP. For each individual DNA, PCR was performed in triplicate. Reactions (50-μl) contained 1× PCR reaction buffer, 5 pmol of biotinylated forward primer TPU1 (5′-AGAGTTTGATCMTGGCTCAG-3′, Escherichia coli 16S rRNA position 8–27 modified from Weisburg et al., ref. 7), 15 pmol of nonbiotinylated reverse primer RTU334a (5′-TGCCACCCGTAGGTGTATGG-3′, E. coli positions 334–353, this study), used only for yet-uncultured bacteria B1-10 and B1–12, or RTU334b (5′ TGCCTCCCGTAGGAGTCTGG 3′, E. coli positions 334 to 353, this study), 1.5 mM MgCl2, 200 μM dATP/dGTP/dCTP, 600 μM dUTP, and 1.25 units of AmpliTaq polymerase. The temperature profile consisted of 5 min initial denaturation at 95°C followed by 35 cycles of 93°C for 1 min, 60°C for 1 min, and 72°C for 1 min followed by a final extension at 72°C for 7 min.
Immobilization of PCR products.
To the crude PCR product, 50 μg of prewashed streptavidin-coated paramagnetic beads (Dynal, Oslo) in 45 μl of 2× BW buffer (10 mM Tris⋅HCl, pH 7.5/1 mM EDTA/2 M NaCl) were added and incubated for 20 min at room temperature.
Preparation of single-stranded templates.
Streptavidin-beads carrying immobilized PCR products were incubated with 0.1 N NaOH for 5 min at room temperature. After removing the supernatant, beads were washed three times with 50 μl of 10 mM Tris⋅HCl, pH 8.0.
Generation of nucleotide-specific abasic sites.
Immobilized single-stranded biotinylated PCR products were redissolved in 15 μl of uracil-DNA-glycosylase buffer (60 mM Tris⋅HCl, pH 7.6/1 mM EDTA). Two units of uracil-DNA-glycosylase (Fermentas, Hanover, PA) were added, and the mixture was incubated for 45 min at 37°C. Beads were washed twice each with 25 μl of 10 mM Tris⋅HCl, pH 8.0.
Elution of products from solid-phase and phosphate backbone cleavage.
To elute solid-phase bound single-stranded DNA, the beads were suspended in 12 μl of aqueous NH3, incubated at 60°C for 10 min, and cooled to 4°C. The supernatant was transferred to a new tube and subsequently heated to 95°C for 10 min followed by 80°C for 11 min with an open lid.
MALDI-TOF MS analysis.
Samples were analyzed from SpectroCHIP arrays. Fifteen nanoliters of analyte were deposited onto a SpectroCHIP (SEQUENOM) by using a nanopipette with a piezoelectric pump. All spectra were recorded by using a Bruker Biflex DE mass spectrometer (Bruker, Billerica, MA).
Sample processing (96- and 384-microtiterplate format) and analysis by MALDI-TOF MS can be completed within 2 h post-PCR by using the described protocol. With further advances in molecular biologic techniques, this time can be shortened substantially.
All experiments were carried out in triplicate to validate the reproducibility of the described principle.
Computational Analyses.
Standard comparative 16S rDNA sequence analysis and reconstruction of phylogenetic trees were done by using the ARB software package (8). All mass spectra analysis and automated data interpretation were performed by using in-house software developed by SEQUENOM.
Results
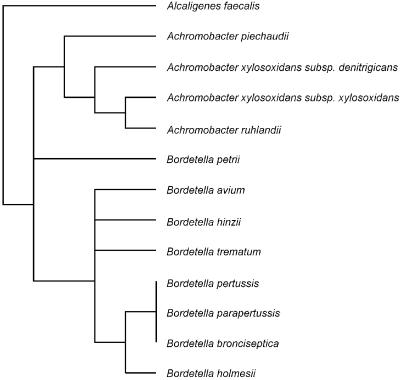
An ≈350-bp region of 16S rDNAs corresponding to E. coli position 8–353 were PCR-amplified from each of three Bordetella species and six yet-uncultured bacteria. The phylogenetic affiliations of the Bordetella species and yet-uncultured bacterial species used in this study are shown in Figs. 1 and 2.
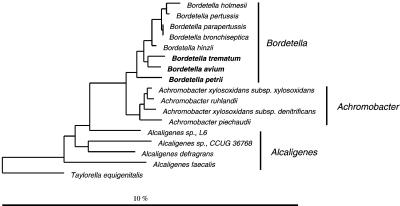
Figure 1.
16S rDNA-based trees showing the phylogenetic relationship of bacteria of the Alcaligenes–Achromobacter–Bordetella (AAB) complex. (Scale bar, 10% estimated sequence divergence.)
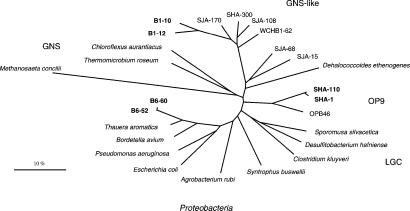
Figure 2.
16S rDNA-based trees showing the phylogenetic relationship of yet-uncultured B and SHA bacteria from anaerobic bioreactors used for biological treatment of chlorinated compounds. The species used for MALDI/TOF MS analysis of base-specific 16S rDNA fragmentation patterns are shown in bold. GNS, green nonsulfur bacteria; GNS-like, domain-level phylogenetic cluster related to green nonsulfur bacteria; OP9, domain-level phylogenetic cluster OP9; LGC, Gram-positives with a low DNA G+C content. (Scale bar, 10% estimated sequence divergence.)
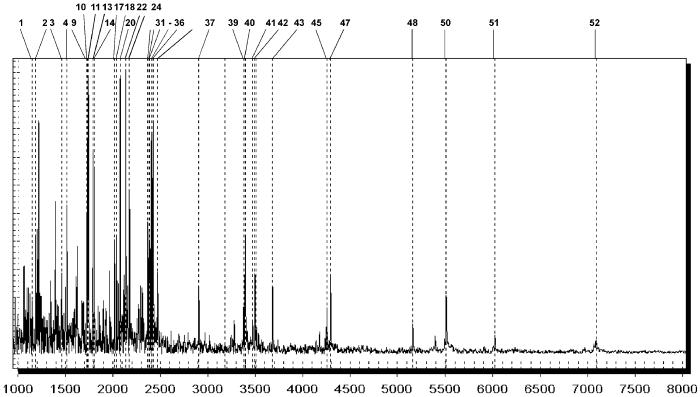
Fig. 3 depicts a representative mass spectrum of a 16S rDNA fragmentation pattern specific for the as-yet-uncultured bacterium B6-52. To facilitate interpretation of the spectrum, all expected mass signals of terminal and internal fragments calculated from the database reference sequence by “virtual” cleavage at all positions of thymine and uracil, respectively, have been cross-referenced with a number corresponding to the mass signals listed in Fig. 4.
Figure 3.
Mass spectrum displaying all signals generated by T-specific fragmentation of the 16S rDNA amplicon of strain B6-52. All expected mass signals corresponding to main cleavage products calculated from the reference sequence are marked with a dotted line. To simplify identification, the expected mass signals have been cross-referenced with the corresponding peak numbers in Fig. 4. All expected mass signals have been detected and allowed unambiguous identification of this bacteria strain. One nonreferenced dotted line overlays a low-intensity signal, which corresponds to the last fragment of the amplicon. Because of a nontemplated addition at the 3′ end of the PCR amplicon, this mass signal is shifted by the mass of the respective nucleotide added (see also Fig. 4).
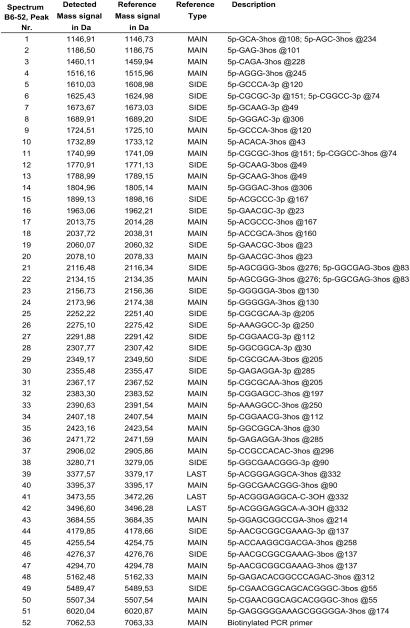
Figure 4.
Detected mass signals for a B6-52 fragmentation pattern corresponding to the mass spectrum displayed in Fig. 3. For all signals, the composition of the corresponding cleavage fragments was assigned automatically. Their exact sequence and location within the uncleaved PCR amplicon is provided in the description column. Different reference fragments were used in congruence with the possible cleavage reaction products. Main, main cleavage product; Side, side products of phosphate backbone cleavage, which occur with significantly lower intensity but can be assigned unambiguously; Last, products at the 3′ end of the amplicon, which often are elongated by one nucleotide because of 3′-terminal transferase activity of Taq; hos, hydrolysis products of phosphate backbone cleavage (main product); bos, β-elimination products of phosphate backbone cleavage. Fragments ending on a 3′-terminal phosphate group generated by δ elimination are marked with 3 P (both termed Side).
Fig. 4 displays all mass signals that could be detected automatically in this spectrum correlating them with their expected sequence (provided in the description column), location within the untreated PCR amplicon (described as @position), reference mass, and chemical nature at the 3′ end. The latter refers to the fact that under the described conditions, the phosphate backbone cleavage is a sequential process that can proceed via hydrolysis and elimination reactions (D.v.d.B., N.S., R. Worl, S. Kammerer, A. Braun, S.B., A. Ruppert, and C.J., unpublished results). Our protocol was optimized to generate 90% of the products as the hydrolysis product. All side products generated during phosphate backbone cleavage, however, can be identified unambiguously from their molecular masses. They are classified as “Side” in Fig. 4 and usually are much lower in intensity but nevertheless are considered during the simulation so that they do not interfere with unambiguous discrimination of strains.
The informative mass signal pattern for strain B6-52 consists of 32 expected terminal and internal cleavage products. All the corresponding mass signals were observed experimentally in the MALDI-TOF mass spectrum and thus allowed unambiguous identification. Twenty additional mass signals listed in Fig. 4, seemingly increasing the complexity of the mass spectrum (as evident from the comparison of dotted lines with actual mass signals), have been identified unambiguously as side products of the phosphate backbone cleavage and thus do not interfere with automated interpretation of the mass spectrum. One further mass signal separately classified in Fig. 4 is called “Last TR”, which refers to the 3′-terminal fragment of the PCR amplicon (the last fragment in the amplicon). A known feature of Taq polymerases is their tendency for nontemplated addition of nucleotides to the 3′ end. We observed this phenomenon. In addition to the mass signal corresponding to the 3′ fragment, which usually is the lowest in intensity, we also observed a fragment suggesting the addition of dAMP (3′ fragment + 313.2 Da) and dCMP (3′ fragment + 289.2 Da). The addition of dUMP or dGMP was less frequent, and the overall degree of nontemplated addition varied with the sequence context. All of these nontemplated additions have been included automatically in identification of fragments and thus are listed correspondingly. In the spectrum depicted in Fig. 3, three types of “Last” fragments have been identified (+U, +C, and +A), whereas the nonelongated fragment is barely visible (marked by the unreferenced dotted line preceding peak number 39). Of course the removal of uracil also proceeds when dUMP is added nontemplated at the 3′ end of the PCR product and the mass signal of the corresponding product thus is not grouped with the two other fragments (cross-compare peaks 39, 41, and 42 in Figs. 3 and 4).
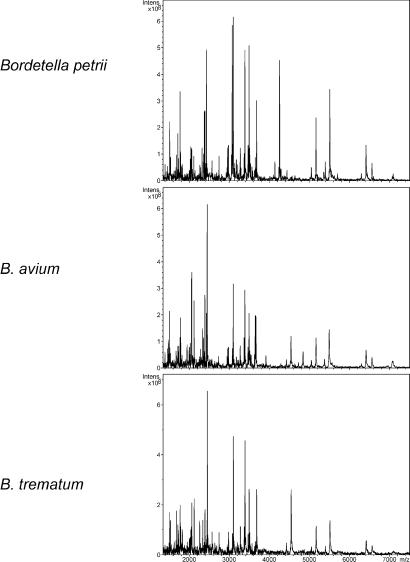
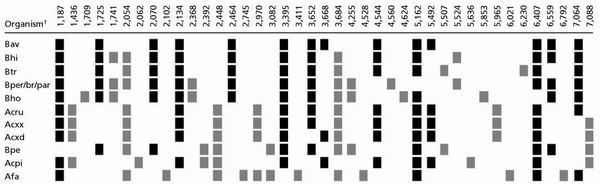
Further examples of typical mass spectra are shown in Figs. 5 and 6. Base-specific fragmentation of B. avium, B. trematum, and B. petrii 16S rDNA amplicons resulted in unique mass spectra despite the high sequence similarities of 97.3–99.1%. To avoid identification problems associated with single species-specific rDNA mass peaks (see Discussion), only patterns of mass peaks were used. Experimentally determined mass signal patterns were compared with mass signal patterns calculated from 50 published 16S rDNA sequences including 13 closely related species of the Alcaligenes–Achromobacter–Bordetella (AAB) complex (9). Bacteria then were ranked according to the number of nonidentical (additional or missing) mass peaks. The resulting theoretical peak patterns for the Bordetella species under study are shown in Table 1. For better visualization, only those peaks were included that are not present in all bacteria of the list (discriminating peaks). Compared with B. avium the closest species, Bordetella hinzii, showed four additional as well as two missing peaks. For B. trematum the most similar mass spectrum again is that of B. hinzii (three additional and two missing peaks). B. petrii clearly is more separated from other AAB species as compared with the former two Bordetella species. The more closely related species, B. trematum, showed six additional and six missing peaks. All calculated mass pattern differences have been validated experimentally.
Figure 5.
Mass spectra of base-specific 16S rDNA fragments of B. petrii, B. avium, and B. trematum. For display, mass spectra were smoothed by a five-point average (Savitzky–Golay method) and baseline-subtracted.
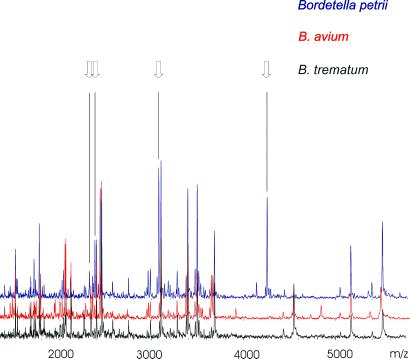
Figure 6.
Overlay of mass spectra of B. petrii (blue), B. avium (red), and B. trematum (black). Peaks specific for B. petrii are marked by the arrows. For display, mass spectra were smoothed by a five-point average (Savitzky–Golay method) and baseline-subtracted.
Table 1.
Ranking of base-specific 16S rDNA fragmentation mass patterns of AAB species according to the number of identical and nonidentical mass peaks compared with the mass spectrum of B. avium
Only discriminating mass peaks >1,186 Da are shown. Mass peaks identical to those of B. avium are marked in black. Additional mass peaks are marked in grey.
Bav, B. avium; Bhi, B. hinzii; Btr, B. trematum; Bper/br/par, B. pertussis/B. bronchiseptica/B. parapertussis; Bho, B. holmesii; Acru, A. ruhlandii; Acxx, Achromobacter xylosoxidans subspecies xylosoxidans; Acxd, A. xylosoxidans subspecies denitrificans; Bpe, Bordetella petrii; Acpi, A. piechaudii; Afa, Alcaligenes faecalis.
For Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica identical mass spectra were calculated because of 100% sequence identity within the 16S rDNA sequence region used for MS. This result is consistent with previous studies concluding that these organisms should be considered as strains or subspecies of a single species rather than separate species (9).
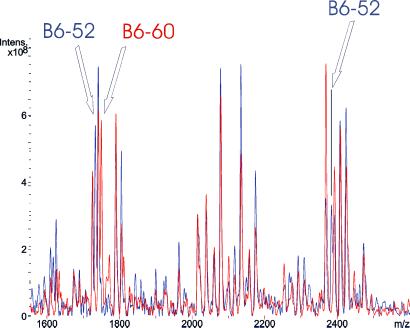
MALDI-TOF MS analysis of base-specific 16S rDNA fragmentation patterns also was tested with further yet-uncultured bacteria (SHA and B) represented by 16S rDNAs directly PCR-amplified and cloned from dechlorinating bioreactors (5, 6). These six selected rDNA sequences displayed pairwise sequence similarities of 92.0 (B1-10 and B1-12), 95.9 (SHA-1 and SHA-110), and 99.4% (B6-52 and B6-60; Fig. 2). Base-specific fragmentation and subsequent MS analysis generated specific spectra for all rDNA amplicons. Even strains B6-52 and B6-60, which differ in only two nucleotide positions, could be identified unambiguously on the basis of their unique mass signal pattern (Fig. 7). A comparison of experimentally observed mass spectra with the above-mentioned data set of 50 simulated mass spectra resulted in unambiguous identification of all SHA and B sequences (data not shown).
Figure 7.
Overlay of base-specific 16S rDNA fragmentation mass spectra of yet-uncultured bacteria B6-52 and B6-60 (B6-52 spectrum is indicated in blue, and B6-60 is indicated in red). Partial 16S rDNA sequences (354-bp) differ in only two nucleotide positions. Fragmentation patterns of the respective bacteria differ in three mass signals. B6-52 shows base-specific fragmentation mass signals at 1,733.1 and 2,383.5 Da, both missing in B6-60. In contrast, B6-60 shows a fragmentation mass signal at 1,749.1 Da, not present in the respective B6-52 spectrum. The arrows indicate the described mass peaks allowing discrimination of the strains.
Discussion
The goal of this study was to combine MS with 16S rDNA-based genotyping to develop a rapid technique for bacterial identification. The described technique avoids the time-consuming and expensive steps inherent in standard 16S rDNA sequencing. Previous work has shown the potential of MS in bacterial genotyping. In those studies MS was used to distinguish bacterial species by size determination of PCR-amplified specific marker genes (10, 11). However, for accurate bacterial identification this approach is limited, because length heterogeneities of specific marker genes do not provide sufficient discriminatory power. Instead, the technique described here makes use of sequence variations among 16S rDNAs of different species as revealed by base-specific fragmentation of 16S rDNA amplicons before MALDI-TOF MS.
To allow 16S rDNA amplification from a wide range of bacterial species, primers were selected that targeted highly conserved regions of 16S rDNA. Because of its length, a full analysis of 16S rDNA would generate too many cleavage fragments and complicate mass spectrometric analysis. To keep the mass signal complexity within the capability limits of conventional MALDI-TOF MS, 16S rDNA sequence regions rather than full-length genes were PCR-amplified. The forward primer TPU1 in combination with reverse primers RTU334a or RTU334b amplified a sequence region corresponding to position 8–353 of the E. coli 16S rRNA. This region was chosen because it includes highly variable areas V1 and V2 (12), allowing discrimination even among closely related bacterial species (13, 14). As described, MALDI-TOF MS analysis of base-specific fragmented 16S rDNA amplicons was tested on two classes of bacteria for which 16S rDNA sequencing currently is the method of choice for identification.
(i) Bordetella species that belong to a group of closely related pathogenic and environmental bacteria including members of the genera Alcaligenes and Achromobacter (AAB complex; Fig. 1). At present, comparative 16S rDNA sequencing is the most reliable method for the identification of AAB species because of the lack of specific phenotypic traits (9).
(ii) Yet-uncultured bacteria of selected anaerobic, organochlorine-reducing microbial consortia (Fig. 2). Currently, these bacteria are represented only by their 16S rDNA, which was PCR-amplified directly and cloned from bioreactors used for the biological treatment of chlorinated compounds (5, 6).
Despite high sequence similarities of 92.0 to 99.4%, MALDI-TOF MS analysis of base-specific 16S rDNA fragments resulted in unique mass spectra for each of the bacteria strains. We based the identification of strains on a comparison of mass peak patterns, because identification based on single species-specific mass peaks is limited; the number of species-specific unique mass peaks will decrease with a growing number of reference spectra/species. An analysis based on single species-specific peaks could lead to an erroneous assignment of an acquired sample spectrum as one of the reference species, especially if they randomly have the one species-specific peak in common.
We were interested also in the potential of this method for clustering and classification of unknown bacteria. As a preliminary assessment, we used the simulated peak pattern to construct most-parsimonious phylogenies of the 13 studied bacteria. The peak patterns in Table 1 were considered as a binary code, and the complete space of phylogenies was searched for the most-parsimonious trees that could explain the observed sequence information. This calculation was done by using the program PENNY, which is part of the PHYLIP software package (15). Alcaligenes faecalis was used as the outgroup. In total, 10 most-parsimonious trees were found, and the strict consensus of these most-parsimonious trees can be found in Fig. 8 (being artificially rooted at the branching point between its outgroup and the remaining species). All branching points found in the phylogenetic tree of Fig. 8 are comparable to the phylogenetic relationships described earlier, and unresolved edges (such as the placement of B. petrii) correspond to speciation events that are not completely decided in microbiology either, which suggests that at least for this example almost all the “phylogenetic information” contained in the DNA sequences of the examined species (see Figs. 1 and 2) can be found also in their fragmentation spectra. It therefore might be feasible to cluster and classify new isolates based on the fragmentation pattern.
Figure 8.
Most-parsimonious tree data based on the mass signal patterns generated by T-specific fragmentation of 16S rRNA PCR amplicons.
This technique uses well established genotypic markers, the 16S rDNA or regions thereof, allowing universal identification of both cultured and as-yet-uncultured bacteria (1, 8). The fact that only a minor part of the bacterial diversity can be cultivated in the laboratory and the increasing importance of yet-uncultured bacteria in both medical and environmental microbiology (1, 16) makes MS analysis of 16S rDNA specific fragmentation patterns superior to protein- or whole cell-based identification by MS requiring in vitro cultivation of bacteria before analysis. Similar to many other phenotypic identification methods, the latter approach is prone to growth-dependent variations resulting in problems with reproducibility (17, 18). In contrast, MALDI-TOF MS analysis of base-specific 16S rDNA fragmentation patterns (as well as most other DNA-based identification methods) is not biased by the cultivation conditions (17, 18). The absence of any time-consuming electrophoresis or chromatography step is an important advantage of MS analysis when compared with conventional 16S rDNA sequencing or rDNA fingerprinting by restriction analysis (19). Other sequencing techniques such as Sanger sequencing by MS (2) or pyrosequencing (3) have been proposed recently as viable alternatives to gel- or capillary-DNA sequencing. However, their short read lengths of 20–30 nucleotides strongly limit the use for 16S rDNA-based bacterial identification, because additional typing steps are required, e.g., the use of taxon-specific PCRs (20). Compared with these techniques, the principle of base-specific fragmentation of PCR amplicons combines the possibility of scanning larger sequence regions for bacterial typing with the speed and accuracy of MALDI-TOF MS.
The experimental procedure required for PCR amplification and subsequent base-specific fragmentation is rather simple and could make this method an ideal candidate for high-throughput identification in clinical diagnostics and environmental microbiology. It has great potential for bacterial and fungal pathogens that cause problems in routine conventional diagnostics, e.g., mycobacteria, Chlamydia, and dermatophytes. The MS-based identification approach is not restricted to 16S rDNA but can be expanded to other genotypic markers, e.g., typing of viral strains or typing of MDRR regions, which will broaden its applicability further.
Recently, other methods for generating base-specific fragmentation patterns of PCR amplicons have been described. They require either a subsequent post-PCR primer extension reaction to introduce a cleavable nucleotide (21) or rely on transcription of the PCR amplicon in RNA molecules that are cleaved by the means of a base-specific RNase (R. Hartmer, N.S., S.B., C.P. Rodi, F. Hillenkamp, C.J., and D.v.d.B., unpublished results). Because they follow the same principle, they as well could be applied to bacterial typing, and further studies may verify their suitability.
Abbreviations
- rDNA
rRNA gene
- MALDI-TOF
matrix-assisted laser desorption ionization–time-of-flight
- AAB
Alcaligenes–Achromobacter–Bordetella
References
- 1.Amann R I, Ludwig W, Schleifer K H. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nordhoff, E., Luebbert, C., Thiele, G., Heiser, V. & Lehrach, H. (2000) Nucleic Acids Res.28, e86. [DOI] [PMC free article] [PubMed]
- 3.Ronaghi M. Anal Biochem. 2000;286:282–288. doi: 10.1006/abio.2000.4808. [DOI] [PubMed] [Google Scholar]
- 4.von Wintzingerode F, Schattke A, Rösick U, Siddiqui R, Göbel U B, Gross R. Int J Syst Evol Microbiol. 2001;51:1257–1265. doi: 10.1099/00207713-51-4-1257. [DOI] [PubMed] [Google Scholar]
- 5.von Wintzingerode F, Landt O, Ehrlich A, Göbel U B. Appl Environ Microbiol. 2000;66:549–557. doi: 10.1128/aem.66.2.549-557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlötelburg C, von Wintzingerode F, Hauck R, Hegemann W, Göbel U B. Int J Syst Evol Microbiol. 2000;50:1505–1511. doi: 10.1099/00207713-50-4-1505. [DOI] [PubMed] [Google Scholar]
- 7.Weisburg W G, Barns S M, Pelletier D A, Lane D J. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K H. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 9.Gerlach G, von Wintzingerode F, Middendorf B, Gross R. Microbes Infect. 2001;3:61–72. doi: 10.1016/s1286-4579(00)01353-8. [DOI] [PubMed] [Google Scholar]
- 10.Hurst G B, Weaver K, Doktycz M J, Buchanan M V, Costello A M, Lidstrom M E. Anal Chem. 1998;70:2693–2698. doi: 10.1021/ac980044e. [DOI] [PubMed] [Google Scholar]
- 11.Muddiman D C, Wunschel D S, Liu C, Pasa-Tolic L, Fox K F, Fox A, Anderson G A, Smith R D. Anal Chem. 1996;68:3705–3712. doi: 10.1021/ac960689j. [DOI] [PubMed] [Google Scholar]
- 12.Van de Peer Y, Chapelle S, De Wachter R. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kataoka M, Ueda K, Kudo T, Seki T, Yoshida T. FEMS Microbiol Lett. 1997;151:249–255. doi: 10.1111/j.1574-6968.1997.tb12578.x. [DOI] [PubMed] [Google Scholar]
- 14.Kullen M J, Sanozky-Dawes R B, Crowell D C, Klaenhammer T R. J Appl Microbiol. 2000;89:511–516. doi: 10.1046/j.1365-2672.2000.01146.x. [DOI] [PubMed] [Google Scholar]
- 15. Felsenstein, J. (1993) PHYLIP, Phylogeny Inference Package (Dept. of Genetics, Univ. of Washington, Seattle), Version 3.5c.
- 16.Relman D A. Science. 1999;284:1308–1310. doi: 10.1126/science.284.5418.1308. [DOI] [PubMed] [Google Scholar]
- 17.Lay J O J. Trends Anal Chem. 2000;19:507–516. [Google Scholar]
- 18.van Baar B L M. FEMS Microbiol Rev. 2000;24:193–219. doi: 10.1016/S0168-6445(99)00036-4. [DOI] [PubMed] [Google Scholar]
- 19.Heyndrickx M, Vauterin L, Vandamme P, Kersters K, De Vos P. J Microbiol Methods. 1996;26:247–259. [Google Scholar]
- 20.Monstein H, Nikpour-Badr S, Jonasson J. FEMS Microbiol Lett. 2001;199:103–107. doi: 10.1111/j.1574-6968.2001.tb10658.x. [DOI] [PubMed] [Google Scholar]
- 21.Shchepinov M S, Denissenko M F, Smylie K J, Worl R J, Leppin A L, Cantor C R, Rodi C P. Nucleic Acids Res. 2001;29:3864–3872. doi: 10.1093/nar/29.18.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]