Abstract
The three proteins that comprise anthrax toxin, edema factor (EF), lethal factor (LF), and protective antigen (PA), assemble at the mammalian cell surface into toxic complexes. After binding to its receptor, PA is proteolytically activated, yielding a carboxyl-terminal 63-kDa fragment (PA63) that coordinates assembly of the complexes, promotes their endocytosis, and translocates EF and LF to the cytosol. PA63 spontaneously oligomerizes to form symmetric ring-shaped heptamers that are capable of binding three molecules of EF and/or LF as competing ligands. To determine whether binding of these ligands depends on oligomerization of PA63, we prepared two oligomerization-deficient forms of this protein, each mutated on a different PA63–PA63 contact face. In solution or when bound to receptors on Chinese hamster ovary K1 cells, neither mutant alone bound ligand, but a mixture of them did. After the two mutants were proteolytically activated and mixed with ligand in solution, a ternary complex was isolated containing one molecule of each protein. Thus EF and LF bind stably only to PA63 dimers or higher order oligomers. These findings are relevant to the kinetics and pathways of assembly of anthrax toxin complexes.
Anthrax toxin comprises three nontoxic monomeric proteins that assemble at the mammalian cell surface to form toxic complexes. Edema factor (EF) and lethal factor (LF) are enzymes that are delivered to the cytosol of mammalian cells by the third protein, protective antigen (PA). EF is an adenylyl cyclase that elicits edema when coinjected with PA into animals and likely serves to impair the immune response to infection (1–3). LF is a zinc protease that cleaves certain mitogen-activated protein kinase kinases (4–6). In combination with PA, LF causes death of animals by a still poorly understood sequence of events (7).
Assembly of the toxic complexes begins when PA binds to a cellular receptor, a widely expressed type 1 membrane protein termed anthrax toxin receptor (8). Receptor-bound PA is cleaved by a member of the furin class of proteases, causing removal of an amino-terminal 20-kDa segment of the protein (PA20), and leaving the carboxyl-terminal 63-kDa fragment (PA63) bound to anthrax toxin receptor (9). Unlike native PA, PA63 oligomerizes to form a ring-shaped heptamer (10). The heptamer binds up to three copies of EF and/or LF competitively and with high affinity (Kd ≈ 1 nM; refs. 11 and 12). Whereas native PA persists on the cell surface, the heptamer is endocytosed (13), presumably because oligomerization aggregates anthrax toxin receptor. The endocytosed toxic complexes are trafficked to endosomal compartments where the low pH causes the PA63 heptamer to insert into the membrane and form a water-filled channel (14, 15). Delivery of EF and LF to the cytosol occurs in concert with channel formation and may involve passage of these proteins through the channel (16).
Comparison of the crystal structures of PA and the PA63 heptamer showed that removal of the PA20 moiety relieves a steric constraint to oligomerization (17). Association of PA63 monomers occurs spontaneously and is mediated by interactions of domains 1′ and 2 of one monomer with domains 3 and 2, respectively, of an adjacent monomer. The hypothesis that EF and LF bind to the surface of domain 1′ exposed by removal of PA20 is supported by identification of mutations within domain 1′ that affect ligand binding (18). No crystallographic structure of complexes of PA63 with EF and/or LF has been solved, however, and the specific contacts involved in this interaction have remained undefined.
In an earlier study we discovered that an oligomerization-deficient mutant of PA (PA–D512A) did not bind as much ligand as wild-type PA, suggesting that ligand binding might depend on PA63 oligomerization (19). To investigate this question we prepared two complementary PA mutants with lesions on different PA63–PA63 contact faces. The presence of a mutated oligomerization face would be expected to block oligomerization of either mutant protein alone. When the proteins are mixed, however, their complementary oligomerization-competent faces should allow mixed dimers to form. Using these mutant proteins singly and in combination, we obtained strong evidence that oligomerization of PA63 is required for ligand binding.
Materials and Methods
Plasmid Construction.
The plasmid pET22b-PA contains the entire PA gene except for the portion that encodes the signal sequence (20). QuikChange mutagenesis was performed according to manufacturer instructions (Stratagene) to introduce site-directed mutations. Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). The oligonucleotides GGTTGATGTCAAAAATGAAAGAACTTTTCTTTCACC, CAATTTTGAAAATGGAGCAGTGGACGTGGATACAGGCTCG, GGTTAATCCTAGTAAACCATTAGAAACG, and their complements were used to introduce the mutations K199E, R468A/R470D, and D512K, respectively.
Preparation of Proteins.
PA, PA63, and LFN, the amino-terminal domain of LF, were purified from Escherichia coli as described (16, 21). Protein concentrations were determined by using Bradford protein assay reagent (Bio-Rad).
Cell-Surface Binding Assay.
Binding assays were performed essentially as described (16). Briefly, Chinese hamster ovary K1 cells were incubated on ice with 2 × 10−8 M trypsin-nicked PA for 2 h. The cells were washed twice with PBS and then incubated on ice with 35S-labeled LFN for 2 h. The cells were washed twice with PBS, and radioactive content was determined by scintillation counting.
Native Gel Electrophoresis.
Trypsin-nicked PA (2 μg) and LFN (3 μg) were mixed, as indicated, in 20 mM Tris⋅HCl (pH 8.0)/150 mM NaCl (in a volume of 8 μl) and incubated for ≈10 min at room temperature. Two microliters of loading dye (bromophenol blue in 50% glycerol) were added to the mixtures, which then were loaded onto 4–20% acrylamide, Tris-glycine gels (BioWhittaker). The running buffer was 25 mM Tris-base/192 mM glycine. The gels were stained with 0.05% Coomassie blue R-250/50% methanol/10% acetic acid and then destained with 10% acetic acid/10% methanol.
Multiangle Laser Light Scattering.
Light scattering was performed as described (11). Trypsin-nicked PA (300 μg from each oligomerization-deficient mutant) was incubated for 30 min at room temperature in the absence or presence of LFN (1 mg). Samples (140 μl) were loaded into a 100-μl loop and injected onto a Shodex KW-803 column equilibrated with 20 mM Tris⋅HCl (pH 8.2)/200 mM NaCl at a flow rate of 0.5 ml/min. The column was connected to a DAWN EOS 18-angle light-scattering detector (Wyatt Technology, Santa Barbara, CA) and an OPTILAB DSP interferometric refractometer (Wyatt Technology). Molecular mass calculations were performed by ASTRA software on normalized voltages from detectors 8–18 by using a dn/dc (refractive index increment) value of 0.185 ml/g. Normalization of the detectors was performed with the isotropic scatterer, BSA, to correct for slight differences in electronic gain among the detectors.
Results
Two Complementary Oligomerization-Deficient PA Mutants.
In a prior study we identified amino acid 512 as being important for oligomerization of PA63 (19). Mutating this residue from aspartic acid to alanine diminished oligomerization of PA63, and unexpectedly also impaired the protein's ability to bind LFN, as a sample ligand. To assess whether ligand binding depends on oligomerization of PA63, we made two other oligomerization-deficient PA mutants. For the first, we replaced D512 with lysine to generate a more severe oligomerization defect on the basis of the fact that D512 is on a loop of domain 3 that inserts into a positively charged cleft in domain 1′ of a neighboring subunit of the heptamer. For the second mutant, we placed mutations on the complementary oligomerization face of PA. We found empirically that multiple mutations on this face were needed to create a strong oligomerization defect and ultimately chose the triple mutant PA–K199E/R468A/R470D for further study.
Ligand Binds Only to Mixtures of the Two Mutants.
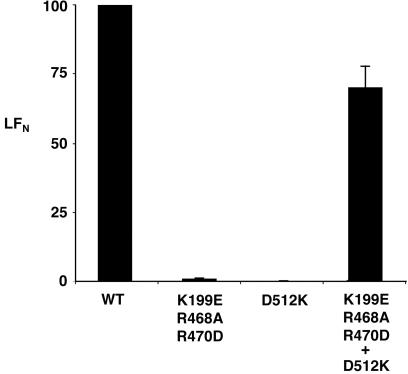
PA–D512K and PA–K199E/R468A/R470D were activated with trypsin and incubated singly or together with Chinese hamster ovary K1 cells at 4°C, a temperature at which endocytosis does not occur. After unbound PA was washed away, the cells were exposed to [35S]LFN, the amino-terminal PA63-binding domain of LF, and bound LFN was quantified by scintillation counting. Cells incubated with either PA–K199E/R468A/R470D or PA–D512K alone did not bind LFN (Fig. 1). In contrast, cells incubated with an equimolar mixture of PA–D512K and PA–K199E/R468A/R470D bound almost as much LFN as cells incubated with wild-type PA. These results support the notion that a high-affinity ligand site was generated when the two mutant forms of PA63 interacted (Fig. 1).
Figure 1.
Neither of two oligomerization-deficient forms of PA63 on cells stably binds LFN, but a mixture of the two does. Trypsin-nicked PA (2 × 10−8 M) wild type (WT), D512K, K199E/R468A/R470D, or a mixture of the two mutant forms (1 × 10−8 M of each) was incubated with Chinese hamster ovary K1 cells at 4°C for 2 h. The cells were washed twice with PBS and incubated with [35S]LFN at 4°C for 2 h. The cells then were washed with PBS, and radioactive content was measured by scintillation counting. The units on the ordinate are percentages of control (wild-type PA.) Nonspecific binding of LFN to cells (less than 10%) was subtracted from the experimental measurements to yield specific binding. The error bars represent SE of the mean.
A Ternary Complex Formed in Solution.
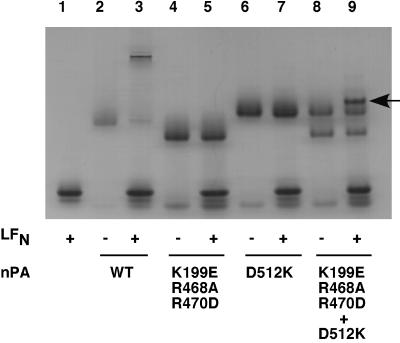
The oligomerization state of PA63 can be assessed by gel electrophoresis under nondenaturing conditions. Trypsin cleaves at the furin site, and in trypsin-nicked PA, the PA20 and PA63 fragments tend to remain associated, thereby blocking oligomerization of PA63. The nicked protein migrates on native gels with high mobility relative to that of liganded PA63 heptamer, formed by incubating nicked PA with LFN (Fig. 2, compare lanes 2 and 3). The electrophoretic mobilities of the nicked PA mutants differ from each other and from that of nicked wild-type PA because of differences in charge. Consistent with their oligomerization defects, neither of the mutants produced a low-mobility species when incubated with LFN (lanes 4–7). However, when a mixture of the two mutants was combined with LFN, a new band was observed with a slightly lower mobility than that of the nicked PA–D512K mutant (compare lanes 8 and 9). We inferred that this band represented a ternary complex generated by interaction of LFN with a PA63 dimer formed from the two mutants.
Figure 2.
Native gel electrophoresis of oligomerization-deficient PA mutants in the presence and absence of ligand. Trypsin-nicked PA (nPA) of wild type (WT) or mutant (2 μg) was incubated in the presence or absence of LFN (3 μg), and the mixture was electrophoresed on a 4–20% acrylamide Tris-glycine gel. The gels were stained with Coomassie blue. A band in lane 9 corresponding to a putative heterotrimer containing a molecule of LFN and a molecule of PA63 from each of the two mutant PAs is indicated by the arrow.
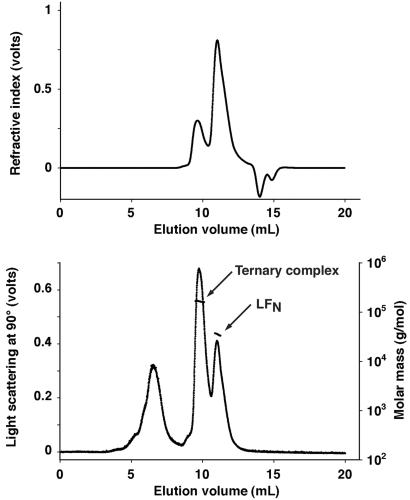
To confirm the identity of this putative ternary complex, we nicked each mutant PA, mixed the two, divided the mixture in half, and added excess LFN to one half (8:1 molar ratio of LFN/PA63 monomer). Each sample then was chromatographed on a Shodex KW-803 size-exclusion column, and the effluent was passed sequentially through a multiangle laser light-scattering instrument and an interferometric refractometer. The latter served as a continuous monitor of protein concentration. Combining the data from these two instruments permitted calculation of the particle masses of protein peaks in the column effluent. As shown in Fig. 3, the sample containing LFN gave two protein peaks with measured molecular masses of 163 ± 1.9 and 36.1 ± 0.4 kDa. The first value corresponds closely to the predicted mass (160 kDa) of a PA63 dimer complexed with a single molecule of LFN, and the second corresponds well with the predicted mass of the unbound LFN (34 kDa). The presence of a small amount of PA20 at the trailing edge of the LFN peak probably accounts for the negative slope of the molecular mass values across this peak. These results support our interpretation of the additional band seen in lane 9 of Fig. 2 as representing a ternary complex as described.
Figure 3.
Use of multiangle laser light scattering to identify a ternary complex of LFN and the two oligomerization-deficient forms of PA63. A mixture of LFN (1 mg) with trypsin-nicked PA–D512K (300 μg) and PA–K199E/R468A/R470D (300 μg) was chromatographed over a Shodex KW-803 gel filtration column connected to a light-scattering instrument (Lower) and an interferometric refractometer (Upper). The values of molecular mass determined in volume increments across each peak are shown (arrows). The light-scattering peak at ≈7 ml elution volume represents a very small amount of contaminating high molecular mass material (note the absence of interferometric refractometer signal). The negative refractive index values at 14–15 ml elution volume represent differences between the composition of the sample and elution buffers.
When the two oligomerization-deficient mutants were mixed in the absence of LFN and chromatographed, only a single peak was seen, with a measured mass of 83.4 ± 2.1 kDa, corresponding to PA63 complexed with PA20 (data not shown), which correlates with the fact that we did not observe a band by PAGE, corresponding to a heterodimer of the two mutant forms of PA63 (Fig. 2). This result suggests that PA63 dimers are inherently unstable and form only transiently during the course of heptamerization. It is noteworthy in this regard that no subheptameric intermediates are seen in chromatographic fractionation of PA63. Thus only the fully formed heptamer of PA63, in which every subunit is bound to two neighboring subunits, is apparently stable in the absence of ligand.
Discussion
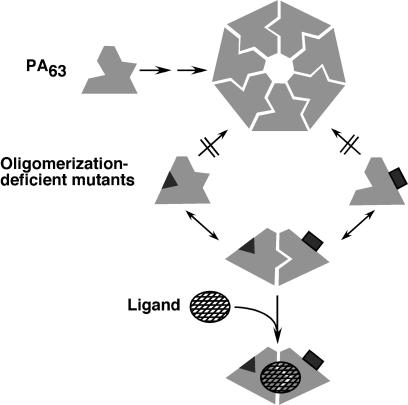
Our results indicate that the enzymatic moieties of anthrax toxin bind stably only to dimeric or oligomeric forms of PA63 (Fig. 4). Dependence of ligand binding on oligomerization would be expected if the ligand site contained contributions from two adjacent monomers or if subunit–subunit interactions stabilized a binding-competent conformation. In an accompanying paper, evidence is presented that the binding site of LFN spans the interface between PA63 subunits of the dimer (18), which substantiates the first of these possibilities.
Figure 4.
Complex formation by wild type and oligomerization-deficient mutants of PA63. Only one potential pathway of formation of the ternary complex is illustrated.
That LFN does not associate stably with monomeric PA63 is at variance with the conclusion of an earlier study reporting that LF bound to PA that had been adsorbed to a plastic surface and then nicked with trypsin (22). The adsorbed PA may have oligomerized in this experiment by diffusion on the plastic surface or via other unrecognized pathways. Alternatively, the observed binding may have reflected combined interactions of LF with both monomeric PA63 and the plastic surface.
Our results have implications regarding the pathways by which anthrax toxin complexes assemble from their component parts. Earlier results have demonstrated that PA63 can assemble to the homoheptamer in the absence of ligand, and that the heptamer can bind up to three molecules of ligand (10, 11). Our results imply that ligand can bind to and stabilize subheptameric oligomers of PA63, creating stable heterooligomeric intermediates. The stability of such intermediates would be expected to accelerate formation of toxic complexes containing heptameric PA63.
Fig. 4 illustrates one pathway by which liganded ternary complex containing dimeric PA63 could form. However, two such pathways are possible: (i) ligand could bind to a transient intermediate of two PA63 molecules, or (ii) ligand could bind to monomeric PA63 followed by interaction of this complex with a second molecule of PA63. The kinetics of these pathways depend on the relative stabilities of the PA63–PA63 and PA63–ligand intermediates, which are unknown. Values of Kd and the on- and off-rate constants of the interactions of the enzymatic moieties for oligomeric PA63 have been measured (12), but other parameters affecting the assembly process have not. Regardless of the pathway by which it formed, the liganded dimer of the mixed oligomerization-deficient mutant PAs is sufficiently stable in solution to be identified as a peak from a size-exclusion column and a band on PAGE.
Acknowledgments
This work was supported by National Institute of Allergy and Infectious Disease Grant R37-A122021. J.M. was supported in part by a postdoctoral fellowship from the Canadian Institutes of Health Research. M.M. was supported in part by a postdoctoral fellowship from the Direction des Systèmes de force et de la Prospective and the Philippe Foundation. D.B.L was supported by a fellowship from the Helen Hay Whitney Foundation. R.J.C. has financial interest in AVANT Immunotherapeutics and PharmAthene, Inc.
Abbreviations
- EF
edema factor
- LF
lethal factor
- PA
protective antigen
- PAn
n-kDa fragment of protective antigen
- LFN
amino-terminal domain of LF
References
- 1.Leppla S H. Proc Natl Acad Sci USA. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Brien J, Friedlander A, Dreier T, Ezzell J, Leppla S. Infect Immun. 1985;47:306–310. doi: 10.1128/iai.47.1.306-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoover D L, Friedlander A M, Rogers L C, Yoon I-K, Warren R L, Cross A S. Infect Immun. 1994;62:4432–4439. doi: 10.1128/iai.62.10.4432-4439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duesbery N S, Webb C P, Leppla S H, Gordon V M, Klimpel K R, Copeland T D, Ahn N G, Oskarsson M K, Fukasawa K, Paull K D, Woude G F V. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 5.Vitale G, Pellizzari R, Recchi C, Napolitani G, Mock M, Montecucco C. Biochem Biophys Res Commun. 1998;248:706–711. doi: 10.1006/bbrc.1998.9040. [DOI] [PubMed] [Google Scholar]
- 6.Vitale V, Bernardi L, Napolitani G, Mock M, Montecucco C. Biochem J. 2000;352:739–745. [PMC free article] [PubMed] [Google Scholar]
- 7.Friedlander A M. J Biol Chem. 1986;261:7123–7126. [PubMed] [Google Scholar]
- 8.Bradley K A, Mogridge J, Mourez M, Collier R J, Young J A T. Nature (London) 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 9.Klimpel K R, Molloy S S, Thomas G, Leppla S H. Proc Natl Acad Sci USA. 1992;89:10277–10281. doi: 10.1073/pnas.89.21.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milne J C, Furlong D, Hanna P C, Wall J S, Collier R J. J Biol Chem. 1994;269:20607–20612. [PubMed] [Google Scholar]
- 11.Mogridge J, Cunningham K, Collier R J. Biochemistry. 2002;41:1079–1082. doi: 10.1021/bi015860m. [DOI] [PubMed] [Google Scholar]
- 12.Elliott J L, Mogridge J, Collier R J. Biochemistry. 2000;39:6706–6713. doi: 10.1021/bi000310u. [DOI] [PubMed] [Google Scholar]
- 13.Beauregard K, Collier R J, Swanson J A. Cell Microbiol. 2000;2:251–258. doi: 10.1046/j.1462-5822.2000.00052.x. [DOI] [PubMed] [Google Scholar]
- 14.Blaustein R O, Koehler T M, Collier R J, Finkelstein A. Proc Natl Acad Sci USA. 1989;86:2209–2213. doi: 10.1073/pnas.86.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milne J C, Collier R J. Mol Microbiol. 1993;10:647–653. doi: 10.1111/j.1365-2958.1993.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 16.Wesche J, Elliott J L, Falnes P Ø, Olsnes S, Collier R J. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]
- 17.Petosa C, Collier R J, Klimpel K R, Leppla S H, Liddington R C. Nature (London) 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham K, Lacy D B, Mogridge J, Collier R J. Proc Natl Acad Sci USA. 2002;99:7049–7053. doi: 10.1073/pnas.062160399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogridge J, Mourez M, Collier R J. J Bacteriol. 2001;183:2111–2116. doi: 10.1128/JB.183.6.2111-2116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson E L, Huynh P D, Finkelstein A, Collier R J. Biochemistry. 1998;37:3941–3948. doi: 10.1021/bi972657b. [DOI] [PubMed] [Google Scholar]
- 21.Miller C J, Elliott J L, Collier R J. Biochemistry. 1999;38:10432–10441. doi: 10.1021/bi990792d. [DOI] [PubMed] [Google Scholar]
- 22.Singh Y, Klimpel K R, Goel S, Swain P K, Leppla S H. Infect Immun. 1999;67:1853–1859. doi: 10.1128/iai.67.4.1853-1859.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]