Abstract
Chemoreceptors of the methyl-accepting chemotaxis protein family form clusters, typically at the cell pole(s), in both Bacteria and Archaea. To elucidate the architecture and signaling role of receptor clusters, we investigated interactions between the serine (Tsr) and aspartate (Tar) chemoreceptors in Escherichia coli by constructing Tsr mutations at the six hydrophobic and five polar residues implicated in “trimer of dimers” formation. Tsr mutants with proline replacements could not mediate serine chemotaxis, receptor clustering, or clockwise flagellar rotation. Alanine and tryptophan mutants, although also nonchemotactic, formed receptor clusters, and some produced clockwise flagellar rotation, indicating receptor-coupled activation of the signaling CheA kinase. The alanine and tryptophan mutants evidently assemble defective receptor complexes that cannot modulate CheA activity in response to serine stimuli. In cells containing wild-type Tar receptors, tryptophan replacements in Tsr interfered with Tar function, whereas four Tsr mutants with alanine replacements regained Tsr function. These epistatic and rescuable phenotypes imply interactions between Tsr and Tar dimers in higher-order signaling teams. The bulky side chain in tryptophan mutants may prevent stimulus-induced conformational changes in the team, whereas the small side chain in alanine mutants may permit signaling control when teamed with functional receptor molecules. Direct physical interactions between Tsr and Tar molecules were observed by in vivo chemical crosslinking. Wild-type Tsr crosslinked to Tar, whereas a clustering-defective proline replacement mutant did not. These findings indicate that bacterial chemoreceptor clusters are comprised of signaling teams, seemingly based on trimers of dimers, that can contain different receptor types acting collaboratively.
Keywords: chemotaxis‖receptor clustering‖epistasis‖functional rescue‖crosslinking
Motile Bacteria and Archaea use transmembrane chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs) to mediate many of their adaptive locomotor behaviors. MCP molecules typically have a periplasmic ligand-binding domain for monitoring attractant or repellent levels in the environment and a cytoplasmic signaling domain for communicating with the motor apparatus (1). The MCP signaling domain, highly conserved in structure, forms ternary complexes with two cytoplasmic proteins, CheA, a histidine kinase, and CheW, which couples CheA activity to chemoreceptor control (2, 3). Changes in receptor ligand occupancy trigger conformational changes in the signaling domain that in turn modulate the flux of phosphoryl groups from CheA to effector proteins that elicit the behavioral response (4, 5). MCPs are capable of detecting chemoeffector concentration changes of only a few parts per thousand over more than a five-log concentration range (6–9). The amplification mechanisms responsible for the high-gain signaling characteristics of bacterial chemoreceptors are still poorly understood but may rely on novel signaling principles that will prove to be widely used in biological signal transduction systems.
Like many membrane receptors, MCP molecules are not uniformly distributed but rather clustered, typically at the cell poles (10). The CheA and CheW proteins also localize to these receptor clusters and are largely responsible for their integrity (10), suggesting that bacterial chemoreceptors form a two-dimensional lattice that is held together by bridging connections to CheA and CheW. Bray and colleagues (11–13) have theorized that receptors in such an array might, through conformational coupling, communicate their signaling states to neighboring receptors to produce a large gain in detection sensitivity. Experimental work, primarily with Escherichia coli, provides growing support for this notion.
E. coli uses five MCP-family receptors to promote chemotactic movements toward different attractant compounds: Tar (aspartate and maltose), Tsr (serine), Tap (dipeptides), Trg (ribose and galactose), and Aer (oxygen and other electron acceptors). Tap, Trg, and, most likely, Aer are present in the cell at roughly 10% the levels of Tsr and Tar (14). Several lines of evidence indicate that high- and low-abundance receptors might signal collaboratively, and that clustering enhances their detection sensitivity. First, the ability of low-abundance receptors to mediate chemotactic responses implies that they are able to exert control over a substantial fraction of the CheA signaling molecules associated with the receptor array. Second, high-abundance receptors assist one another (15) and low-abundance receptors (16–18) in achieving the methylation changes needed to adapt to sensory stimuli. Third, a multivalent galactose ligand that promotes clustering of Trg (19) also served to recruit Tar and Tsr molecules to the cluster and greatly enhanced their detection sensitivity, implying that communication between receptors in a cluster produces signal amplification (20).
In vitro studies of receptors and receptor fragments indicate that more than one receptor signaling domain is needed to form a ternary complex that activates and controls the CheA kinase. The optimal coupling stoichiometries vary from one system to another, but invariably specify receptors or receptor fragments in excess of CheA, for example, 8–10 dimers of the Tsr signaling domain (21) or 7 dimers of the Tar signaling domain (22) per CheA dimer. Intact receptors in ternary complexes also exhibit cooperative inhibition of CheA in response to attractant ligands, consistent with the presence of several interacting receptor molecules in the functional signaling unit (23, 24). Ternary complex formation, CheW binding, and CheA control reside in an 80-aa segment at the membrane-distal tip of the MCP signaling domain (ref. 25; P.A. and C. C. Downs, unpublished results). The signaling tip also contains the contact sites for a trimer of dimers arrangement observed in the crystal structure of a Tsr fragment (26) that has signaling activity both in vivo and in vitro (P.A., unpublished results), suggesting that this higher-order structure might represent an active signaling form of MCPs.
To explore the physiological role of Tsr trimer contacts, we created a series of mutations to alter trimer formation or geometry and examined their effects on receptor signaling and clustering. We found that nearly all mutational changes at the trimer contacts disrupted Tsr signaling ability. Some mutant receptors could not cluster, whereas others formed clusters that failed to either activate or modulate the CheA kinase. Many of the cluster-proficient lesions enabled the mutant Tsr molecules to inactivate other receptors, presumably through formation of mixed signaling complexes. On the basis of these findings and of supporting studies of receptor crosslinking in vivo, we propose that MCPs can form mixed signaling teams, most likely based on trimers of dimers, that share control of the same CheA molecule(s). These receptor signaling teams appear to be important architectural and functional components of chemoreceptor clusters and may, through mechanisms not yet understood, confer their high-gain signaling properties.
Materials and Methods
Bacterial Strains.
All strains were derivatives of E. coli K12 RP437 (27): RP3098 [Δ(flhD-flhB)4] (28); RP9352 [Δtsr-7028 Δ(tar-tap)5201 Δtrg-100 ΔcheZ-6725] (29); UU1248 [Δtsr-7028 Δ(tar-cheB)2234 Δtrg-100 Δaer-1]; UU1250 [Δtsr-7028 Δ(tar-tap)5201 Δtrg-100 Δaer-1]; KO607 [Δtsr-7028 Δ(tar-tap)5201 Δtrg-100 recA56] (30).
Plasmids.
Plasmids used were pCJ30, an IPTG- (isopropylβ-d-thiogalactopyranoside) inducible expression vector derived from pBR322 (31) that confers ampicillin resistance (32); pJC3, a relative of pCJ30 that carries wild-type tsr and confers optimal serine chemotaxis at 20 μM IPTG (33); pLC112, a salicylate-inducible expression vector that confers chloramphenicol resistance [constructed from portions of pACYC184 (34) and pKMY297 (35)]; pLC113, a relative of pLC112 that carries wild-type tar and confers optimal aspartate chemotaxis at 0.7 μM sodium salicylate; pCS66, a derivative of pLC113 that makes a functional Tar with an Arg-Ser-(His)6 addition at its C terminus; pPA114, a relative of pLC112 that carries wild-type tsr and confers optimal serine chemotaxis at 0.6 μM sodium salicylate; and pVS49, which makes a YFP-CheZ fusion protein under inducible arabinose control (36).
Construction of Tsr Mutants.
Mutations at the trimer contact residues of Tsr were constructed in pJC3 by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Candidate mutants were verified by sequencing the entire tsr coding region to avoid any with collateral damage.
Tsr Expression Level.
The steady-state levels of mutant Tsr proteins expressed from pJC3 derivatives were measured in host UU1250. Strains were grown at 35°C to mid-log phase in tryptone broth (37) containing 50 μg/ml of ampicillin and 20 μM IPTG. Samples (1 ml) of each culture were pelleted by centrifugation (6,000 × g), washed once with an equal volume of cold 10 mM Tris⋅HCl (pH 8.0)/1 mM EDTA, and resuspended and lysed by boiling in 100 μl of sample buffer (38). Lysate proteins were analyzed by electrophoresis in SDS-containing 16% polyacrylamide gels. Gels were electroblotted to polyvinylidine difluoride membranes, which were then treated with a mixture of anti-Tsr (21) and anti-CheY sera, each diluted 1/1,000. Antibody-reactive bands were visualized with 35S-labeled staphylococcal protein A (Amersham Pharmacia) and a Molecular Dynamics PhosphorImager, and quantified with imagequant software (iqmac, Ver. 1.2, Molecular Dynamics). The relative amounts of Tsr protein in different samples were compared by using as an internal standard a chromosomally encoded protein that crossreacted with the anti-cheY serum.
Behavioral Assays.
Chemotaxis to serine and aspartate was assessed by colony size and morphology on tryptone semisolid agar plates (37). Flagellar rotation patterns of antibody-tethered cells were analyzed as described (29). The overall percent of time spent in clockwise (CW) rotation was computed as a weighted sum: the percent of cells that rotated exclusively CW, plus 0.75 × the percent of cells rotating predominantly CW, plus 0.5 × the percent of cells reversing frequently, plus 0.25 × the percent of cells rotating predominantly, but not exclusively, counterclockwise.
Receptor Clustering Assay.
Mutant pJC3 derivatives were transferred into strain RP9352 carrying pVS49 to evaluate receptor clustering by Tsr* mutants. Cells were grown at 30°C in tryptone broth containing 12.5 μg/ml of chloramphenicol, 50 μg/ml of ampicillin, 20 μM IPTG, and 0.005% l(+)-arabinose. Cells were collected at mid-log phase and examined by fluorescence microscopy, essentially as described (36). Cell fields were photographed and inspected by eye to determine the proportion of individuals with a distinct bright spot of fluorescence indicative of a receptor cluster. At least 100 cells were scored for each mutant.
Complementation Tests.
Mutant pJC3 derivatives were transferred into strain KO607 carrying pPA114 to evaluate the dominance of Tsr* mutants. Individual transformant colonies were inoculated into tryptone semisolid agar plates containing 50 μg/ml of ampicillin, 12.5 μg/ml of chloramphenicol, 0.6 μM sodium salicylate, and IPTG at 0, 10, 20, or 40 μM. Plates were scored after incubation at 35°C for 7–8 h, and Tsr* mutants were classified as either recessive (no reduction in wild-type serine response at any induction level) or dominant (impaired serine chemotaxis at some induction level). Dominance was classified in severity according to the threshold concentration of IPTG inducer needed to produce the effect: D3 (0 μM), D2 (10 μM), D1 (20 μM), or D0 (40 μM).
Epistasis tests were done in an identical manner except that mutant pJC3 derivatives were transferred to strain KO607 carrying pLC113 and the sodium salicylate concentration in the assay plates was 0.7 μM. Plates were scored after incubation for 8–9 h, and mutants were classified as epistatic if they impaired aspartate chemotaxis at some induction level: E3 (0 μM), E2 (10 μM), E1 (20 μM), or E0 (40 μM). Some nonepistatic mutants regained serine chemotaxis ability in these tests and were classified as functionally rescuable (by wild-type Tar).
Receptor Crosslinking.
Strains UU1248 and RP3098 carrying pCS66 and pJC3 (or one of its derivatives) were grown at 35°C in tryptone broth containing 100 μg/ml of ampicillin and 50 μg/ml of chloramphenicol. At early log phase, IPTG (20 μM) and sodium salicylate (0.6 μM) were added to induce Tsr and Tar-(His)6 expression to their normal cellular levels. At mid-log phase, cells were harvested by centrifugation (6,000 × g), washed twice with KEP [10 mM potassium phosphate (pH 7.0)/0.1 mM EDTA], and resuspended in the same buffer at an OD600 of 5. The crosslinking agent dithiobis(succinimidylpropionate) (Pierce) was added to 10 ml of cell suspension to a final concentration of 0.5 mM. After 2 h at room temperature, the reaction was quenched by addition of Tris⋅HCl (pH 8.0) to a final concentration of 0.1 M. After an additional 15 min at room temperature, the cells were collected by centrifugation, washed with 50 mM Tris⋅HCl (pH 8.0), and stored at −20°C.
Cell samples were thawed and resuspended in 10 ml of TK buffer [10 mM Tris⋅HCl (pH 8.0)/40 mM KCl] supplemented with protease inhibitors (2 μM pepstatin/2 μM leupeptin/1 mM EDTA/0.6 mM phenylmethylsulfonyl fluoride). Cells were broken by two passes through a French press at 10,000 psi, and cell debris was removed by 15-min centrifugation at 12,000 × g. Membranes were pelleted from the supernatant by centrifugation at 150,000 × g for 1 h and resuspended in 0.9 ml of TK supplemented with 0.6 mM phenylmethylsulfonyl fluoride. To solubilize membrane proteins, sodium lauryl maltoside was added to a final concentration of 1% and the sample was shaken at 4°C for 45–60 min. The sample was then diluted 4× with TK and centrifuged at 150,000 × g for 1 h to remove insoluble material.
Solubilized membranes were incubated for 2 h at 4°C with Ni-NTA column matrix (Qiagen, Chatsworth, CA), which had been preequilibrated with TKLM buffer ([0.1% lauryl maltoside, 10 mM Tris⋅HCl, pH 8.0/40 mM KCl] (39). The resin was pelleted by centrifugation (6,000 × g), washed twice (5 min each) with TKLM buffer, then 4 times (5 min each) with TKLM buffer containing 20 mM imidazole. The bound material was then eluted in two 15-min washes with TKLM buffer containing 0.1 M EDTA. The two supernatants were combined and stored at −20°C until analysis.
Samples were boiled for 10 min in sample buffer, then DTT was added to 50 mM final concentration. Samples were incubated at 37°C for an additional 30 min to break dithiobis(succinimidylpropionate) crosslinks. MCP bands were resolved by SDS/PAGE, as described (40), and visualized by immunoblotting with an antiserum that reacts with the highly conserved signaling domains of both Tsr and Tar (21).
Results
Tsr Trimer Contact Sites and Mutants.
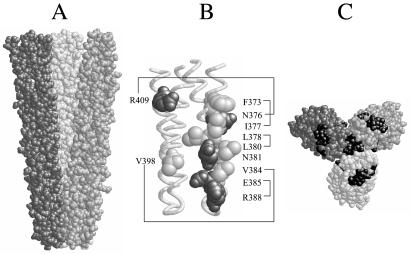
The trimer of dimers arrangement observed in a crystallized fragment of the Tsr signaling domain (26) is shown in Fig. 1A. Each dimer comprises a four-helix bundle of two coiled-coil subunits that make hairpin turns at the membrane-distal tip of the Tsr cytoplasmic domain (Fig. 1B). The dimers associate laterally through a contact region near their tips that involves surface residues from both subunits of each dimer (Fig. 1B). Six residues participate in pairwise hydrophobic interactions, four in pairwise polar interactions. One contact residue, N381, lies buried at the center of the contact region and forms a hydrogen-bonded network with its counterparts in the other two dimers. Although the two subunits in each dimer are structurally identical, they make different contributions to trimer stability. One subunit contains nine contact residues at the trimer interface, the other only two. Note also that the counterparts of the trimer contact residues in the opposing subunits of the dimers lie on the outside faces of the trimer, a very different structural environment (Fig. 1C).
Figure 1.
Structural features of the Tsr trimer of dimers. (A) Space-filling view of Tsr residues 350–430 in the trimer of dimers arrangement, corresponding to the cytoplasmic tip of the Tsr signaling domain. Each dimer is shaded differently to emphasize their convergence at the tip, where the contact sites are located. (B) α-Carbon backbone of the tip of one dimer, with the trimer contact residues shown in space-filling view. Hydrophobic residues are light gray, polar residues are dark gray. Lines indicate pairing interactions with contact residues in the other two dimers; N381 forms a hydrogen-bonded network with its two counterparts. (C) An end-on, space-filling view of the tip of the trimer of dimers. The trimer contact residues in all six monomeric subunits are shaded black to indicate the two different environments (at the trimer interface and on the exterior surface) in which they reside.
To explore possible signaling functions of the Tsr trimer contact residues, we created a set of proline, alanine, and tryptophan replacements at each of the contact sites. Because the target residues lie on the surface of the receptor dimer, none of the mutations seemed likely to perturb subunit packing interactions within the dimer. We expected that proline replacements would locally destabilize the α-helical secondary structure of the trimer contact region and abolish any function(s) it performs. In contrast, we expected that alanine replacements would preserve local secondary structure and simply eliminate a side chain interaction that contributes to trimer stability. Finally, we expected that tryptophan replacements would also preserve local secondary structure, but that their bulky side chain might interfere drastically with trimer binding contacts and packing interactions. The tryptophan lesions could cause substantial functional defects, if the trimer contacts play important roles in vivo. To facilitate discussion of the mutants, we will use the same shorthand notation to identify the mutant proteins and their mutational changes. For example, F373W denotes (a Tsr protein with) a tryptophan replacement at phenylalanine residue 373. Tsr* will refer to any such mutant protein; Tsr+ will indicate the wild-type protein.
Mutations were created in an IPTG-regulatable Tsr plasmid to permit control of mutant gene expression. The Tsr* plasmids were introduced into an E. coli host deleted for all five MCP genes to evaluate function of the mutant receptors in the absence of other chemoreceptors. We first examined their steady-state expression levels and ability to mediate serine chemotaxis, receptor clustering, and CW flagellar rotation. We then constructed partially diploid strains to investigate interactions between the Tsr* defects and wild-type Tsr or Tar receptors. The results of all tests are summarized in Table 1 and discussed below.
Table 1.
In vivo properties of Tsr trimer contact mutants
| Tsr residue | Proline mutants
|
Alanine mutants
|
Tryptophan mutants
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Expression level† | Serine chemotaxis‡ | Cluster formation§ | CW time¶ | Complementation‖ | Expression level† | Serine chemotaxis‡ | Cluster formation§ | CW time¶ | Complementation‖ | Expression level† | Serine chemotaxis‡ | Cluster formation§ | CW time¶ | Complementation‖ | |
| Hydrophobic | |||||||||||||||
| F373 | 0.7 | 0.1 | 0.2 | 0.0 | D2 | — | — | — | — | — | 0.9 | 0.2 | 0.7 | 0.1 | E2 |
| I377 | 1.1 | 0.1 | 0.0 | 0.0 | D2 | 1.1 | 0.2 | 0.6 | 0.15 | E2 | 1.4 | 0.1 | 0.7 | 0.0 | E2 |
| L378 | 1.4 | 0.1 | 0.0 | 0.0 | D1 | 2.4 | 0.1 | 0.7 | 0.0 | E3 | 0.9 | 0.15 | 0.6 | 0.1 | E3 |
| L380 | 1.2 | 0.1 | 0.2 | 0.0 | D1 | 1.2 | 0.3 | 0.7 | 0.2 | E1 | — | — | — | — | — |
| V384 | 0.7 | 0.1 | 0.1 | 0.0 | R | 0.9 | 0.25 | 0.5 | 0.05 | R* | 0.9 | 0.1 | 0.6 | 0.0 | E1 |
| V398 | 1.4 | 1.2 | 0.65 | 0.75 | WT | 1.3 | 0.65 | 0.85 | 0.2 | WT | 1.4 | 0.3 | 0.7 | 0.1 | E0 |
| Polar | |||||||||||||||
| N376 | 1.1 | 0.2 | 0.3 | 0.0 | D2 | 1.1 | 0.35 | 0.65 | 0.0 | D1* | 0.8 | 0.15 | 0.85 | 0.0 | E3 |
| R409 | 0.5 | 0.15 | 0.2 | 0.0 | D2 | 1.25 | 1.0 | 0.8 | 0.1 | WT | 0.85 | 0.1 | 0.45 | 0.0 | E1 |
| E385 | 0.45 | 0.2 | 0.0 | 0.0 | R | 1.3 | 0.15 | 0.85 | 0.1 | D2* | 0.7 | 0.2 | 0.6 | 0.25 | E3 |
| R388 | 0.55 | 0.1 | 0.0 | 0.0 | R | 1.0 | 0.2 | 0.65 | 0.1 | R* | 0.85 | 0.2 | 0.4 | 0.75 | E2 |
| N381 | 0.6 | 0.15 | 0.05 | 0.0 | E1 | 0.85 | 0.1 | 0.05 | 0.0 | E1 | 0.6 | 0.1 | 0.4 | 0.0 | E1 |
Data are rounded to the nearest 5% value. Mutants F373A and L380W were not made.
Steady-state level of mutant protein relative to level of wild-type Tsr. Proteins were measured by quantitative immunoblotting (see Materials and Methods).
Colony size on semisolid tryptone agar relative to the wild-type Tsr control.
Fraction of cells displaying one or more receptor patches detectable with a YFP-CheZ reporter. Under the conditions used (see Materials and Methods), 85% of wild-type control cells formed patches.
Calculated fraction of time that tethered cells spent in CW rotation. Under the conditions used (see Materials and Methods), wild-type control cells spent 25–30% of their time in CW rotation.
Complementation pattern on semisolid agar at 35°C: wild-type (WT); recessive (R); dominant (D); epistatic (E). The degree of dominance or epistasis was scored from low (0) to high (3) on the basis of the expression level (IPTG concentration) at which the effect was first observed (see Materials and Methods). The Tsr function of the recessive and dominant mutants marked with an asterisk was restored by wild-type Tar (see Results).
Functional Defects of Tsr Trimer Contact Mutants.
The steady-state intracellular levels of all mutant proteins proved similar to wild-type Tsr (Table 1), so function tests were done at the inducer concentration optimal for chemotaxis by Tsr+. On tryptone soft agar plates, all but three of the Tsr* strains were defective in serine chemotaxis, spreading at 35% or less of the wild-type rate (Table 1). The three mutants with chemotactic ability (V398P, V398A, and R409A) indicate that V398 and R409 play a less critical role in Tsr function than do the other contact residues. Moreover, the fact that a proline replacement is tolerated at V398 could mean that α-helicity in this region of Tsr is not critical for function (see Fig. 1B).
The ability of the mutant Tsr proteins to activate the CheA kinase, presumably through ternary complex formation, was evaluated by measuring the extent of CW flagellar rotation in tethered cells. In the absence of functional transducers, the uncoupled autophosphorylation activity of CheA is too low to produce CW rotation. When coupled to Tsr+, CheA activity is much higher and about 30% of the cell's time is spent in CW rotation (Fig. 2, “plus” symbol). Receptor clustering was evaluated with a fluorescent reporter protein (YFP-CheZ) that interacts with CheA (36). Tsr+ forms clusters that contain CheW and CheA, which in turn attract reporter molecules to the cluster, resulting in one or more bright spots in the cell (see example in Fig. 5). In contrast, some Tsr* strains lacked these spots (see example in Fig. 5), indicating that those mutant receptors either could not cluster or could not recruit CheW and CheA to the cluster.
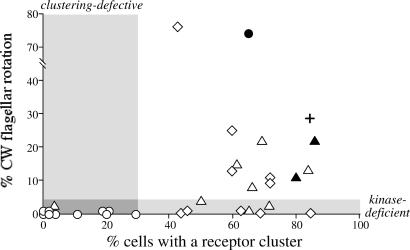
Figure 2.
Receptor clustering and flagellar rotation phenotypes of Tsr trimer contact mutants. These experimental data (in their unrounded form) correspond to the rounded values presented in Table 1. Proline replacement mutants (circles); alanine replacement mutants (triangles); tryptophan replacement mutants (diamonds). The plus indicates the wild-type control. Filled symbols: Tsr mutants with at least 65% of wild-type serine chemotaxis ability; open symbols: mutants with less than 35% of the wild-type ability. Shaded areas mark cutoff criteria for classifying Tsr* mutants as deficient in receptor clustering ability (less than 30% of cells with a cluster) and/or in activation of the CheA kinase (less than 5% of time spent in CW rotation).
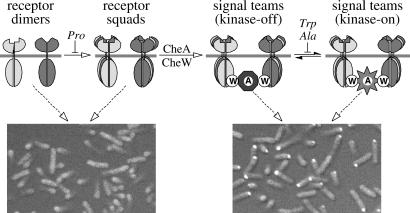
Figure 5.
Model of chemoreceptor cluster assembly and organization. Receptors of different detection specificities (indicated by shading differences) can associate, most likely through their common trimer contact residues, to form mixed receptor squads, which then bind CheA and CheW to form collaborative signaling teams. Component stoichiometries in the receptor complexes are not yet known, but squads and signal teams may be based on trimers of dimers. Proline mutations at the trimer contact sites of Tsr generally block squad formation, whereas alanine and tryptophan mutations allow assembly of signal teams that are locked in the kinase-on or kinase-off state. The connections between signaling teams that lead to macroscopic cluster formation are not explicitly shown. The light micrographs at the bottom of the figure show examples of the two extreme receptor-clustering phenotypes seen in this work. Mutant receptors unable to form signal teams, e.g., proline mutants, failed to assemble clusters detectable with a YFP-CheZ reporter (Left, Tsr-I377P). Mutant receptors able to assemble signal teams, regardless of their ability to respond to serine stimuli, e.g., alanine and tryptophan mutants, formed clusters (Right, Tsr-I377W).
Comparison of the CW rotation and receptor-clustering abilities of the mutant receptors revealed three principal classes of functional defects (Fig. 2). With only one exception (V398P), the proline replacement mutants exhibited few or no cells with receptor clusters and no CW rotation episodes. These properties are consistent with the expected null phenotype of the proline mutants. In contrast, all but one of the alanine and tryptophan mutants exhibited proficient receptor clustering (Table 1; Fig. 2). However, about half of them produced less than 5% CW rotation, indicating that the mutant receptors in this group make signaling complexes that are defective in activating CheA. The remainder of the alanine and tryptophan receptor mutants produced at least 8% CW rotation, but only two of them (V398A, R409A) mediated serine chemotaxis. The nonchemotactic mutants in this group evidently make signaling complexes that are able to activate CheA, but they seem to be locked in a stimulus-unresponsive state. These receptors either cannot detect serine stimuli or cannot down-regulate CheA activity in response to such stimuli.
Complementation Properties of Trimer Contact Mutants.
To gain further insight into the functional defects of Tsr trimer contact mutants, particularly those with “locked” signaling properties, we investigated their effects on signaling by wild-type Tsr and Tar. Partial diploids were constructed by introducing Tsr* plasmids into cells carrying a compatible plasmid with wild-type Tsr or Tar under control of an independently regulatable promoter. The expression of the wild-type receptor was set at its optimal level for chemotaxis and the relative amount of the mutant protein was varied by adjusting its inducer concentration in the soft agar assay plates.
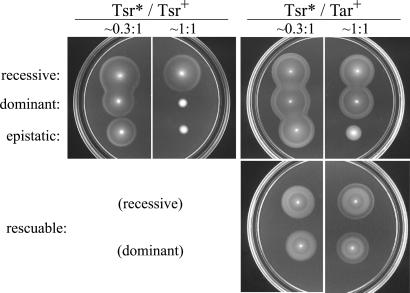
The Tsr* mutations defined four different complementation patterns (Fig. 3). Recessive mutations had no effect on Tsr+ or Tar+ function at any expression level. Dominant mutations impaired Tsr+ function, but had no effect on Tar+ function at any expression level. Epistatic mutations showed dominance over Tsr+, but also impaired Tar+ function. Mutations in the fourth group were either recessive or dominant to Tsr+, but regained ability to mediate serine chemotaxis in the presence of wild-type Tar, an effect we term “functional rescue” to distinguish it from conventional complementation between alleles of the same gene.
Figure 3.
Complementation behaviors of Tsr trimer contact mutants. Strains with a Tsr* plasmid and a compatible Tsr+ or Tar+ plasmid were tested for serine and aspartate chemotaxis on tryptone soft agar plates at different relative expression levels. Note the outer (serine) ring in colonies of “rescuable” mutants at 1:1 coexpression with wild-type Tar. Tsr* mutants shown in these examples: V384P (recessive); I377P (dominant); I377W (epistatic); R388A (recessive, rescuable); E385A (dominant, rescuable).
Most of the dominant mutants had proline replacements (Table 1), a behavior consistent with a null defect in a dimeric protein. Proline-containing Tsr* subunits would be expected to form dimers with Tsr+ subunits, because the Tsr periplasmic domain, whose structure is probably impervious to mutations at the tip of the cytoplasmic domain, should suffice for dimer formation. If Tsr*/Tsr+ dimers lack function, the mutant protein would exert dominance by “spoiling” wild-type subunits through mixed dimer formation.
All tryptophan replacement mutations were dominant to Tsr+ and epistatic to Tar+ (Table 1). Their dominance can be explained by subunit spoiling, but their epistasis most likely occurs by a different mechanism because Tar and Tsr are not known to form heterodimers (41). If they could, then the dominant proline mutations would most likely have been epistatic as well. Instead, epistasis must reflect a functional property that the dominant proline mutations lack. We propose that epistasis occurs through formation of mixed receptor complexes whose function is “spoiled” by the presence of a defective Tsr* dimer. According to this model, proline replacement mutations prevent complex formation and so are not epistatic. In contrast, tryptophan replacements permit complex formation but distort the geometry of the complex so that it cannot stimulate or control CheA. Any Tar dimers incorporated into those defective complexes would also not be able to signal properly. In the Discussion, we make the case that these receptor complexes are based on trimers of dimers, but any higher-order complex that contains receptors of different types would account for the epistatic properties of the tryptophan mutants.
The four Tsr* mutants that were functionally rescued by wild-type Tar had alanine mutations (Table 1). These mutant receptors were adept at forming clusters (Table 1), so we propose that functional rescue, like epistasis, occurs through formation of mixed, higher-order receptor signaling complexes that contain both Tsr* and Tar+ dimers. Unlike tryptophan, alanine introduced at these sites must not distort the overall geometry of the mixed complex, enabling its Tar members to function and to assist the Tsr* members in activating and modulating CheA in response to serine stimuli. Evidently, the Tsr* proteins with alanine replacements are more conformationally malleable than those with tryptophan replacements at the same sites. The alanine mutants respond productively when teamed with functional receptors, whereas the tryptophan mutants lock the signaling complex in an unresponsive state.
Chemical Crosslinking of Tsr to Tar.
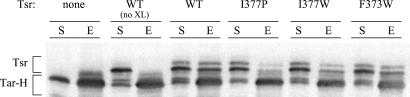
Our explanation for epistatic and rescuable Tsr* mutants predicts that wild-type Tsr and Tar molecules form close functional associations, possibly based on trimers of dimers, in vivo. If so, then it should be possible to crosslink Tsr and Tar molecules within cells. To accomplish this, we coexpressed Tsr and a (His)6-tagged, but normally functional, derivative of Tar in cells having no other MCPs. The cells were treated with a membrane-permeable crosslinker that reacts with lysine residues and has a 12-Å spacer with a disulfide linkage. Crosslinked membrane proteins were solubilized in detergent and passed over a Ni-NTA resin to enrich for molecules with (His)6 affinity tags. The eluted material was treated with reducing agent to break crosslinks, then analyzed by gel electrophoresis and immunoblotting with an MCP-specific antibody that detects Tsr and Tar molecules equally well. This experimental scheme revealed Tsr bands that copurified with Tar-(His)6 only in samples treated with crosslinking agent (Fig. 4). Two Tsr* proteins with epistatic defects (I377W and F373W) also crosslinked to Tar-(His)6, albeit at a somewhat reduced level (Fig. 4), consistent with our proposal that epistasis results from formation of mixed complexes of Tsr* and Tar dimers. In contrast, a Tsr* protein (I377P) that is dominant, but not epistatic, failed to crosslink to Tar-(His)6 (Fig. 4), implying that the trimer contact region is important for forming mixed receptor complexes and for the Tsr-Tar interaction detectable by crosslinking.
Figure 4.
In vivo crosslinking of Tsr to (His)6-tagged Tar (Tar-H). Tsr and Tar-H bands were visualized by immunoblotting with an MCP-specific antibody. Lanes labeled “S” show Tsr and Tar-H proteins in the solubilized membrane fraction of the cells; “E” lanes indicate solubilized membrane proteins eluted from the Ni-NTA resin used to purify the (His)6-tagged Tar. All samples were treated with reducing agent to cleave disulfide crosslinks before gel electrophoresis. Samples in the lanes labeled “no XL” were not treated with crosslinking agent.
These crosslinking experiments were conducted in a host strain lacking the MCP-modifying enzymes, CheB and CheR, to simplify the banding patterns and to augment mobility differences between Tar and Tsr molecules. We observed a similar extent of crosslinking in a strain also lacking CheA and CheW (data not shown). These results indicate that crosslinking does not depend on the CheA, CheB, CheR, or CheW functions and most likely arises through a direct unassisted interaction between Tsr and Tar molecules, presumably at their trimer contact regions.
Discussion
Genetic analysis of the trimer contacts in Tsr is complicated by the fact that the contact residues reside in two different structural environments in the trimer of dimers (Fig. 1C). Although we cannot be certain that the mutant effects we observed were due to perturbations of the trimer interface rather than functions of the contact residues in different contexts, the phenotypes of trimer contact mutants, combined with the demonstration of receptor interactions by chemical crosslinking, provide strong circumstantial evidence for an important signaling role of receptor trimers in vivo.
Amino acid replacements at the trimer contact sites almost invariably destroyed the ability of Tsr to mediate chemotactic responses. Receptors with proline replacements, ostensibly null mutations, were unable to form clusters or activate the CheA kinase. In contrast, all but one of the receptors with alanine or tryptophan replacements formed clusters but failed to modulate CheA activity in response to serine stimuli. Because our clustering assay required that the mutant receptors bind CheA and CheW, the signaling defects of the alanine and tryptophan mutants most likely arise from perturbation of dimer–dimer interactions within a higher-order receptor complex. Moreover, in the presence of wild-type Tar receptors, the alanine and tryptophan mutants produced epistatic or rescuable behaviors consistent with formation of mixed receptor complexes containing Tsr* and Tar dimers. Finally, our crosslinking experiments demonstrated that Tsr and Tar molecules form close associations in vivo that were affected by lesions at the trimer contact sites of Tsr, but not by the absence of the CheA, CheB, CheR, or CheW signaling proteins.
These results define several organizational units within chemoreceptor clusters (Fig. 5). We propose that after insertion into the cytoplasmic membrane and dimerization, chemoreceptor molecules coalesce into squads that can contain members with different detection specificities. Receptor squads in turn recruit CheA and CheW molecules to form signaling teams that modulate CheA activity in response to the stimuli detected by their various receptor members. We suggest that chemoreceptor squads and signaling teams are based on trimer of dimer interactions between receptor molecules, as envisioned in the clustering model proposed by Shimizu et al. (42). Proline replacements at the Tsr trimer contacts abrogate squad formation, whereas alanine and tryptophan replacements permit squad and team formation, but hinder the conformational changes needed to modulate team signaling activity in response to serine stimuli (Fig. 5). Because the trimer contacts are identical in all MCPs of E. coli, we predict that low-abundance receptors will be found to reside mainly in mixed signaling teams with high-abundance receptors, which might provide Tap, Trg, and Aer with functionally crucial signaling assistance from Tar and Tsr. Further in vivo crosslinking studies, by using the approaches and mutants described in this report, should serve to clarify the structures of receptor squads and signaling teams.
Receptor clusters probably represent a dynamic array of signaling teams with shared connections to CheA and CheW. How might receptor teams produce signal amplification in chemoreceptor clusters? If the receptors in a signaling team collaborate to activate a shared CheA molecule, as suggested by previous work with receptor signaling fragments (15, 21) and by the epistatic and rescuable phenotypes of Tsr trimer contact mutants, they should exhibit cooperative signaling similar to that demonstrated with receptor signaling complexes in vitro (23, 24). However, cooperative behavior within a signaling team may not account entirely for the detection sensitivity of chemoreceptor clusters. Signaling interactions between receptor teams might play an important role as well (11, 12). The mechanisms of communication within and between receptor signaling teams undoubtedly hold the key to understanding the high detection sensitivity of bacterial chemoreceptor networks.
Acknowledgments
We thank Bob Bourret (Univ. of North Carolina Medical School) for strain KO607, Phil Matsumura (Univ. of Illinois Medical School) for CheY antiserum, Victor Sourjik (Harvard Univ.) for plasmid pVS49, Ed King for assistance with fluorescence confocal microscopy, and Sung-Hou Kim for encouragement, atomic coordinates, and helpful information at the outset of this project. This work was supported by Research Grant 5-R37-GM19559 from the National Institutes of Health. The Protein–DNA Core Facility at the University of Utah receives support from National Cancer Institute Grant CA42014 to the Huntsman Cancer Institute.
Abbreviations
- MCP
methyl-accepting chemotaxis protein
- CW
clockwise
- IPTG
isopropyl-β-d-thiogalactopyranoside
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Falke J J, Hazelbauer G L. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borkovich K A, Kaplan N, Hess J F, Simon M I. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gegner J A, Graham D R, Roth A F, Dahlquist F W. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 4.Borkovich K A, Simon M I. Methods Enzymol. 1991;200:205–214. doi: 10.1016/0076-6879(91)00140-r. [DOI] [PubMed] [Google Scholar]
- 5.Ninfa E G, Stock A, Mowbray S, Stock J. J Biol Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 6.Segall J E, Block S M, Berg H C. Proc Natl Acad Sci USA. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jasuja R, Lin Y, Trentham D R, Khan S. Proc Natl Acad Sci USA. 1999;96:11346–11351. doi: 10.1073/pnas.96.20.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim C, Jackson M, Lux R, Khan S. J Mol Biol. 2001;307:119–135. doi: 10.1006/jmbi.2000.4389. [DOI] [PubMed] [Google Scholar]
- 9.Sourjik V, Berg H C. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maddock J R, Shapiro L. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 11.Bray D, Levin M, Morton-Firth C. Nature (London) 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 12.Duke T A, Bray D. Proc Natl Acad Sci USA. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duke T A, Novère N L, Bray D. J Mol Biol. 2001;308:541–553. doi: 10.1006/jmbi.2001.4610. [DOI] [PubMed] [Google Scholar]
- 14.Hazelbauer G L, Engstrom P, Harayama S. J Bacteriol. 1981;145:43–49. doi: 10.1128/jb.145.1.43-49.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Li G, Weis R. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- 16.Hazelbauer G L, Park C, Nowlin D M. Proc Natl Acad Sci USA. 1989;86:1448–1452. doi: 10.1073/pnas.86.5.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng X, Lilly A A, Hazelbauer G L. J Bacteriol. 1999;181:3164–3171. doi: 10.1128/jb.181.10.3164-3171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weerasuriya S, Schneider B M, Manson M D. J Bacteriol. 1998;180:914–920. doi: 10.1128/jb.180.4.914-920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gestwicki J E, Strong L E, Kiessling L L. Chem Biol. 2000;7:583–591. doi: 10.1016/s1074-5521(00)00002-8. [DOI] [PubMed] [Google Scholar]
- 20.Gestwicki J E, Kiessling L L. Nature (London) 2002;415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- 21.Ames P, Parkinson J S. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Levit M, Lurz R, Surette M G, Stock J B. EMBO J. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bornhorst J A, Falke J J. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G, Weis R M. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 25.Ames P, Yu Y A, Parkinson J S. Mol Microbiol. 1996;19:737–746. doi: 10.1046/j.1365-2958.1996.408930.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim K K, Yokota H, Kim S H. Nature (London) 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 27.Parkinson J S, Houts S E. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith R A, Parkinson J S. Proc Natl Acad Sci USA. 1980;77:5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J D, Parkinson J S. Proc Natl Acad Sci USA. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oosawa K, Mutoh N, Simon M I. J Bacteriol. 1988;170:2521–2526. doi: 10.1128/jb.170.6.2521-2526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolivar F, Rodriguez R, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 32.Bibikov S I, Biran R, Rudd K E, Parkinson J S. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J. Ph.D. thesis. Salt Lake City: Univ. of Utah; 1992. [Google Scholar]
- 34.Chang A C Y, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yen K M. J Bacteriol. 1991;173:5328–5335. doi: 10.1128/jb.173.17.5328-5335.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sourjik V, Berg H C. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 37.Parkinson J S. J Bacteriol. 1976;126:758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell D M, Gennis R B. FEBS Lett. 1995;368:148–150. doi: 10.1016/0014-5793(95)00626-k. [DOI] [PubMed] [Google Scholar]
- 40.Feng X, Baumgartner J W, Hazelbauer G L. J Bacteriol. 1997;179:6714–6720. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chelsky D, Dahlquist F W. Biochemistry. 1980;19:4633–4639. doi: 10.1021/bi00561a015. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu S T, Le Novère N, Daniel Levin M, Beavil A J, Sutton B J, Bray D. Nat Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]