Abstract
Many species of purple photosynthetic bacteria repress synthesis of their photosystem in the presence of molecular oxygen. The bacterium Rhodobacter capsulatus mediates this process by repressing expression of bacteriochlorophyll, carotenoid, and light-harvesting genes via the aerobic repressor, CrtJ. In this study, we demonstrate that CrtJ forms an intramolecular disulfide bond in vitro and in vivo when exposed to oxygen. Mutational and sulfhydryl-specific chemical modification studies indicate that formation of a disulfide bond is critical for CrtJ binding to its target promoters. Analysis of the redox states of aerobically and anaerobically grown cells indicates that they have similar redox states of approximately −200 mV, thereby demonstrating that a change in midpoint potential is not responsible for disulfide bond formation. In vivo and in vitro analyses indicate that disulfide bond formation in CrtJ is insensitive to the addition of hydrogen peroxide but is sensitive to molecular oxygen. These results suggest that disulfide bond formation in CrtJ may differ from the mechanism of disulfide bond formation used by OxyR.
Synthesis of the purple bacterial photosystem is regulated in response to redox state with reducing conditions stimulating photosystem synthesis over that of oxidizing growth conditions (1). Several overlapping regulatory circuits have been described that regulate transcription of photosynthesis genes in response to alterations in oxygen tension (2, 3). An aerobic repression circuit has been identified that uses the repressor CrtJ to aerobically suppress bacteriochlorophyll (bch), carotenoid (crt), and light harvesting-II (puc) gene expression (2–4). Biochemical studies have demonstrated that CrtJ binds to target promoters with a higher affinity when CrtJ is first preincubated with oxygen-saturated buffer (5). Interestingly, analyses for the presence of cofactors that might be involved in redox sensing show no evidence for the presence of metals, heme, or flavins associated with CrtJ (5). These results prompted us to consider that CrtJ might have the capability of sensing redox changes through the formation of a disulfide bond similar to that described for the hydrogen peroxide defense regulator, OxyR. Studies have indicated that exposure of OxyR to hydrogen peroxide catalyzes the formation of an intramolecular disulfide bond both in vitro and in vivo (6–8). It is thought that the formation of a disulfide bond causes a conformational change in OxyR that stimulates activation of genes involved in defense against reactive-oxygen species (9). In this study, we show that binding of CrtJ to target promoters depends on the formation of an intramolecular disulfide bond. We also show that disulfide bond formation in CrtJ appears to involve sensitivity to molecular oxygen, rather than sensitivity to hydrogen peroxide, as is the case of OxyR.
Materials and Methods
Overexpression and Purification of CrtJ.
An S-tagged CrtJ overexpression vector was constructed by polymerase chain amplifying (PCR) the CrtJ coding sequence by using a forward primer containing an NcoI restriction site (5′-CCCAT GGCGACGGGAGGCTTGCA-3′), and a reverse primer containing an XbaI restriction site (5′-CCTCTAGACGGTGC CTTTCCCGTTTCG-3′). The PCR-amplified DNA segment was cloned into pBluescript II SK(±), and subsequently subcloned into NcoI and XbaI sites in pET-29(a)(+) (Novagen) to generate the plasmid, pET29∷crtJ. pET29∷crtJ was transformed into Escherichia coli BL21(DE3) pLysS, and the protein was then overexpressed by induction of a 500-ml culture with 0.1 mM isopropyl-β-d-thiogalactopyranoside at 25°C for 4 h. Cells were harvested and resuspended in 30 ml of ice-cold TPAE buffer composed of 50 mM Tris⋅HCl (pH 8.0), 200 mM potassium acetate, and 1 mM EDTA, and lysed by three passes through a chilled French pressure cell operated at 18,000 psi (1 psi = 6.89 kPa). The lysate was clarified by centrifugation at 28,000 × g for 30 min at 4°C. The supernatant was then transferred to a fresh tube with contaminating proteins precipitated by gradual addition of ammonium sulfate over a 30-min period at 4°C to a final concentration of 1 M. After 30 min of gentle mixing, soluble and insoluble proteins were fractionated by centrifugation at 28,000 × g for 30 min. The supernatant was decanted to a fresh tube to which additional ammonium sulfate was added to bring the final ammonium sulfate concentration to 1.5 M. An enriched CrtJ protein precipitant was then collected by centrifugation at 28,000 × g for 30 min, followed by washing the pellet twice with 1.5 M ammonium sulfate. The protein pellet was resuspended in TPAE buffer containing 10 mM DTT, from which the undissolved proteins were removed by centrifugation. The soluble supernatant fraction was further clarified by filtration through a 0.45 μm Acrodisc (Gelman). The protein preparation was size separated on a FPLC Superose 12 size-filtration column (Pharmacia), equilibrated with TPAE buffer at a flow rate of 1 ml/min, with CrtJ containing fractions collected and dialyzed overnight at 4°C against TPAE buffer containing 20% glycerol. Isolated CrtJ, which was greater than 98% pure as based on Coomassie blue staining of an SDS polyacrylamide gel, was stored at −80°C. For some experiments, a His-tagged variant of CrtJ was also used that was isolated as described by Ponnampalam et al. (4).
Isulfhydryl Alkylation and Thiol Trapping with AMS.
Iodoacetamide alkylation of thiol groups was performed with purified CrtJ as described (10). Before alkylation, CrtJ was first treated with either oxygen (air) or 20 mM β-mercaptoethanol at room temperature for 30 min, and then treated with iodoacetamide as described in ref (10). The DNA-binding activity of alkylated CrtJ was then assayed by gel mobility shift assay using a 32P-labled DNA probe containing the bchC promoter region what was prepared by PCR amplification as described by Ponnampalam et al. (4).
For in vitro AMS modification of free thiol groups, purified CrtJ protein was first treated with oxygen, or freshly prepared hydrogen peroxide (0.001 mM, 0.01 mM, 0.1 mM, and 1.0 mM) or 20 mM β-mercaptoethanol at room temperature for varying times (10–30 min). The protein was then precipitated by the addition of trichloroacetic acid to 10% to fix its redox state and to remove excess hydrogen peroxide or β-mercaptoethanol. The precipitants were then dissolved in 50 mM Tris⋅HCl (pH 8.0) buffer containing 0.1% SDS and 15 mM 4-acetamido-4′-maleimidylstibene-2′,2′-disulfonic acid (AMS) for 1 h. The protein was then separated by nonreducing SDS/PAGE, with CrtJ detected by Western blot analysis using AP monoclonal antibodies (LumiBlot kit; Novagen).
For in vivo analysis of disulfide bonds, a strain of Rhodobacter capsulatus was used (ES7 CrtJ-flag) that expressed a chromosomally encoded S-Tag variant of CrtJ (11). Spectral and gel mobility shift data indicated that addition of an S tag at the amino terminus does not affect repressing properties of Crt J (data not shown). The ES7 CrtJ-flag construct was grown in RCV+ (12) media with gentamycin added to a final concentration of 5 μg/ml. Cells were grown aerobically in a partially filled Erlenmeyer flask that was shaking at 300 rpm or anaerobically in the dark in a completely filled screw capped test tube to a density of 65 Klett units (≈1.2 × 106 cells per ml). Aerobically grown cells were treated with 1/10 volume of 100% tricloroacetic acid (TCA) and incubated on ice for 20 min. Anaerobically grown screw-capped cultures were transferred to an anaerobic chamber containing an atmosphere of 85% N2/10% CO2/5% H2 and then divided into aliquots, with a portion of the cells kept under strictly anaerobic (reducing) conditions and another portion of the cells anaerobically treated with varying concentrations (0.001 mM, 0.01 mM, 0.1 mM, and 1.0 mM) of freshly prepared hydrogen peroxide (H2O2). After various incubation times (2–20 min), the cells were treated with 1/10 volume of 100% TCA and then incubated on ice for 20 min. In one set of assays, anaerobically grown cell samples were transferred to an Erlenmeyer flask and bubbled with O2 (air) while shaking at 400 rpm for 10 or 30 min. After exposure to O2 the cells were with 1/10 volume of 100% TCA and incubated on ice for 20 min.
The TCA treated samples were then AMS modified as follows. Precipitated proteins were collected by centrifugation for 10 min at 6,000 × g. After complete removal of the supernatant the pellet was washed with an equal volume of 500 mM Tris⋅HCl (pH 7.9) and centrifuged as before, washed with double distilled H2O (ddH2O) to remove excess salt and centrifuged again. Pellet was ultimately resuspended in 25 μl of 50 mM Tris⋅HCl (pH 7.9)/0.1% SDS/15 mM AMS (with the exception of the non-AMS-modified sample which was resuspended in the same buffer without AMS). AMS modifications were performed at 34°C for a minimum of 2 h. Alkylated samples were loaded on SDS/7.5% PAGE gels (Bio-Rad).
After electrophoresis, proteins were blotted to nitrocellulose filters and probed with anti-flag M2 monoclonal antibody (Sigma) according to product instructions. The bound antibodies were detected by using the enhanced chemiluminescence of SuperSignal West Dura substrate (Pierce) according to the manufacturer's instructions.
Redox Titration.
A measurement of relative percent disulfide bond formation at various redox potentials was measured by assaying fluorescence emission of CrtJ when incubated in the presence of monobromobimane (mBBr). Previous studies (13–15) have indicted that mBBr fluorescence increases significantly when it forms a covalent linkage with a Cys-sulfhydral. Thus, the level of mBBr fluorescence provides a good measurement of free sulfhydrals versus disulfides that are present in protein preparations that have been equilibrated in buffers containing different redox potentials. For this assay, a series of reactions were set up under oxidizing (air) or reducing (argon) conditions at different redox levels by varying ratios of oxidized (GSSG) and reduced (GSH) glutathione at a total glutathione concentration of 5 mM in 100 mM Hepes (pH 7.0 or 8.0) containing 100 μg of CrtJ. Eh values for different GSH/GSSG ratios at pH 7.0 were calculated from the Nernst Equation, by using the ratio of oxidized to reduced glutathione and literature values for the midpoint potential (Em) of glutathione at pH 7.0 as described (15). Eh values at pH 8.0 were calculated by taking into account the pKa of the thiol group of glutathione, as described (15). After incubation for 2 h to allow CrtJ to reach equilibrium at the desired redox potential, mBBr (Calbiochem) was then added to a final concentration of 300 mM and allowed to react with the Cys-sulfhydryls for 20 min. The protein was then precipitated by addition of 100 μl of 20% TCA and then incubated on ice for 30 min. The protein was pelleted by centrifugation, washed once with 1% TCA, and then resuspended in 100 mM Tris⋅HCl, pH 8.0/1% SDS. The fluorescence level of the CrtJ–mBBr preparation was measured using an Aminco–Bowman Series 2 Luminescence spectrometer with excitation at 380 nm and emission at 480 nm. The data were plotted and fit to a two-electron Nernst curve by using sigma plot software with maximum fluorescence set at the most reduced sample. Em values obtained were independent of time, over times ranging from 0.5 to 2.0 h and independent of total glutathione concentration, over a range from 2.5 to 10.0 mM, as would be expected if true redox equilibrium has been attained at all Eh values (13–15).
Site-Directed Mutagenesis.
A chromosomal deletion of crtJ was constructed to facilitate insertion of individual Cys to Ala mutations into the crtJ allele. Disruption of the chromosomal copy of crtJ was performed by gene-transfer-agent (GTA)-mediated recombination of a plasmid-encoded crtJ∷km construct into the chromosome. The generated crtJ-deleted strain, CD2–4, contains a deletion that extends from 46 bp upstream of CrtJ to 64 bp downstream of CrtJ. Individual Cys to Ala mutations were constructed in CrtJ by using the two-step PCR method (16) using a set of overlapping internal primers that incorporated the appropriate Cys to Ala codon changes. The PCR-generated fragments were cloned into the suicide vector, pZJD3, checked by sequence analysis, and integrated into the chromosome of strain CD2–4 by conjugation. A second recombination event was then identified by screening for loss of the gentamycin-resistance marker, leading to the generation of C22A, C249A, and C420A mutant strains. Proper allelic replacement was also analyzed by PCR and Western blot analysis. Mutant derivatives of the CrtJ overexpression vectors were also constructed by replacement of appropriate restriction sites.
Results
CrtJ Forms an Intramolecular Disulfide Bond Under Aerobic Conditions.
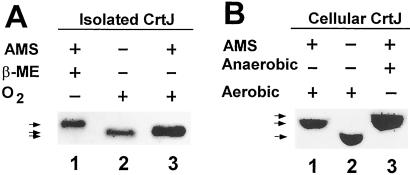
To test whether CrtJ forms a disulfide bond, we undertook several in vivo and in vitro assays for the presence or absence of disulfide bonds when CrtJ is exposed to oxidizing or reducing conditions, respectively. The formation of an intermolecular disulfide bond was ruled out by observing no stable dimers or higher oligomers in oxidized CrtJ preparations as assayed by SDS/PAGE under nonreducing conditions (data not shown). However, in vitro thiol trapping experiments did show evidence for an intramolecular disulfide bond in oxidized CrtJ. In this analysis, Cys sulfhydryl groups were covalently modified with AMS, which retards electrophoretic mobility. Because disulfide bonds are resistant to AMS modification, the presence of an intramolecular disulfide bond in oxidized CrtJ would result in fewer Cys sulfhydryl groups available to react with AMS than would be present in reduced CrtJ. Consequently, if there was an intramolecular disulfide bond forming in CrtJ, then oxidized CrtJ would migrate faster during SDS/PAGE than would reduced CrtJ after AMS modification. As shown in Fig. 1A, AMS treatment of reduced (β-mercaptoethanol exposed) CrtJ resulted in significantly slower electrophoretic migration (lane 1) than is observed with a CrtJ preparation that was exposed to oxygen saturated buffer before treatment with AMS (lane 3). AMS treated oxidized CrtJ also migrates slightly slower than untreated CrtJ (lane 2). Because CrtJ contains three Cys residues, we interpret these results as evidence that oxidized CrtJ may only have one Cys that is available for AMS modification, whereas reduced CrtJ would have three Cys available for AMS modification.
Figure 1.
Examination of disulfide bond formation in CrtJ by AMS modification. (A) In lanes 1 and 3, purified CrtJ was either reduced by exposure to β-mercaptoethanol (β-ME) or oxidized by exposure to oxygen for 20 min before AMS treatment, respectively. Lane 2 is untreated CrtJ. (B) Lanes 1 and 3 are TCA extracts from aerobically and anaerobically grown cells, respectively, that were modified by AMS under anaerobic or aerobic conditions. Lane 2 is untreated extracts from aerobically grown cells. The mobility of CrtJ was assayed by Western blot analysis. Note that the gel system was modified slightly between A and B, which increases separation of unmodified CrtJ.
We also undertook in vivo analysis of disulfide bond formation in CrtJ. For this analysis, we first grew R. capsulatus cells under anaerobic (photosynthetic) or aerobic (heterotrophic) growth conditions. At mid-log phase, the aerobically grown cells were treated with 10% TCA to denature the cellular proteins which effectively “fixes ” CrtJ sulfhydryls into their oxidized or reduced states. In parallel, a mid-log phase anaerobic culture was placed in an anaerobic chamber containing no oxygen (85% N2/5% H2/10% CO2) and similarly treated with TCA. Precipitated denatured proteins were then neutralized with a buffer, washed, treated with AMS, and then size fractionated by SDS/PAGE and then analyzed for CrtJ migration by Western blot analysis.
As shown by the results in Fig. 1B, AMS treated anaerobically grown cell extracts (lane 3) exhibited significantly slower migration of CrtJ than did AMS treated aerobic extracts (lane 1). Indeed, the migration patterns of CrtJ in the AMS treated aerobic and anaerobic cell extracts show electrophoretic patterns identical to those observed with purified preparations of AMS treated oxidized and reduced CrtJ (Fig. 1A). These thiol-trapping experiments clearly demonstrate that CrtJ forms an intramolecular disulfide bond and that the disulfide bond is present in aerobically but not anaerobically grown R. capsulatus cells.
Disulfide Bond Formation Occurs in Response to Molecular Oxygen.
Another issue is the mechanism of disulfide bond formation in CrtJ. In wild-type E. coli, there is little change in the redox-state of the cytoplasm when cells are shifted from aerobic to anaerobic growth conditions. Measurements of oxidation and reduction states of glutathione indicates that the E. coli cellular redox potential remains between −260 and −280 mV in aerobic and in anaerobic cells (6, 7). Consequently, most cytoplasmic proteins remain in a reduced state, even when wild-type E. coli is grown aerobically. This finding has raised the question regarding how a disulfide bond forms in the transcription factor OxyR, which is known to have a midpoint potential for formation of its intramolecular disulfide bond of −185 mV (7). It has been shown that the disulfide bond formation in OxyR is stimulated by trace amounts of hydrogen peroxide, even under reducing conditions (7). Indeed, E. coli mutations that increase hydrogen peroxide levels just 2-fold over those found in wild-type cells cause constitutive in vivo disulfide bond formation in OxyR (7). Because oxidized OxyR is responsible for activating expression of reactive-oxygen defense genes, it appears that it has evolved a mechanism to form a disulfide bond by direct interaction with hydrogen peroxide.
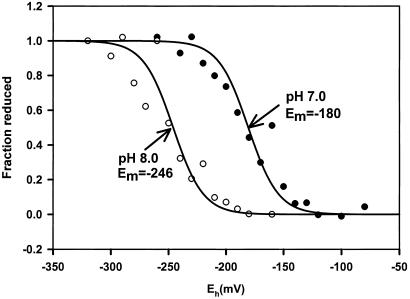
To address the issue of disulfide bond formation in CrtJ, we first undertook an analysis of the redox-state of aerobically and anaerobically grown R. capsulatus cells. For this analysis, the cytosolic redox potential was estimated by measuring the ratio of glutathione (GSH) and glutathione disulfide (GSSG) by using the glutathione reductase recycling method (17, 18). The results of this assay indicate that anaerobically grown cells have a potential of −224 mV, and that aerobically grown cells have a potential of −222 mV. We can conclude, therefore, that R. capsulatus cells grown under either condition both have ambient potentials of approximately −220 mV, which is just slightly less reducing then that reported for E. coli (6, 7). We next measured the midpoint potential of disulfide bond formation of CrtJ in vitro by assaying the amount of fluorescence emission from the covalent adduct formed by the reaction of CrtJ with mBBr. Redox titrations using mBBr fluorescence to monitor the level of disulfide reduction have proven to be a reliable method for measuring midpoint potential (Em) values (13–15). The data shown in Fig. 2 gives a good fit to an n = 2 Nernst equation for a single component with a Em midpoint of −180 mV at pH 7.0. These results are precisely as expected for a two-electron event such as that which would be occurring for a reversible cleavage/formation of a single disulfide bond. The −180 Em value measured for CrtJ at pH 7.0, is very similar to that reported for OxyR (7). A similar redox titration performed at pH 8.0 also gives a similar curve with a good fit to an n = 2 Nernst equation (Fig. 2). The Em at this pH was a more reducing −226 mV. This pH alteration of CrtJ redox potential is predicted for a reaction that involves an uptake of two protons per disulfide bond reduction (13–15).
Figure 2.
Redox titration of disulfide bond formation in CrtJ. The presence of reactive thiols was measured by analysis of the fluorescence emission level of CrtJ after incubation with mBBr at various redox values under anaerobic (argon) conditions. The filled circles represent the fluorescence emission at pH 8.0 and the open circles is at pH 7.0. The amplitudes are on a scale where the amplitude measured at the most negative redox value is a value of 1.0. The line represents a fit of the data to a two-electron Nernst curve. Eh (mV) denotes actual potentials.
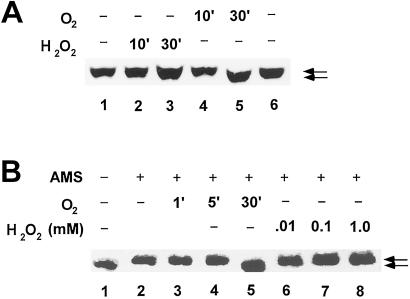
We also addressed whether reduced CrtJ that is present in anaerobically grown cells are susceptible to oxidation by the addition of low concentrations of hydrogen peroxide to anaerobic cultures, as is the case for OxyR. In the case of OxyR, it has been observed that addition of as little as 5 μM of H2O2 to E. coli cultures will lead to about 50% conversion of reduced OxyR to oxidized OxyR with addition of 10 μM leading to full conversion to an oxidized state (7). For R. capsulatus, we observed that addition of a wide range of hydrogen peroxide concentrations ranging from 0.001 to 1.0 mM to anaerobically grown cells did not lead to detectable in vivo formation of a disulfide bond in CrtJ as measured by AMS modification analysis. An example of the lack of detectable oxidation is shown in Fig. 3A, lanes 2 and 3, where incubation of a high level of H2O2 (1 mM) shows no evidence of disulfide bond formation. In contrast, we did observe that addition of oxygen to anaerobically grown cells did result in conversion of CrtJ from a reduced to a oxidized state after a 30-min exposure (Fig. 3A, lane 5).
Figure 3.
Effect of oxygen on disulfide bond formation in CrtJ in vivo and in vitro. (A) In vivo analysis of CrtJ disulfide bond formation after exposure to oxygen. Lanes 1 and 6 depict the SDS/PAGE migration of CrtJ from anaerobically grown cells that were treated with AMS. Lanes 2 and 3 are of anaerobically grown cells that were exposed to 1.0 mM H2O2 for 10 and 30 min, respectively, before treatment with AMS. Lanes 4 and 5 are of anaerobically grown cells that were exposed to O2 for 10 or 30 min, respectively, before treatment with AMS. (B) In vitro analysis of CrtJ disulfide bond formation after exposure to oxygen. Lanes 3, 4, and 5 show effect of exposure of CrtJ to oxygen for 1, 5, and 10 min before treatment with AMS, respectively. Lanes 6–8 show the effect of a 5 min treatment of CrtJ to 0.01, 0.1, and 1.0 mM H2O2 before treatment with AMS, respectively. Lane 1 is a control that shows the migration of CrtJ that has not been exposed to AMS, whereas lane 2 is a control that shows the migration of reduced CrtJ that has been exposed to AMS. For optimal separation between oxidized and reduced CrtJ, the isolated protein used for this analysis contained a Cys → Ala mutation at position 22, which is a residue that does not undergo disulfide bond formation.
We also addressed whether disulfide bond formation in isolated CrtJ was stimulated in vitro by exposure to oxygen and/or H2O2. For this analysis, we exposed reduced CrtJ to oxygen (air) for varying lengths of time, precipitated the protein with TCA, and then assayed CrtJ for AMS modification. The results of this analysis indicate that exposure of CrtJ to air for ≈30 min (Fig. 3B, lane 5) results in oxidation of CrtJ. In contrast, no oxidation was observed with exposure of CrtJ to H2O2 at various concentrations ranging from 10 μM to 1 mM (Fig. 3B, lanes 6–8). Indeed, no oxidation of CrtJ occurred during variation of both H2O2 concentration (10 μM to 1 mM) as well as time of exposure (2–30 min). These results indicate that disulfide bond formation in CrtJ differs from that of OxyR in that it appears to response to an increase in molecular oxygen rather than to the presence of an reactive-oxygen species such as H2O2.
Disulfide Bond Formation Is Required for CrtJ Binding to the bchC Promoter.
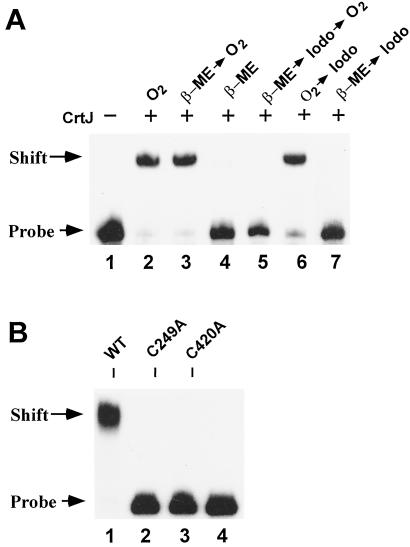
We next addressed whether formation of an intramolecular disulfide bond affects DNA-binding properties of CrtJ. For this analysis, we performed gel mobility shift assays with a DNA segment of the bchC promoter region that contains two CrtJ DNA-binding palindromes (5). Lanes 2–4 in Fig. 4A are control reactions that show the reversible effect of redox on the DNA-binding activity of CrtJ. Specifically, lane 2 shows that exposure of CrtJ to atmospheric oxygen results in the ability of CrtJ to form a stable bchC–CrtJ complex. In contrast, lane 4 shows that reduction of similar amounts of CrtJ with 20 mM β-mercaptoethanol inhibits the formation of a stable DNA–CrtJ complex. Lane 3 demonstrates that the inhibitory effect of β-mercaptoethanol is reversible, because DNA-binding activity is restored when the β-mercaptoethanol-treated CrtJ is subsequently exposed to pure oxygen for 20 min before addition of the DNA probe.
Figure 4.
Inhibition of reversible redox responsive DNA-binding of CrtJ by iodoacetamide and site-directed mutagenesis. (A) 32P-labeled bchC promoter probe was incubated with purified wild-type CrtJ under various reaction conditions and then size fractionated by gel electrophoresis. Lane 1, DNA probe only. Lane 2, oxygen oxidized CrtJ incubated with the DNA probe. Lane 3, CrtJ incubated with 20 mM β-mercaptoethanol (β-ME) for 20 min and then oxidized by bubbling with pure oxygen for 20 min before addition of the DNA probe. Lane 4, CrtJ incubated with β-mercaptoethanol for 20 min before adding the DNA probe. Lane 5, CrtJ sequentially treated for 20 min with 20 mM β-mercaptoethanol, 50 mM iodoacetamide (Iodo), and then pure oxygen. Lane 6, oxygen-treated CrtJ was incubated with 50 mM iodoacetamide for 20 min followed by addition of the DNA probe. Lane 7, CrtJ sequentially incubated for 20 min with β-mercaptoethanol followed by 50 mM iodoacetamide. Lanes 2–7 each contained 9 pmol of CrtJ in the DNA-binding reactions. (B) Loss of in vitro DNA-binding activity of Cys mutants. Lane 1 is wild type CrtJ incubated with the bchC DNA probe, lane 2 is C249A mutant CrtJ, lane 3 is C420A mutant CrtJ, and lane 4 is only the bchC promoter probe. Each of the protein preparations were treated with oxygen for 20 min before addition of the DNA probe.
The involvement of Cys in affecting DNA-binding was also addressed by assaying whether alkylation of CrtJ Cys sulfhydryls affects DNA-binding activity. Specifically, Fig. 4A, lane 5, is a DNA-binding assay in which CrtJ was first reduced by exposure to 20 mM β-mercaptoethanol, alkylated with iodoacetamide, and finally oxidized by exposure to pure oxygen. Unlike lane 3, where reduced CrtJ is capable of being reactivated by exposure to oxygen, the reduced and alkylated CrtJ can no longer be activated. Importantly, DNA-binding activity is not affected when CrtJ is first oxidized and then treated with iodoacetamide (lane 6). Because iodoacetamide does not react with disulfide bonds, these results indicate that oxidized CrtJ is capable of forming a disulfide bond that subsequently promotes DNA binding.
Mutational Analysis of Cys Residues in CrtJ.
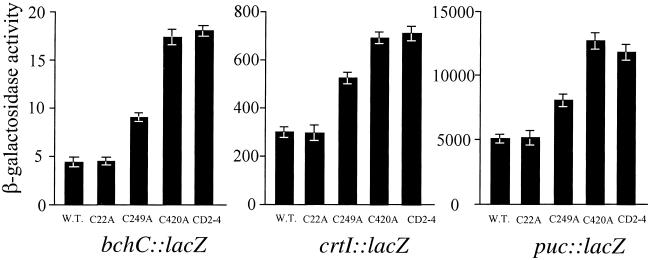
We also undertook mutational analysis of Cys residues in CrtJ to determine which Cys residues form an intramolecular disulfide bond, as well as to address the functional importance of forming a disulfide bond in vivo. Sequence analysis indicates that R. capsulatus CrtJ contains three Cys residues located at amino acids 22, 249, and 420. Cys-249 and -420 were identified as attractive targets for mutational analysis because they are also conserved in a CrtJ homolog from Rhodobacter sphaeroides (19). Furthermore, Cys-420 is also very near the C-terminal helix-turn-helix DNA-binding motif located at amino acid residues 426–463 (19). For our in vivo studies, we constructed individual Cys to Ala mutants at all three locations and recombined the mutations into the chromosomal copy of crtJ. Analysis of expression patterns of CrtJ repressed genes using β-galactosidase-based reporter plasmids indicates that the Cys to Ala mutation at position 22 (strain C22A) had no effect on CrtJ-mediated aerobic repression of the pucB, bchC, and crtI promoters, as evidenced by aerobic expression levels that are the same as that observed with wild-type R. capsulatus (strain SB1003) (Fig. 5). In contrast, a Cys to Ala mutation at codon 420 (strain C420A) elevated expression of the reporter plasmids to a level that was the same as observed with the crtJ deleted strain CD2–4. This finding indicates that the Cys to Ala mutation at this position abolished CrtJ in vivo repressing activity (Fig. 5). The Cys to Ala mutation at codon 249 (strain C249A) also elevates aerobic expression of the reporter plasmids, but not nearly to the same extent as does the C420A mutation. A similar pattern is observed with OxyR, in which a mutation in one of the disulfide bond forming Cys (Cys-199) exhibits a much more severe in vivo phenotype than does a mutation in the second disulfide bond forming Cys (Cys-208) (9). Western blot analysis also indicates that there are comparable levels of CrtJ in wild type and in the C420A and C249A mutant strains, indicating that the C420A and C249A mutations are not simply affecting protein stability (data not shown).
Figure 5.
Measurement of aerobic expression of the pucB, crtI, and bchC promoters in the wild-type strain SB1003, the crtJ deletion strain CD2–4, and in the C22A, C249A, and C420A crtJ mutant strains. Reporter plasmids and β-galactosidase assays were as described in ref. 3. β-galactosidase activity refers to the amount of O-nitrophenyl-β-d-galactoside hydrolyzed per minute per milligram of protein. β-galactosidase activity is the average of three independent assays.
Mutating Cys-249 and Cys-420 to Ala Abolishes CrtJ DNA Binding.
To confirm the in vivo expression analysis of photosynthesis genes, we also conducted in vitro DNA-binding studies with purified C249A and C420A mutant CrtJ proteins. As shown in the gel mobility shift assays in Fig. 4B, oxidized C249A and C420A mutant CrtJ protein preparations both exhibit defects in binding to the bchC promoter probe, relative to that observed with oxidized wild-type CrtJ. Furthermore, acetylation of the purified mutant proteins with AMS showed no evidence of altered electrophoretic mobility with protein that was pre-exposed to β-mercaptoethanol or to oxygen (data not shown). Together with the in vivo mutational data, these results implicate the conserved Cys-249 and Cys-420 residues as being involved in the formation of an intramolecular disulfide bond.
Discussion
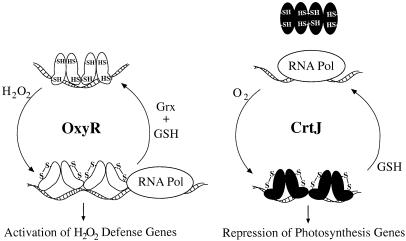
Previous studies demonstrated that CrtJ is a redox-responding repressor of photosystem synthesis in R. capsulatus (4, 5). In vitro DNA-binding assays indicated that oxidized CrtJ bound to target promoters at a much higher affinity than did reduced CrtJ (5). The in vitro studies correlate with in vivo genetic studies, which indicates that CrtJ functions as an aerobic repressor of bacteriochlorophyll biosynthesis (4). Visible spectral, electron paramagnetic resonance and inductively coupled plasma-mass spectrophotometer analyses of purified CrtJ have ruled out the presence of a redox-responsive metal center that could be involved in the DNA-binding activity of CrtJ (unpublished data). This finding lead us to investigate whether oxidized CrtJ forms a disulfide bond in response to oxidizing conditions. The in vivo and in vitro results of this study demonstrate that CrtJ does indeed reversibly form an intramolecular disulfide bond, and that formation of this bond is required for DNA-binding activity of CrtJ (Fig. 6).
Figure 6.
A comparative depiction of CrtJ and OxyR oxidation and reduction cycles that affect DNA-binding and subsequent repression or activation of target genes. GSH indicates reduced glutathione, which, we have demonstrated, is capable of reducing CrtJ disulfide bonds in vitro.
In E. coli the antioxidant defense regulator, OxyR, forms a disulfide bond in response to slight increases (2-fold) in hydrogen peroxide in vitro and in vivo (6, 7, 20). Surprisingly, hydrogen peroxide stimulates disulfide bond formation in OxyR even though the midpoint potential of disulfide bond formation is −185 mV and the midpoint potential of the E. coli cytosol is −260 to −280 mV both aerobically and anaerobically (6, 7). Our observation that aerobically and anaerobically grown R. capsulatus cells also retain similar cytosolic redox states (−222 and −224 mV, respectively), indicates that aerobic formation of a disulfide bond in CrtJ is also not stimulated by a change in the cytosolic redox poise. In fact, our Em value at pH 7 of −180 mV indicates that if CrtJ is in redox equilibrium with the cytosolic Eh, then CrtJ disulfide should be 95% reduced under both aerobic or anaerobic conditions. Thus, it appears that disulfide bond formation in CrtJ may be directly stimulated by the presence of molecular oxygen.
Different sensitivities of OxyR and CrtJ to oxygen and hydrogen peroxide is not surprising given the different functions of the regulators. OxyR, which is a member of the well characterized LysR family of prokaryotic transcription factors, has a role in controlling induction of reactive-oxygen defense genes such as catalase and superoxide dismutase in response to the presence of submicromolar amounts of H2O2 (20). In contrast, CrtJ, which has no identifiable homology to OxyR or to any other proteins in the GenBank database, has a role in repressing photosynthesis gene expression in response to the presence of oxygen. Furthermore, the fact that CrtJ and OxyR share no sequence similarity, and react in unique ways to these different oxygen species, indicates that CrtJ and OxyR may have independently evolved mechanisms formation of a disulfide bond.
Even though CrtJ and OxyR both regulate gene expression in response to the formation of a disulfide bond, there are some interesting differences regarding how these proteins interact with DNA. The ability of CrtJ to bind DNA, and subsequently repress transcription, is directly stimulated by the formation of a disulfide bond. This is in contrast to OxyR, which binds to target DNA under both oxidizing and reducing conditions and, according to DNase I protection patterns, simply undergoes a conformational change (21). This finding indicates that only oxidized OxyR is capable of interacting with RNA polymerase in a manner that stimulates transcription. Thus, the oxidation and reduction of disulfide bonds in CrtJ and OxyR appear to have very different effects on the DNA-binding capabilities of these proteins.
Acknowledgments
We thank Zeyu Jiang and Lisa Ragatz for critical discussions and comments during the preparation of the manuscript. This work was supported by National Institutes of Health Grant GM53940 (to C.E.B.), Robert A. Welch Foundation Grant D-0710 (to D.B.K.), and a postdoctoral fellowship from the Japan Society for the Promotion of Science (to S.M.).
Abbreviations
- AMS
4-acetamido-4′-maleimidylstibene-2′,2′-disulfonic acid
- mBBr
monobromobimane
- TCA
tricloroacetic acid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cohen-Bazire G, Sistrom W R, Stanier R Y. J Cell Comp Physiol. 1957;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 2.Bauer C E, Bird T H. Cell. 1996;85:5–8. doi: 10.1016/s0092-8674(00)81074-0. [DOI] [PubMed] [Google Scholar]
- 3.Bauer C E. In: Regulation of Photosynthesis. Aero E-M, Andersson B R, editors. Dordrecht, The Netherlands: Kluwer Academic; 2001. pp. 67–83. [Google Scholar]
- 4.Ponnampalam S N, Buggy J J, Bauer C E. J Bacteriol. 1995;177:2990–2997. doi: 10.1128/jb.177.11.2990-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponnampalam S N, Bauer C E. J Biol Chem. 1998;272:18391–18396. doi: 10.1074/jbc.272.29.18391. [DOI] [PubMed] [Google Scholar]
- 6.Zheng M, Åslund F, Storz G. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 7.Åslund F, Zheng M, Beckwith J, Storz G. Proc Natl Acad Sci USA. 1999;96:6161–1665. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, Ryu S. Cell. 2001;105:103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 9.Kullik I, Toledano M B, Tartaglia L A, Storz G. J Bacteriol. 1995;177:1275–1284. doi: 10.1128/jb.177.5.1275-1284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K. Proc Natl Acad Sci USA. 1997;94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong C, Elsen S, Swem L R, Bauer C E. J Bacteriol. 2002;184:2805–2814. doi: 10.1128/JB.184.10.2805-2814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yen H-C, Marrs B. Arch Biochem Biophys. 1977;181:411–418. doi: 10.1016/0003-9861(77)90246-6. [DOI] [PubMed] [Google Scholar]
- 13.Krimm I, Lemaire S, Ruelland E, Miginiac-Maslow M, Jacquot J-P, Hirasawa M, Knaff D B, Lancelin J-M. Eur J Biochem. 1998;255:185–195. doi: 10.1046/j.1432-1327.1998.2550185.x. [DOI] [PubMed] [Google Scholar]
- 14.Hirasawa M, Schürmann P, Jacquot J-P, Keryer E, Hartman F C, Knaff D B. Biochemistry. 1999;38:5200–5205. doi: 10.1021/bi982783v. [DOI] [PubMed] [Google Scholar]
- 15.Setterdahl A T, Goldman B S, Hirasawa M, Jacquot P, Smith A J, Kranz R G, Knaff D B. Biochemistry. 2000;39:10172–10176. doi: 10.1021/bi000663t. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Malcolm B A. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 17.Anderson M E. Methods Enzymol. 1985;113:548–555. doi: 10.1016/s0076-6879(85)13073-9. [DOI] [PubMed] [Google Scholar]
- 18.Hwang C A, Sinskey J, Lodish H F. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 19.Penfold R J, Peemberton J M. J Bacteriol. 1994;176:2869–2876. doi: 10.1128/jb.176.10.2869-2876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer C E, Elsen S, Bird T H. Annu Rev Microbiol. 1999;53:495–523. doi: 10.1146/annurev.micro.53.1.495. [DOI] [PubMed] [Google Scholar]
- 21.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]