Abstract
We investigated the role of the CXCR4 chemokine receptor in development of the mouse hippocampus. CXCR4 mRNA is expressed at sites of neuronal and progenitor cell migration in the hippocampus at late embryonic and early postnatal ages. mRNA for stromal cell-derived factor 1 (SDF-1), the only known ligand for the CXCR4 receptor, is expressed close to these migration sites, in the meninges investing the hippocampal primordium and the primordium itself. In mice engineered to lack the CXCR4 receptor, the morphology of the hippocampal dentate gyrus (DG) is dramatically altered. Gene expression markers for DG granule neurons and bromodeoxyuridine labeling of dividing cells revealed an underlying defect in the stream of postmitotic cells and secondary dentate progenitor cells that migrate toward and form the DG. In the absence of CXCR4, the number of dividing cells in the migratory stream and in the DG itself is reduced, and neurons appear to differentiate prematurely before reaching their target. Our findings indicate a role for the SDF-1/CXCR4 chemokine signaling system in DG morphogenesis. Finally, the DG is unusual as a site of adult neurogenesis. We find that both CXCR4 and SDF-1 are expressed in the adult DG, suggesting an ongoing role in DG morphogenesis.
Chemokines (chemotactic cytokines) are a family of small proteins that orchestrate the diverse functions of cells of the immune system (1). The effects of these proteins are transduced by a family of G protein-coupled receptors (1). In addition to their roles in the control of normal leukocyte trafficking, chemokines and their receptors also play important roles in the pathogenesis of inflammatory disease and of AIDS. In the latter case, some chemokine receptors, particularly CCR5 and CXCR4, have been shown to act as cellular binding sites for the HIV-1 virus (2).
In addition to their widespread expression in the immune system, it now has been shown that chemokines and their receptors are expressed throughout the central nervous system. All of the major cell types in the brain, including neurons, glia, and microglia, have been shown to express many of the known receptors for chemokines (3, 4). Furthermore, many cells in the brain can synthesize and secrete chemokines under a variety of circumstances. It has been speculated widely that the chemokines and their receptors may be important in the genesis of the inflammatory component associated with diverse brain disorders and also with the pathogenesis of the widespread neurological symptoms associated with infection by the HIV-1 virus (3, 4). However, it is less clear which roles chemokines might play with respect to the normal functions of the brain. One possibility that has been raised, by analogy with their effects on leukocytes, is that chemokines and their receptors have an important chemotactic function in the migration of developing neurons during embryogenesis (5).
The CXCR4 receptor is one of the most highly expressed chemokine receptors both during development and in the mature brain (6–8). Deletion of the gene for the CXCR4 receptor, or of its only known ligand stromal cell-derived factor 1 (SDF-1), results in embryonic lethality. The brains of mouse embryos homozygous for the absence of CXCR4 receptors have severe abnormality in the development of cerebellar morphology (5, 9). Cells appear to migrate incorrectly from the proliferative external granule cell layer of the cerebellum, resulting in the ectopic positioning of granule cells in the Purkinje cell layer. We now have examined the development of the neocortex and hippocampus in CXCR4 mutant mice and have demonstrated that these mice exhibit a severe abnormality in the development of the DG. Our findings indicate that CXCR4 receptors play a widespread role in brain morphogenesis.
Materials and Methods
Mice.
CD-1 timed-pregnant mice were obtained from Charles River Breeding Laboratories. CXCR4 mutant mice (5) were obtained as heterozygotes from D. R. Littman (New York University Medical Center, New York) and interbred to generate homozygote embryos. Tail samples were collected and genotyped by PCR by using primers derived from the second exon of the CXCR4 gene: forward primer (5′-gctgactggtactttgggaa-3′) and reverse primer (5′-ctggctgacgtcggcaaaga-3′). Noon of the day on which a vaginal plug is discovered is termed embryonic (E) day 0.5; the day of birth, postnatal (P) day 0.
Histology and in Situ Hybridization.
Timed-pregnant mice were killed by CO2 inhalation. Embryonic brains were fixed and sectioned coronally at 40 μm with a microtome (Leica) and processed for in situ hybridization (10, 11). DNA template for SDF-1α riboprobe was generated by reverse transcription–PCR by using RNA isolated from adult mouse (12–14). A 538-bp mouse SDF-1α cDNA fragment was amplified with the forward primer 5′-acgccaaggtcgtcgccgtgctgg-3′ and the reverse primer 5′-gttagggtaatacaattccttaga-3′. Within this 538-bp cDNA fragment, a 263-nt sequence also is found in the SDF-1β cDNA sequence. The resultant SDF-1 probe likely detects both SDF-1α and SDF-1β mRNA. The rat CXCR4 cDNA was cloned as described (15), and a 870-bp SacI/ClaI fragment from CXCR4/pcDNA3.1 construct was used as CXCR4 riboprobe, which can crossreact with mouse CXCR4.
For Nissl staining, brains were fixed in 4% (wt/vol) paraformaldehyde in PBS, cryoprotected, embedded in OCT, and sectioned at 20 μm with a cryostat. Sections were stained with cresyl violet acetate (Sigma).
Bromodeoxyuridine (BrdUrd) Labeling.
For BrdUrd labeling of dividing cells at E18.5, pregnant mice were injected i.p. with 100 μg/g BrdUrd/body weight (Roche Diagnostics) and killed 4 h later. To label dividing cells in the adult DG, mice were given four injections separated by 3 h and killed 24 h after the first injection. BrdUrd incorporation was detected with anti-BrdUrd-FITC antibody (Becton Dickinson), followed by anti-FITC-AP, and detected with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate as the chromogen mix (Boehringer Mannheim) (10). BrdUrd labeling and in situ hybridization were combined as described, using the diaminobenzidine substrate (Vector Laboratories) to detect BrdUrd (16).
To compare the number of dividing cells contributing to the mutant versus wild-type DG, BrdUrd+ cell counts were made at E18.5 in two areas: the DG itself and the migratory stream of cells that leave the ventricular zone and move toward the DG. Counts were made in fields from two sections per embryo; each section contained two hippocampi (total of eight fields per embryo: four DG fields and four migratory stream fields). Counts were made from three mutants and three littermates from three independent experiments.
Preparation of Acutely Isolated Hippocampal Neurons.
Hippocampal neurons were prepared as described (17, 18). Cells were plated onto poly-l-lysine-coated coverslips at 4 × 104 cells per coverslip.
Intracellular Calcium Measurement.
Acutely isolated hippocampal neurons were loaded with 3 μM fura-2 AM, and intracellular free calcium concentration was measured as described (19). SDF-1α (50 nM, R & D Systems) was applied for 3–4 min. The number of cells responding to SDF-1α was counted. The identity of neurons was confirmed by their responsiveness to high potassium.
Immunocytochemistry and Confocal Microscopy.
Acutely isolated hippocampal neurons were fixed and processed for immunocytochemistry by using anti-human CXCR4 antibody that crosses to react with the murine CXCR4 (C-20; Santa Cruz Biotechnology) as described (19). An antibody selectively recognizing the phosphorylated active form of cAMP response element binding protein (CREB; Upstate Biotechnology, Lake Placid, NY) was used on neurons exposed to the chemokine SDF-1α. Immunostaining with control IgG or with peptide-preabsorbed antiserum was used as a negative control. Cells were imaged by confocal scanning laser microscopy as described (20). The identity of neurons was confirmed by immunostaining with a neuron-specific enolase antibody (Polysciences).
Hippocampal Slice Cultures.
Slice cultures were prepared as described (10). The telencephalon of E18.5 brain was sectioned coronally at 350 μm on a McIIwain tissue chopper (Brinkmann), and the hippocampus was cut away with a surgical microblade. Slices were placed on Millicell-CM inserts (Millipore) and maintained in B-27-supplemented NeuroBasal Medium (Life Technologies, Gaithersburg, MD). For in situ hybridization, slices were fixed overnight in 4% (wt/vol) paraformaldehyde at 4°C.
Results
Expression of CXCR4 and Its Ligand SDF-1 in Embryonic and Neonatal Cerebral Cortex.
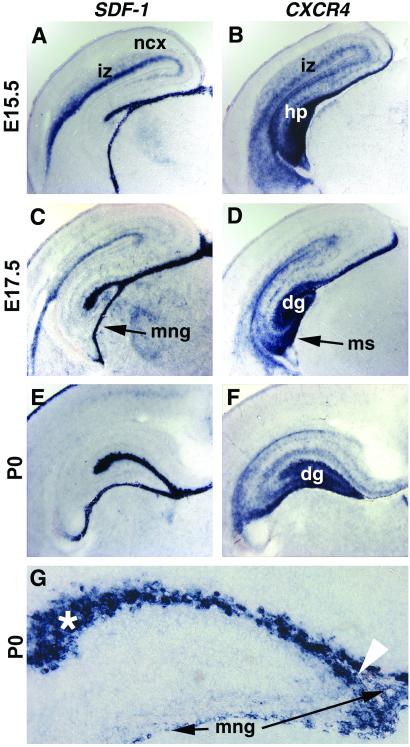
We first examined the expression of mRNA for CXCR4 and its ligand SDF-1 in the mouse cerebral hemispheres at different developmental stages. On E15.5 and E17.5, SDF-1 was expressed in the intermediate zone of the neocortex, through which newborn neurons migrate. SDF-1 expression was marked in the meninges overlying the hippocampus and in the hippocampal primordium itself near the hippocampal fissure, overlying the developing DG (Fig. 1 A and C). CXCR4 message also was expressed in the hippocampal and neocortical intermediate zone, with strongest expression reserved for the DG/CA3 pole of the hippocampus (Fig. 1 B and D). By P0, CXCR4 message was particularly striking at the dentate pole of the hippocampus, where neuronal proliferation and migration continues into postnatal life (Fig. 1F). By P0, SDF-1 expression was weak in neocortex (Fig. 1E) but still marked in the meninges overlying the hippocampus (Fig. 1 E and G) and in cells near the hippocampal fissure (Fig. 1G), overlapping a region of dense reelin expression (V. Banuchi, S. Assimacopoulos, and E.A.G., unpublished observations). The latter region notably contains reelin-expressing Cajal–Retzius cells, previously implicated in DG cell migration (21). Thus, both the temporal and spatial patterns of CXCR4 and SDF-1 expression suggest an involvement in neuronal migration in the neocortex and hippocampus. CXCR4 was expressed at sites of neuronal migration in the hippocampus and neocortex, whereas SDF-1 was expressed both within the cortical primordium and in the meninges investing the developing hippocampus. The latter observation suggests an intriguing signaling action on the developing hippocampus by the meninges overlying the DG.
Figure 1.
Expression of SDF-1 and CXCR4 in developing neocortex and hippocampus. (A–G) Coronal sections through mouse forebrains on E15.5, E17.5, and the day of birth (P0) processed for in situ hybridization. G is a higher magnification of the DG (dg) region. SDF-1 is expressed in the intermediate zone (iz) of the neocortex (ncx) (A), in the meninges (mng) overlying the hippocampus (hp) (A, C, E, and G), and in a zone close to the hippocampal fissure also occupied by Cajal–Retzius cells (A, C, E, and G; see text). White arrowhead in G points to the opening of the hippocampal fissure. White asterisk in G marks some of the SDF-1-expressing cells lining the hippocampal fissure. CXCR4 also is expressed in the iz at E15.5 (B). At all ages, strongest CXCR4 expression appears in the developing dg and in a migratory stream (ms) of cells moving toward the dg (D and F).
CXCR4 Receptors Are Functional in Intracellular Signaling in Embryonic and Neonatal Hippocampal Neurons.
Next, we confirmed CXCR4 protein expression in developing hippocampal neurons by indirect immunofluorescent antibody staining of acutely dissociated embryonic and neonatal hippocampal neurons. The majority of hippocampal neurons were immunoreactive for the CXCR4 receptor (Fig. 2).
Figure 2.
Acutely isolated hippocampal neurons express CXCR4 receptors. (A–D) Confocal microscopy images of E17.5 and P0 hippocampal neurons immunostained for CXCR4 antibody. (A and B) Anti-CXCR4 antibody staining. Immunostaining was diminished after preadsorption of the CXCR4 antibody with its peptide antigen (C and D). (E and F) Differential interference contrast microscopy images of the same field of cells as C and D.
The CXCR4 receptors expressed by embryonic and neonatal hippocampal neurons are functional as assessed by activation of intracellular signaling pathways. In response to SDF-1α, both embryonic and neonatal hippocampal neurons exhibited increased activation of CREB (Fig. 3 A–D). This result is consistent with previous results from hippocampal neuronal cultures (14). However, we show that CXCR4 receptors are functional as early as E17.5 in the mouse. In addition, we used Fura-2-based Ca2+ imaging to investigate the presence of functional chemokine receptors in acutely dissociated embryonic and neonatal hippocampal neurons. We found that SDF-1α could induce Ca2+ mobilization in a subset of cells (Fig. 3 E and F).
Figure 3.
Effect of SDF-1α on CREB activation and calcium mobilization in acutely isolated E17.5 and P0 hippocampal neurons. (A–D) Fluorescence microscopy image of neurons immunoreactive for the activated form of CREB. Neurons were treated with PBS (control) or SDF-1α for 10 min in Eagle's minimal essential medium. (A and B) No CREB activation in neurons treated with PBS. (C and D) Activation of CREB in neurons treated with 100 nM SDF-1α. (E and F) SDF-1α increase [Ca2+]i in hippocampal neurons. SDF-1α was applied for the times indicated by the bars (3–4 min). Representative traces from single neurons are shown in E and F. On E17.5, 8 of 127 hippocampal neurons were responsive to SDF-1α. By P0, 11 of 194 neurons were responsive to SDF-1α.
Abnormal Development of the DG in Mice Deficient in CXCR4.
Previous reports indicate that mice that are deficient in either SDF-1 or its receptor CXCR4 show major defects in cell migration in the developing cerebellar cortex (5, 9). Here, we analyzed development of the cerebral cortex in CXCR4 mutant mice, focusing on the hippocampus. In all cases, we compared development of CXCR4 homozygote mutants with their wild-type and heterozygote littermates. Although the cerebral cortex overall is somewhat smaller in homozygote CXCR4 mutants than in wild-type mice, no obvious difference was detected in cortical layer formation at E18.5, the day before birth (Fig. 4 A–D). Notably, in Nissl-stained sections, the developing pyramidal cell layer in Ammon's horn of the hippocampus appears similar in breadth and overall morphological development in CXCR4 mutants (Fig. 4D) and their wild-type littermates (Fig. 4C). By contrast, the DG is difficult to identify at all in homozygous mutant sections at E18.5, appearing small and morphologically immature compared with wild-type mice (Fig. 4 B and D). This morphological observation was confirmed by analyzing expression of the basic helix–loop–helix gene, NeuroD, expressed in DG cells and cells migrating toward the DG (16, 22). In homozygote CXCR4 mutant mice, the target DG is underpopulated with NeuroD-expressing cells and is indistinct (Fig. 4F). What primary defects could lead to this abnormal morphology?
Figure 4.
Abnormal development of the DG in mice deficient in CXCR4. (A–D) Nissl-stained coronal sections through E18.5 wild-type (A and C) or homozygote CXCR4 mutant forebrains (B and D). C and D are higher-magnification views of A and B. Gross morphology appears similar between wild-type and mutant brains in the developing neocortex (A and B) and the developing CA fields (ca1, ca3) of the hippocampal pyramidal cell layer (C and D). By contrast, the developing dg is distinct in the wild type (C) but almost undetectable in the mutant (asterisk in D). (E and F) Coronal sections processed with in situ hybridization to show expression of NeuroD. The developing dg is distinct in a wild-type mouse (E) but not in a homozygote CXCR4 mutant (asterisk in F). (G and H) Hippocampal slices prepared from an E18.5 wild-type (G) or CXCR4 mutant mouse (H) and maintained for 7 days in vitro (7DIV). Expression of Prox1 reveals a classic, V-shaped dg in the wild-type slice. By contrast, the CXCR4 mutant slice shows a small, poorly formed dg (asterisk in H).
Abnormal Development of the DG in Hippocampal Slice Cultures.
A possibility is that the CXCR4 mutant DG is delayed in its development and could recover a normal morphology if allowed to develop longer. Because the CXCR4 mutation is embryonic lethal, it is impossible to evaluate the postnatal development of the DG in mutant mice in vivo. We therefore prepared hippocampal slices from E18.5 CXCR4 mutant and wild-type mice and maintained them in vitro for 1–2 weeks, analyzing DG development by expression of Prox1. Expression of the homeobox gene Prox1 is a marker of differentiated dentate granule neurons in the perinatal and postnatal mouse brain (16, 23). After 7–13 days in vitro, slices from wild-type mice develop a classic, V-shaped DG, with both external and internal blades in evidence (Fig. 4G). By contrast, even after 7–13 days in vitro, the CXCR4 mutant DG still has failed to form correctly, remaining underpopulated by dentate granule cells (Fig. 4H).
Defects in the Secondary Proliferative Cell Population That Forms the DG.
Early in the generation of the DG, a primary dentate neuroepithelium adjacent to the lateral ventricle gives rise to a secondary proliferative cell population that streams along the ventral surface of the developing hippocampus, close to the meninges, to take up a position in the hilus of the DG and form a tertiary germinal matrix (24, 25). Normal DG formation would be disrupted if the proliferating cells of the migratory stream fail to reach the dentate hilus in sufficient numbers. It is noteworthy, therefore, that CXCR4 is expressed both in the migratory stream of dividing cells as well as in the forming DG itself, and that the gene encoding the CXCR4 ligand, SDF-1, is expressed not only in the meninges underlying the route of migration but also surrounding the hippocampal fissure, adjacent to the DG, the target of the migration (Fig. 1 A, C, E, and G).
Therefore, we focused on possible defects in the migratory stream in CXCR4 mutant mice compared with wild-type mice. A striking difference appeared in the expression of Prox1. In wild-type mice at E18.5, Prox1 is expressed, as expected, in the two forming blades of the DG (Fig. 5 A and C). In homozygote CXCR4 mutants, by contrast, Prox1 is expressed in cells along the entire migratory stream (Fig. 5 B and D). Again, this abnormality does not appear to represent a developmental delay in the expression of Prox1 in the mutant; similar differences in expression patterns were observed at E16.5 and E17.5 between the mutant and wild-type DG (data not shown). In sections processed to show both misplaced Prox1-expressing cells and dividing cells by BrdUrd labeling, the two cell populations appear largely nonoverlapping (Fig. 5E). Thus, Prox1 does not appear to be abnormally up-regulated by dividing cells in the migratory stream; rather, the migratory stream now is filled with cells that are differentiating before reaching their final position.
Figure 5.
Defects in the secondary proliferative cell population that forms the DG. (A–D) Coronal sections through the E18.5 hippocampus of wild-type (A and C) or CXCR4 mutant mice (B and D), processed to show expression of Prox1 (blue) or Prox1 together with BrdUrd-labeled dividing cells (orange). In the wild-type mouse, Prox1 is expressed in the forming dg (A and C). By contrast, in the mutant, Prox1 is expressed in the vestigial dg but also along the migratory stream (ms) (arrows in D) of dividing cells running along the ventral surface of the hippocampus into the dg. BrdUrd-labeled cells of the ms can be seen between the two arrows in C. (E) A higher magnification of the migrating stream of cells shown in D. Numerous blue, Prox1-expressing cells appear among the brown, BrdUrd-labeled cells, but the populations appear largely distinct. Arrows in E point to two single-labeled cells. (F and G) High-magnification views of BrdUrd-labeled dividing cells (dark blue) coursing through the ms in a wild-type (F) and a CXCR4 mutant mouse (G). About 30% fewer BrdUrd-labeled cells appear in the ms of the mutant (G) than in the wild type (F).
Counts of BrdUrd-labeled cells both in the migratory stream and the DG itself at E18.5 revealed a further difference (two typical fields from which counts were made in the migratory stream are shown in Fig. 5 F and G). In the migratory stream of homozygote CXCR4 mutant mice, we observed a 31% reduction in BrdUrd+ cells as compared with wild-type littermates (P < 0.01, Student's t test, sigmaplot 5.0 software). In the mutant DG itself, we saw a 39% reduction in BrdUrd-labeled cells compared with wild-type mice (P < 0.01). The reduction of DG progenitor cells, specifically, cannot be determined with absolute precision because the migratory stream also contains glia (15). Moreover, glia have been shown to express chemokine receptors including CXCR4 (26) and are responsive to chemokines such as GRO-1 (27) and SDF-1 (26). Thus, although most BrdUrd-labeled cells in the migratory stream of both wild-type and CXCR4 mutant mice are likely to be DG neuronal progenitor cells (24, 25), some may be glia en route to the DG. The implications of this possibility are discussed further below.
Ongoing Expression of CXCR4 and SDF-1 in the Adult DG.
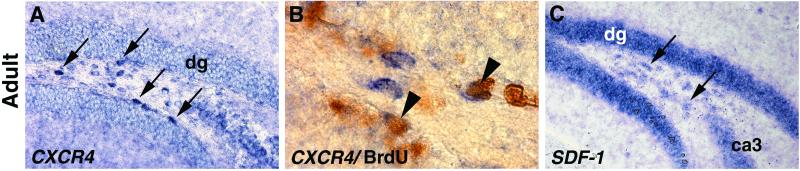
The DG is unusual in that granule cell neurogenesis continues into early adult life (24, 25). Therefore, we analyzed expression of CXCR4 and SDF-1 in CD-1 mice at 6–10 weeks of age. Late dentate granule neurons arise from dividing cells in the DG hilus or along the inner surfaces of the DG blades (25, 28). In the adult, we found that SDF-1 now is expressed strikingly in the DG blades (Fig. 6C), not a site of perinatal expression (Fig. 1), and CXCR4 is expressed in a few cells in the hilus and along the inner surface of each blade (Fig. 6A). A few CXCR4-expressing cells could be labeled with BrdUrd (Fig. 6B), suggesting that some of the CXCR4-expressing cells are dividing and may be granule neuron progenitors. The patterns of expression of CXCR4 and SDF-1 in the adult DG therefore indicate a continuing role in the genesis and guidance of late-born granule neurons.
Figure 6.
Expression of SDF-1 and CXCR4 in the adult mouse DG. (A–C) Coronal sections through the hippocampus of CD-1 adult mice. CXCR4 is expressed in scattered cells in the dg hilus and lining the dg blades (arrows point to four such cells) (A). Double labeling with BrdUrd (brown) suggests that some of the CXCR4-expressing (blue) cells are still dividing (B). Arrowheads in B indicate double-labeled cells; singly BrdUrd-labeled or CXCR4-expressing cells appear in the same field. SDF-1 is expressed in the blades of the dg, and scattered SDF-1-expressing cells (arrows) also appear in the hilus of the dg (C).
Discussion
Our findings indicate that, without functional CXCR4, morphogenesis of the hippocampal DG fails. This failure appears partly or entirely from defects in the migratory stream of cells that forms a secondary germinal matrix for the DG. The cells of this stream normally express CXCR4 and migrate along a route lined with meninges expressing the only known endogenous CXCR4 ligand SDF-1. Cells adjacent to the developing blades of the DG, the target of the migratory stream, also express SDF-1. In the absence of CXCR4, the number of dividing cells in the migratory stream and in the DG itself is reduced, and cells appear to differentiate prematurely. Thus, progenitor cells fail to reach the forming DG in sufficient numbers, which remains underpopulated and immature. Even when the migratory stream is allowed to develop past embryonic stages in slice culture, DG morphogenesis does not catch up.
Several alternative or perhaps complementary explanations may account for the defects in the migratory stream. One possibility is that SDF-1/CXCR4 signaling is required to maintain the secondary dentate progenitor cell population in mitosis. Without the signal, progenitor cells drop out of division prematurely and differentiate. A second possibility is that SDF-1/CXCR4 signaling regulates movement of the migratory stream. In the absence of the signal, cells along the migratory stream are slowed or stalled and differentiate ectopically. Support for the latter view comes from the pattern of SDF-1 expression, both beneath the migratory stream and in the target, the developing DG (Fig. 1G). Levels of SDF-1 protein would be expected to increase closer to the target, consistent with a chemotactic role for SDF-1. Informatively, the tissues we show to express SDF-1 have been implicated previously in dentate granule cell migration (21). Selective destruction of medial cerebral meningeal cells, with secondary degeneration of the Cajal–Retzius cells lining the hippocampal fissure, leads to a defect in DG morphogenesis, apparently via a disruption of granule cell migration (21, 28). Our present findings indicate that the loss of SDF-1/CXCR4 signaling could, at least in part, mediate this migratory defect.
A third intriguing possibility, supported by recent, elegant work in the cerebellum, is that SDF-1 may act both as a chemotactic signal and to promote mitosis. In the cerebellum, SDF-1 cooperates with Sonic hedgehog (SHH) to enhance granule cell proliferation and functions as a chemotactic cue to hold granule precursors in the external granule layer, close to the SHH source (29). DG progenitors show a different behavior from cerebellar precursors by leaving the primary proliferative matrix and dividing as they migrate. Thus, the developing cerebellum and DG may show both similarities and differences in their deployment of SDF-1/CXCR4 signaling. In one scenario of DG development, SDF-1/CXCR4 signaling attracts the cells of the migratory stream toward their target (rather than holding the cells in a static proliferative layer as in the cerebellum). Meanwhile, as the cells migrate, SDF-1, expressed in cells underlying the migratory stream, also promotes mitosis. SHH itself is not expressed in hippocampus; thus, another mitogen may cooperate with SDF-1.
In the DG, granule cell neurogenesis continues into adult life (24, 25). It therefore is notable that the adult expression of SDF-1 and CXCR4 differs from the embryonic patterns but remains consistent with continued functions in granule cell mitogenesis and/or chemotaxis. In the adult, CXCR4 is expressed where granule neuron progenitors are found, whereas SDF-1 is expressed in the target of the newborn cells, the blades of the DG.
Still other potential SDF-1-mediated effects on early DG morphogenesis prompt further investigation. For example, the migratory stream contains glia, a cell type shown to express CXCR4 receptors and to release glutamate in response to SDF-1 (26). Given that neuronal migration can be influenced by glutamatergic activation (30), could SDF-1-evoked glutamate release from glia in the migratory stream influence the migration of DG progenitor cells? In leukocyte chemotaxis, the SDF-1/CXCR4 and Slit/Robo signaling pathways interact antagonistically (31). Do similar interactions come into play in the formation of the DG, which also has been shown to express Slit and Robo family members (32)? Future studies are needed to elucidate the potentially complex functions of SDF-1/CXCR4 signaling in both developing and adult DG. Finally, because the CXCR4 receptor is one of the major binding sites for the HIV-1 virus (2), it is possible that interference with the developmental functions of SDF-1 and CXCR4 receptors plays a significant role in pediatric aspects of HIV-1-related neuropathology.
Acknowledgments
We thank C. Yu, D. Ren, C. Mauer, S. Assimacopoulos, and T. Fukuchi-Shimogori for technical help, S.A. for critical comments on the data, P. Gruss and D. Lowenstein for Prox1 and NeuroD cDNAs, and Y. Zou and D. Littman for providing us with the CXCR4 heterozygous mice. This work was supported by National Institutes of Health Grants DA13141, MH40165, and DK44840 (to R.J.M.) and Grant MH59962, and a March of Dimes Research Grant (to E.A.G.).
Abbreviations
- SDF-1
stromal cell-derived factor 1
- DG
dentate gyrus
- CREB
cAMP response element binding protein
- En
embryonic day n
- Pn
postnatal day n
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rossi D, Zlotnik A. Annu Rev Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Berger E A, Murphy P M, Farber J M. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 3.Bajetto A, Bonavia R, Barbero S, Florio T, Schettini G. Front Neuroendocrinol. 2001;22:147–184. doi: 10.1006/frne.2001.0214. [DOI] [PubMed] [Google Scholar]
- 4.Miller R, Oh S. In: Universes in Delicate Balance; Chemokines in the Nervous System. Ransohoff R, editor. Amsterdam: Elsevier Science; 2002. pp. 273–288. [Google Scholar]
- 5.Zou Y R, Kottmann A H, Kuroda M, Taniuchi I, Littman D R. Nature (London) 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 6.Jazin E E, Soderstrom S, Ebendal T, Larhammar D. J Neuroimmunol. 1997;79:148–154. doi: 10.1016/s0165-5728(97)00117-3. [DOI] [PubMed] [Google Scholar]
- 7.McGrath K E, Koniski A D, Maltby K M, McGann J K, Palis J. Dev Biol. 1999;213:442–456. doi: 10.1006/dbio.1999.9405. [DOI] [PubMed] [Google Scholar]
- 8.Lavi E, Strizki J M, Ulrich A M, Zhang W, Fu L, Wang Q, O'Connor M, Hoxie J A, Gonzalez-Scarano F. Am J Pathol. 1997;151:1035–1042. [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Q, Jones D, Borghesani P R, Segal R A, Nagasawa T, Kishimoto T, Bronson R T, Springer T A. Proc Natl Acad Sci USA. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tole S, Christian C, Grove E A. Development (Cambridge, UK) 1997;124:4959–4970. doi: 10.1242/dev.124.24.4959. [DOI] [PubMed] [Google Scholar]
- 11.Grove E A, Tole S, Limon J, Yip L, Ragsdale C W. Development (Cambridge, UK) 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 12.Lu M, Conzen S D, Cole C N, Arrick B A. Cancer Res. 1996;56:4578–4581. [PubMed] [Google Scholar]
- 13.Lu M, Arrick B A. Oncogene. 2000;19:6351–6360. doi: 10.1038/sj.onc.1204025. [DOI] [PubMed] [Google Scholar]
- 14.Meucci O, Fatatis A, Simen A A, Bushell T J, Gray P W, Miller R J. Proc Natl Acad Sci USA. 1998;95:14500–14505. doi: 10.1073/pnas.95.24.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh S B, Endoh T, Simen A A, Ren D, Miller R J. J Neuroimmunol. 2002;123:66–75. doi: 10.1016/s0165-5728(01)00485-4. [DOI] [PubMed] [Google Scholar]
- 16.Pleasure S J, Collins A E, Lowenstein D H. J Neurosci. 2000;20:6095–6105. doi: 10.1523/JNEUROSCI.20-16-06095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mintz I M, Bean B P. Neuron. 1993;10:889–898. doi: 10.1016/0896-6273(93)90204-5. [DOI] [PubMed] [Google Scholar]
- 18.Gillard S E, Lu M, Mastracci R M, Miller R J. J Neuroimmunol. 2002;124:16–28. doi: 10.1016/s0165-5728(02)00005-x. [DOI] [PubMed] [Google Scholar]
- 19.Oh S B, Tran P B, Gillard S E, Hurley R W, Hammond D L, Miller R J. J Neurosci. 2001;21:5027–5035. doi: 10.1523/JNEUROSCI.21-14-05027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meucci O, Fatatis A, Simen A A, Miller R J. Proc Natl Acad Sci USA. 2000;97:8075–8080. doi: 10.1073/pnas.090017497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Super H, Soriano E, Uylings H B. Brain Res Brain Res Rev. 1998;27:40–64. doi: 10.1016/s0165-0173(98)00005-8. [DOI] [PubMed] [Google Scholar]
- 22.Liu M, Pleasure S J, Collins A E, Noebels J L, Naya F J, Tsai M J, Lowenstein D H. Proc Natl Acad Sci USA. 2000;97:865–870. doi: 10.1073/pnas.97.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliver G, Sosa-Pineda B, Geisendorf S, Spana E P, Doe C Q, Gruss P. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- 24.Altman J, Bayer S A. J Comp Neurol. 1990;301:365–381. doi: 10.1002/cne.903010304. [DOI] [PubMed] [Google Scholar]
- 25.Altman J, Bayer S A. J Comp Neurol. 1990;301:325–342. doi: 10.1002/cne.903010302. [DOI] [PubMed] [Google Scholar]
- 26.Bezzi P, Domercq M, Brambilla L, Galli R, Schols D, De Clercq E, Vescovi A, Bagetta G, Kollias G, Meldolesi J, et al. Nat Neurosci. 2001;4:702–710. doi: 10.1038/89490. [DOI] [PubMed] [Google Scholar]
- 27.Wu Q, Miller R H, Ransohoff R M, Robinson S, Bu J, Nishiyama A. J Neurosci. 2000;20:2609–2617. doi: 10.1523/JNEUROSCI.20-07-02609.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartmann D, Sievers J, Pehlemann F W, Berry M. J Comp Neurol. 1992;320:33–61. doi: 10.1002/cne.903200103. [DOI] [PubMed] [Google Scholar]
- 29.Klein R S, Rubin J B, Gibson H D, DeHaan E N, Alvarez-Hernandez X, Segal R A, Luster A D. Development. 2001;128:1971–1981. doi: 10.1242/dev.128.11.1971. [DOI] [PubMed] [Google Scholar]
- 30.Komuro H, Rakic P. Science. 1993;260:95–97. doi: 10.1126/science.8096653. [DOI] [PubMed] [Google Scholar]
- 31.Wu J Y, Feng L, Park H T, Havlioglu N, Wen L, Tang H, Bacon K B, Jiang Z, Zhang X, Rao Y. Nature (London) 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen Ba-Charvet K T, Brose K, Marillat V, Kidd T, Goodman C S, Tessier-Lavigne M, Sotelo C, Chedotal A. Neuron. 1999;22:463–473. doi: 10.1016/s0896-6273(00)80702-3. [DOI] [PubMed] [Google Scholar]