Abstract
α-Amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor (AMPAR) stability and movement at synapses are important factors controlling synaptic strength. Here, we study the roles of proteins [N-ethylmaleimide-sensitive fusion protein (NSF), glutamate receptor AMPAR binding protein (ABP)-interacting protein (GRIP)/(ABP), and protein interacting with C-kinase-1 (PICK1) that interact with the GluR2 subunit in the control of the surface expression and cycling of AMPARs. Epitope-tagged GluR2 formed functional receptors that exhibited targeting to synaptic sites. Constructs in which binding to NSF, PDZ proteins (GRIP/ABP and PICK1), or GRIP/ABP alone was eliminated each exhibited normal surface targeting and constitutive cycling. The lack of NSF binding, however, resulted in receptors that were endocytosed to a greater extent than wild-type receptors in response to application of AMPA or N-methyl-d-aspartate (NMDA). Conversely, the behavior of the GluR2 mutants incapable of binding to GRIP/ABP suggests that these PDZ proteins play a role in the stabilization of an intracellular pool of AMPARs that have been internalized on stimulation, thus inhibiting their recycling to the synaptic membrane. These results provide further evidence for distinct functional roles of GluR2-interacting proteins in AMPAR trafficking.
The activity-dependent insertion and endocytosis of α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors (AMPARs) play an important role in modulating synaptic strength, such as occurs during certain forms of long-term potentiation (LTP; refs. 1 and 2) and long-term depression (LTD; ref. 3). Thus, the trafficking of AMPARs must be subject to regulation at several steps allowing precise control of the number of AMPARs in the synaptic membrane. AMPARs are heteromeric assemblies of which GluR2 is a particularly important subunit in that it is a component of AMPARs expressed on principal (e.g., pyramidal) cells in adult brain (4) and controls their biophysical properties (5). GluR2 interacts with a number of intracellular proteins that have potential roles in regulating AMPAR trafficking (6–8), including N-ethylmaleimide sensitive fusion protein (NSF), a protein involved in membrane fusion events (9). Interfering with this interaction by using blocking peptides reduces AMPAR expression at synapses (10–13). It is unclear, however, whether this loss of AMPARs is due to a block of their surface delivery or because NSF stabilizes AMPARs in the plasma membrane and limits their endocytosis. The PDZ domain-containing proteins protein interacting with C-kinase-1 (PICK1), glutamate receptor-interacting protein (GRIP), and AMPAR binding protein (ABP), also interact with GluR2 (6–8) and have been suggested to play roles in the clustering and surface expression of AMPARs (14–16). Preventing the interaction of GluR2 with these proteins impairs LTD in both the hippocampus and cerebellum (17–20). These studies, however, suggest conflicting roles for GRIP/ABP and PICK1 in the induction and maintenance of LTD. Thus, like NSF, the exact roles of GRIP/ABP and PICK1 in AMPAR trafficking remain unclear. Here, we explore the role of GluR2-interacting proteins in AMPAR trafficking by assessing the trafficking behavior of wild-type and mutant forms of epitope-tagged GluR2 in hippocampal neurons.
Materials and Methods
Epitope Tagging and Mutagenesis of GluR2.
GluR2/GluRB(flop) cDNA (gift of P. Seeburg, Max-Planck Institute for Medical Research, Heidelberg, Germany) was epitope tagged at the amino terminus with a FLAG sequence (DYKDDDDK) as described previously for FLAG-GluR1 (21). Green fluorescent protein (GFP) cDNA was PCR amplified and subcloned in frame between FLAG and GluR2. This FLAG-GFP-GluR2 construct was inserted into the vector pTet7, putting it under the control of a tetracycline-repressible element. PCR-mediated mutagenesis was used to generate mutant constructs unable to interact with intracellular proteins.
Cell Culture and Transfections.
HEK293 cells [American Type Culture Collection (ATCC)] were plated on poly-d-lysine-coated glass coverslips 1 day before transfection and maintained in DMEM containing 10% FBS and 1% penicillin/streptomycin. Hippocampal cell cultures were prepared as described previously (22). Calcium phosphate-mediated gene delivery was used to transfect cells with constructs and a tetracycline repressor-VP16 fusion protein. HEK293 cells were transfected 1 day after plating, and expression of proteins was allowed to proceed for 48–72 h. Primary neuronal cultures were transfected at 10–11 days in vitro (DIV), and expression of constructs was allowed to proceed for 72 h in the presence of 1 ng/ml doxycycline before experimental manipulations.
Immunocytochemistry.
Surface labeling of constructs in HEK293 cells and neurons was performed by labeling paraformaldehyde-fixed cells with anti-FLAG antibody, followed by A594-conjugated anti-mouse antibody (Jackson ImmunoResearch). For quantification of surface:total ratios of constructs, neurons were fixed with 4% paraformaldehyde, and surface receptors were labeled with anti-FLAG antibody followed by Cy5-conjugated anti-mouse antibody. Cells were subsequently permeabilized with 0.2% Triton X-100, and intracellular receptors were labeled with anti-FLAG antibody followed by A568-conjugated anti-mouse antibody. Methanol-permeabilized cells were used for staining with anti-synaptophysin antibody (Zymed) followed by A594-conjugated secondary antibody.
For receptor cycling assays, surface-expressed receptor constructs were labeled in live neurons with anti-FLAG antibody (Sigma, 7.6 μg/ml in conditioned medium) for 15 min at 37°C. For constitutive cycling experiments, the excess antibody was washed off, and receptors were allowed to cycle for a further 30 min in conditioned medium at 37°C. For AMPA-induced internalization, antibody labeling was performed in the presence of 50 μM d-2-amino-5-phosphonovaleric acid (d-APV), 100 μM LY341495 [to prevent activation of N-methyl-d-aspartate receptors (NMDARs) and mGluRs, respectively], and 50 μM AMPA. NMDA-induced internalization was assayed by adding 20 μM 2,3-dihydroxy-6-nitro-7-sulfamoylbenzo[f]quinoxaline (NBQX), 100 μM LY341495 (to prevent activation of AMPARs and mGluRs, respectively), and 100 μM NMDA to cells after the 15 min antibody treatment for 1 min. The NMDA was washed off, and receptor cycling was allowed to proceed for 10 min in conditioned medium containing 50 μM APV, 20 μM NBQX, and 100 μM LY341495 to prevent further glutamate receptor activation. At the end of these manipulations receptor cycling was stopped by addition of cold PBS on ice. For all internalization assays, treated and control cells were obtained from the same culture preparations and were examined in parallel. Receptors remaining on the cell surface were labeled with Cy5-conjugated anti-mouse antibodies (Jackson ImmunoResearch) at 4°C for 1 h. Cells were fixed and permeabilized by addition of methanol at −20°C for 5 min, and internalized receptors were labeled with A568-conjugated anti-mouse antibodies (Jackson ImmunoResearch). Some experiments were also performed by fixation with 4% paraformaldehyde before labeling of surface receptors, followed by permeabilization with 0.2% Triton X-100. Both methanol and paraformaldehyde/Triton X-100 fixation and permeabilization gave equivalent results; thus, the data presented represent a combination of both protocols. All drugs were obtained from Tocris Cookson (Ballwin, MO). For all manipulations, control experiments were performed to ensure that antibody labeling was saturating at all steps.
Data Acquisition and Analysis.
Hippocampal cells were imaged by using either a ×40 or ×63 objective and acquired on a Zeiss LSM510 confocal microscope. All images were analyzed by using metamorph software (Universal Imaging, West Chester, PA). All imaging and analysis were performed with the researcher blind to the transfected construct and treatment. For individual experiments, images for all conditions were taken by using identical acquisition parameters and analyzed by using the same thresholds. Fluorescence of A568-conjugated antibodies, elicited by excitation at 543 nm, was used to assay internalized receptors. Fluorescence of Cy5-conjugated antibodies, elicited by excitation at 633 nm, was used to assay surface-expressed receptors. The amount of red fluorescent signal was used to quantify internalized protein and was divided by the amount of total fluorescent signal (A568 + Cy5) to calculate internalized:total ratios, thus controlling for any differences in expression levels of constructs. Statistical analysis was performed by using one-way ANOVA and Fisher's LSD tests.
Electrophysiology.
Whole cell voltage clamp recordings were made with an Axopatch 1D amplifier (Axon Instruments, Foster City, CA) using low resistance pipettes (2–6 MΩ). The pipette solution contained (in mM): 120 Cs-gluconate, 8 NaCl, 10 Hepes, 0.5 EGTA, and 2 MgATP adjusted to pH 7.2 with CsOH. The extracellular solution contained (in mM): 120 NaCl, 5 KCl, 5 Hepes, 20 glucose, 25 sucrose, 1.8 CaCl2, and 1 MgCl2 adjusted to pH 7.35 with NaOH. HEK293 cells were held at −70 mV in voltage clamp, and pressure application of agonist was achieved by a puffer pipette positioned adjacent to the cell. Evoked currents were stored by using igor pro software (WaveMetrics, Lake Oswego, OR).
Results
Epitope-Tagged GluR2 Produces Functional Receptors.
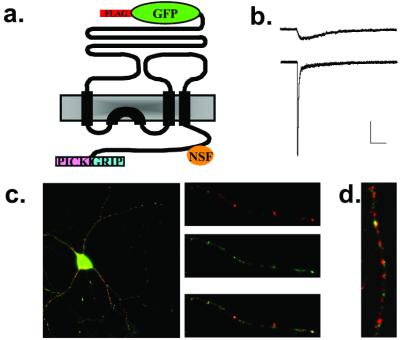
FLAG-GFP-GluR2 (Fig. 1a) was expressed under the control of a tetracycline-repressible element to limit its expression level and thereby minimize the possibility of saturating native interacting protein partners. To assess the function of this construct, HEK293 cells were transfected with a version [FLAG-GFP-GluR2(Q)] that was mutated at the Q/R editing site in the pore-forming region of the subunit (R607Q). This change results in AMPARs that exhibit a larger single channel conductance compared with the R form (23). Both FLAG-GFP-GluR2 and FLAG-GFP-GluR2(Q) were expressed on the surface of HEK293 cells (data not shown). Transfected cells exhibited large, rapidly desensitizing responses to glutamate application and small nondesensitizing responses to kainate application (Fig. 1b). Such responses are characteristic of untagged homomeric GluR2 AMPARs (23), demonstrating that FLAG-GFP-GluR2 forms functional AMPARs that are delivered to the cell surface.
Figure 1.
Expression and functional analysis of FLAG-GFP-GluR2. (a) Schematic drawing of FLAG-GFP-GluR2 indicating the binding sites within the C-terminal domain for NSF and PDZ domain containing proteins. (b) Representative electrophysiological responses from HEK293 cells transfected with FLAG-GFP-GluR2(Q). Cells were held at −70 mV and kainate (1 mM, Upper) or glutamate (1 mM, Lower) was applied. Scale bars = 100 pA, 100 ms. (c) Surface labeling (red) indicates that a proportion of the total FLAG-GFP-GluR2 (green) is delivered to the cell surface. (d) FLAG-GFP-GluR2 (green) colocalizes with synaptophysin (red) in hippocampal neuronal cultures.
Epitope-Tagged GluR2 Is Expressed in the Plasma Membrane of Neurons at Synapses.
Surface labeling of living hippocampal neurons revealed that FLAG-GFP-GluR2 was efficiently delivered to the plasma membrane of dendrites (Fig. 1c) as evidenced by its colocalization with the dendritic marker microtubule-associated protein 2 (MAP2) (data not shown). It also showed a high level of colocalization with synaptophysin (Fig. 1d) indicative of its expression at synaptic sites. Thus, in neurons, FLAG-GFP-GluR2 exhibits a distribution similar to native AMPAR subunits being localized primarily to postsynaptic sites at the cell surface.
GluR2 Surface Expression Does Not Require Binding to NSF or PDZ Proteins.
To determine whether interactions with NSF or PDZ proteins (GRIP/ABP, PICK1) are required for the surface expression of GluR2, we made several mutant constructs: (i) FLAG-GFP-GluR2(ΔNSF) with mutations N851S and P852A to inhibit the interaction between GluR2 and NSF (10); (ii) FLAG-GFP-GluR2(Del3aa) in which the extreme three C-terminal amino acids of GluR2 were deleted to prevent the interaction with GRIP/ABP and PICK1 (14); (iii) FLAG-GFP-GluR2(S880A), which will prevent GRIP/ABP but not PICK1 binding to GluR2 (24, 25); and (d) FLAG-GFP-GluR2(S880D) to mimic phosphorylation at S880, a possible in vivo mechanism for preventing GRIP/ABP but not PICK1 binding (24, 25). All of these constructs were expressed in the plasma membrane of HEK293 cells (data not shown) indicating their efficient assembly and trafficking to the cell surface.
These same constructs were efficiently expressed in the soma and dendrites of cultured hippocampal neurons as evidenced by their colocalization with microtubule-associated protein 2 (data not shown). Because differences in the level of expression of each construct might influence their trafficking, we first assessed the approximate level of expression by measuring total GFP fluorescence per unit area over the entire cell (soma and processes) for a large number of cells. This measure did not differ, indicating approximately equal levels of expression [normalized values: FLAG-GFP-GluR2, 1.00 ± 0.03 (n = 129 cells); FLAG-GFP-GluR2(ΔNSF), 1.03 ± 0.06 (n = 36); FLAG-GFP-GluR2(Del3aa), 1.01 ± 0.06 (n = 40); FLAG-GFP-GluR2(S880A), 1.07 ± 0.04 (n = 41); FLAG-GFP-GluR2(S880D), 1.00 ± 0.04 (n = 52); ANOVA: df = 4, MS = 0.045, F = 0.429, and P = 0.787].
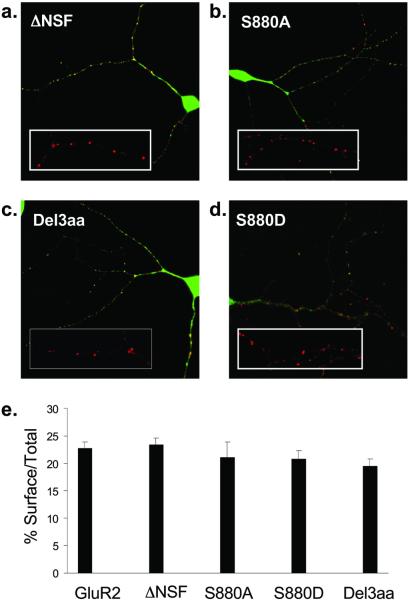
We then examined whether the surface expression of GluR2 in neurons was affected by these mutations. All of the mutant constructs formed surface puncta (Fig. 2 a–d) that colocalized with synaptophysin (data not shown), indicating their delivery to the synaptic plasma membrane. To determine whether the mutations caused quantitative effects on the degree of GluR2 surface expression, we determined the ratio of surface to total GluR2 for each cell, a measure that obviates any cell-to-cell variability in the amount of receptor protein expressed. Surprisingly, the surface expression of FLAG-GFP-GluR2(ΔNSF) was not significantly different from that of FLAG-GFP-GluR2 (wild-type, 22.7 ± 1.2%, n = 59; FLAG-GFP-GluR2(ΔNSF), 23.4 ± 1.3%, n = 25; Fig. 2 a and e). The mutant constructs that eliminate binding of GRIP/ABP alone or GRIP/ABP and PICK1 to GluR2 also showed a normal level of surface expression [FLAG-GFP-GluR2(S880A), 21.1 ± 2.8%, n = 16; FLAG-GFP-GluR2(S880D), 20.8 ± 1.5%, n = 31; FLAG-GFP-GluR2(Del3aa), 19.4 ± 1.3%, n = 30; Fig. 2 b–e]. These results demonstrate that binding of GluR2 to NSF or to its PDZ protein binding partners is not required for normal surface expression of AMPARs in the plasma membrane at synapses
Figure 2.
Surface expression of FLAG-GFP-GluR2 mutant constructs in cultured hippocampal neurons. (a–d) Examples of hippocampal neurons transfected with FLAG-GFP-GluR2 mutant constructs (green) all showing surface expressed receptors (red). (Insets) Higher power view of surface expressed puncta of FLAG-GFP-GluR2 mutant constructs. (a) FLAG-GFP-GluR2(ΔNSF). (b) FLAG-GFP-GluR2(S880A). (c) FLAG-GFP-GluR2(Del3aa). (d) FLAG-GFP-GluR2(S880D). (e) Quantification of the proportion of AMPARs on the cell surface compared with the total shows no difference between constructs (ANOVA: df = 4, MS = 79.5, F = 1.07, P = 0.37).
Interactions with NSF and PDZ Proteins Do Not Influence Constitutive Cycling of GluR2.
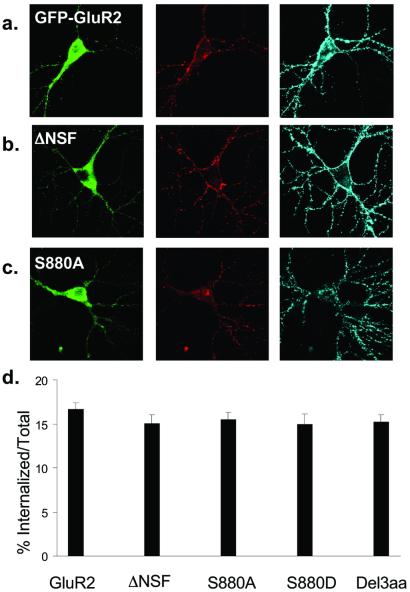
The surface expression of AMPARs is not a static process because they constitutively cycle in and out of the plasma membrane (2, 3, 12). To assess whether binding to NSF or the PDZ proteins affected the constitutive cycling of AMPARs, we labeled surface GluR2 and allowed trafficking to proceed for 30 min. A small proportion of AMPARs were observed in intracellular pools with all constructs (Fig. 3 a–c, red, Center) with numerous receptors still present at the cell surface (Fig. 3 a–c, blue, Right). These surface AMPARs consist of receptors that were not internalized in this time period and receptors that were endocytosed and then cycled back to the plasma membrane. Thus, this assay reflects the relative balance between constitutive endocytosis and recycling. Quantitation of the proportion of total GluR2 that originally was on the surface and remained internalized revealed that there was no difference between any of the mutant constructs [Fig. 3d; FLAG-GFP-GluR2, 16.6 ± 0.7%, n = 39; FLAG-GFP-GluR2(ΔNSF), 15.0 ± 1.1%, n = 23; FLAG-GFP-GluR2(S880A), 15.5 ± 0.9%, n = 17; FLAG-GFP-GluR2 (S880D), 15.0 ± 1.1%, n = 23; FLAG-GFP-GluR2(Del3aa), 15.3 ± 0.8%, n = 33]. These results suggest that the net turnover of GluR2 via constitutive cycling is unchanged by interaction with NSF, GRIP/ABP, or PICK1. They also demonstrate that the lack of difference in the surface expression of different constructs is not due to an effect on constitutive endocytosis that was balanced by an equal effect on the delivery of new AMPARs to the surface.
Figure 3.
Constitutive endocytosis of FLAG-GFP-GluR2 is not influenced by binding to cytoplasmic proteins. (a–c) Representative images of transfected cells in which constitutive endocytosis of surface-expressed AMPARs was imaged. (Left) GFP fluorescence (green). (Center) Receptors that were surface labeled and then constitutively internalized (red). (Right) Remaining surface receptors (blue). (a) FLAG-GFP-GluR2. (b) FLAG-GFP-GluR2(ΔNSF). (c) FLAG-GFP-GluR2(S880A). (d) Quantification of the proportion of the total amount of surface-expressed AMPARs that were internalized shows no significant difference between constructs (ANOVA: df = 4, MS = 12.7, F = 0.60, P = 0.67).
Interactions with NSF and GRIP/ABP Influence Regulated Endocytosis of GluR2.
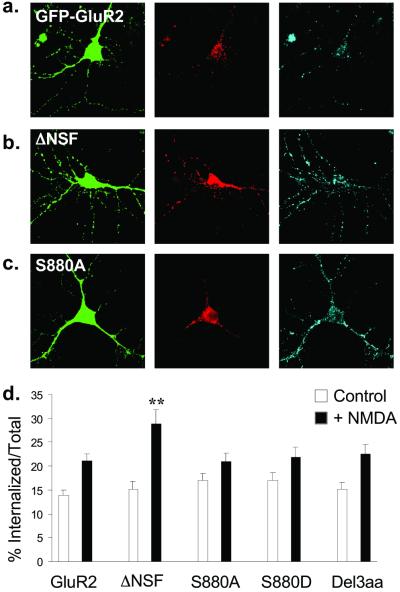
Application of exogenous AMPA or NMDA to hippocampal neurons causes a rapid endocytosis of AMPARs that shares features with LTD (3, 26). To determine whether GluR2 binding to NSF or PDZ proteins is important for this regulated endocytosis, we again compared the behavior of wild-type FLAG-GFP-GluR2 with the mutant constructs. Ten minutes after the application of NMDA (100 μM for 1 min in the presence of LY341495 and NBQX), FLAG-GFP-GluR2(ΔNSF) showed a significantly increased degree of internalization (28.9 ± 2.9%, n = 23; Fig. 4 b and d) compared with FLAG-GFP-GluR2 (21.1 ± 1.4%, n = 50; Fig. 4 a and d). NMDA also induced internalization of the PDZ binding domain mutants, but the magnitude of this internalization was not significantly different from that of FLAG-GFP-GluR2 [FLAG-GFP-GluR2(S880A), 21.0 ± 1.7%, n = 36; FLAG-GFP-GluR2(S880D), 22.0 ± 1.9%, n = 18; FLAG-GFP-GluR2(Del3aa), 22.6 ± 2.1%, n = 19; Fig. 4 c and d].
Figure 4.
NMDA induces endocytosis of FLAG-GFP-GluR2 that is regulated by interaction with NSF. (a–c) Representative images of transfected cells in which endocytosis of AMPARs was triggered by NMDA application. Panels are the same as in Fig. 3. (a) FLAG-GFP-GluR2. (b) FLAG-GFP-GluR2(ΔNSF). (c) FLAG-GFP-GluR2(S880A). (d) Quantification shows all constructs exhibit significant internalization (P < 0.05) after NMDA stimulation (filled columns) compared with control conditions (open columns). There is significantly increased NMDA-induced internalization of FLAG-GFP-GluR2(ΔNSF) (**, P < 0.01) compared with FLAG-GFP-GluR2. There is no significant difference between constructs in the absence of NMDA (open columns). ANOVA: df = 4, MS = 64.7, F = 1.21, P = 0.31.
We also examined the effects of application of AMPA (50 μM in the presence of d-APV and LY341495), which triggers greater AMPAR endocytosis than NMDA application and thereby facilitates examination of events downstream of the initial endocytosis. As expected from the previous experiment, the magnitude of the AMPA-triggered internalization of FLAG-GFP-GluR2(ΔNSF) was greater (control, 18.3 ± 1.5%, n = 31; +AMPA, 38.4 ± 2.5%, n = 31; Fig. 5 b and d) than for the wild-type FLAG-GFP-GluR2 (untreated control, 17.3 ± 1.0%, n = 55; +AMPA, 33.1 ± 1.4%, n = 60; Fig. 5 a and d). In contrast, all of the constructs that could not bind the PDZ proteins GRIP/ABP showed a decrease in the proportion of internalized receptor compared with FLAG-GFP-GluR2 [FLAG-GFP-GluR2(S880A), 28.2 ± 1.3%, n = 22; FLAG-GFP-GluR2(S880D), 25.7 ± 1.6%, n = 26; FLAG-GFP-GluR2(Del3aa), 26.8 ± 1.5%, n = 23; Fig. 5 c and d, filled bars]. Consistent with the previous assays of constitutive endocytosis, in the absence of AMPA, all these constructs showed equal levels of internalization as FLAG-GFP-GluR2 (Fig. 5d, open bars).
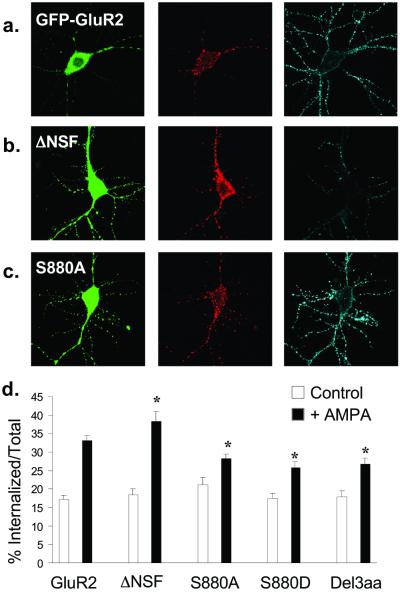
Figure 5.
AMPA-induced endocytosis of FLAG-GFP-GluR2 is regulated by interaction with cytoplasmic proteins. (a–c) Representative images of transfected cells in which AMPA-induced endocytosis of FLAG-GFP-GluR2 constructs was imaged. Panels are the same as in Fig. 3. (a) FLAG-GFP-GluR2. (b) FLAG-GFP-GluR2(ΔNSF). (c) FLAG-GFP-GluR2(S880A). (d) Quantification shows all constructs exhibit significant internalization (P < 0.005) on AMPA stimulation (filled columns) compared with control conditions (open columns). There is significantly increased AMPA-induced internalization of FLAG-GFP-GluR2(ΔNSF) (*, P < 0.05) and decreased internalization of FLAG-GFP-GluR2(S880A), FLAG-GFP-GluR2(S880D), and FLAG-GFP-GluR2(Del3aa) compared with FLAG-GFP-GluR2 (*, P < 0.05). There is no significant difference between constructs in the absence of AMPA (open columns). ANOVA: df = 4, MS = 62.4, F = 0.98, P = 0.42.
These results suggest that the interaction of GluR2 with NSF functions to limit the regulated internalization of AMPARs. On the other hand, the reduction in the internalized:total ratio exhibited by the constructs that cannot bind GRIP/ABP could result from two distinct mechanisms: either a deficit in the degree or rate of triggered endocytosis or a change in the fate of the receptors after endocytosis takes place. This latter mechanism could further involve either an increase in the rate of reinsertion of internalized receptors into the plasma membrane or an increased entry of internalized receptors into a degradative pathway. To distinguish these possibilities, we performed experiments in which, after AMPA treatment, receptors were allowed to cycle for a further hour at 37°C. During this period, no more regulated endocytosis will take place so internalized receptors will either remain in an intracellular pool, be degraded, or return to the synaptic membrane. In the absence of AMPA the internalized:total ratio for each construct did not differ significantly [FLAG-GFP-GluR2, 14.3 ± 0.8%, n = 74; FLAG-GFP-GluR2(S880A), 13.2 ± 1.9%, n = 19; FLAG-GFP-GluR2(S880D), 13.5 ± 1.5%, n = 23; FLAG-GFP-GluR2(Del3aa), 15.0 ± 1.2%, n = 21; Fig. 6, open bars). After the 1 h incubation period after AMPA treatment there was less FLAG-GFP-GluR2 in the intracellular pool compared with the AMPA treatment alone (Fig. 5d), but a significant proportion still remained (FLAG-GFP-GluR2, 19.3 ± 1.0%, n = 75, Fig. 6). However, all of the PDZ domain mutant constructs showed significantly less immunoreactivity in the intracellular pool [FLAG-GFP-GluR2(S880A), 14.8 ± 1.9%, n = 20; FLAG-GFP-GluR2(S880D), 15.9 ± 1.3%, n = 25; FLAG-GFP-GluR2(Del3aa), 15.9 ± 1.1%, n = 27; Fig. 6, filled bars). Indeed, after this period of recycling, the PDZ binding domain mutants showed no significant difference in the ratio of internalized to total receptors compared with the control, untreated condition. These results suggested that GRIP/ABP is necessary to stabilize an intracellular pool of receptors.
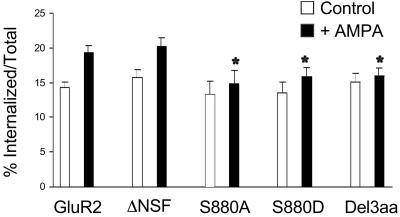
Figure 6.
The intracellular pool of internalized FLAG-GFP-GluR2 unable to interact with GRIP/ABP is dissipated 1 h after stimulation. One hour after AMPA treatment, FLAG-GFP-GluR2 and FLAG-GFP-GluR2(ΔNSF) still show a significant increase in the amount of internalized receptors compared with untreated cells (P < 0.05) whereas the mutants unable to interact with GRIP/ABP or PICK1 do not [FLAG-GFP-GluR2(S880A), P = 0.57; FLAG-GFP-GluR2(S880D), P = 0.25; FLAG-GFP-GluR2(Del3aa), P = 0.60). There is no significant difference in magnitude of AMPAR-induced internalization of FLAG-GFP-GluR2(ΔNSF) (P = 0.64), but decreased internalization of FLAG-GFP-GluR2(S880A), FLAG-GFP-GluR2(S880D), and FLAG-GFP-GluR2(Del3aa) compared with FLAG-GFP-GluR2 (*, P < 0.05). There is no significant difference between constructs in the absence of AMPA (open bars). ANOVA: df = 4, MS = 30.4, F = 0.61, P = 0.65).
We also examined whether the lack of NSF binding had an effect on events subsequent to the triggered internalization of AMPARs by performing this same assay on cells expressing FLAG-GFP-GluR2(ΔNSF). After allowing recycling for 1 h after AMPA treatment, there was still a significant increase in the ratio of internalized:total receptors (control, 15.7 ± 1.4%, n = 40; +AMPA, 20.2 ± 1.2%, n = 20; Fig. 6), which was not significantly different from that obtained with FLAG-GFP-GluR2. This result suggests that the effect of NSF binding is only on the initial internalization of receptors and does not play a critical role in AMPAR trafficking after endocytosis.
Discussion
The detailed molecular mechanisms by which AMPAR expression at synapses is controlled are of great interest because of their importance for synaptic function and plasticity. A number of proteins that directly interact with specific AMPAR subunits have been identified, and evidence has been presented that these may play important, albeit undefined, roles in AMPAR trafficking and targeting (6–8). In this study, we have focused on the role of NSF, GRIP/ABP, and PICK1, the set of proteins known to interact with the key AMPAR subunit GluR2.
Previous work on the role of NSF found that interfering with the NSF–GluR2 interaction by using peptides caused a fairly rapid decrease in synaptic strength (10–12) and, with chronic exposure, an almost complete loss of surface AMPARs (12, 13). However, it was not clear whether this was due to an impairment in the delivery of AMPARs to the synaptic plasma membrane or a decrease in the stability of AMPARs in the membrane after they were successfully delivered. Our results strongly support the latter of these explanations. Furthermore, they suggest that NSF binding is primarily important when synapses are stimulated because only the regulated but not the constitutive cycling of GluR2 was affected by mutating its NSF binding domain. These latter results correlate well with the electrophysiological observation that the rundown of excitatory postsynaptic currents caused by the peptide that interferes with the NSF–GluR2 interaction is activity dependent (12, 13). Binding of NSF to GluR2 thus stabilizes a population of AMPARs in the plasma membrane and makes them resistant to regulated endocytosis.
Preventing the interaction of GluR2 with GRIP/ABP had no effect on its surface expression or constitutive internalization but did reduce the amount of protein that remained internalized after application of AMPA to trigger endocytosis. Furthermore, if after the triggered endocytosis we allowed AMPAR trafficking to continue for an hour, while a significant portion of wild-type GluR2 remained in intracellular pools, the mutant GluR2 constructs that could not bind GRIP/ABP did not. The most straightforward explanation for these results is that, after regulated endocytosis, binding to GRIP/ABP stabilizes the internalized receptors in an intracellular pool and prevents them from being recycled back to the plasma membrane or entering a degradative pathway. This interpretation is consistent with a recent electrophysiological study (17) in which peptides that interfere with the GluR2–GRIP/ABP interaction caused a run-up of excitatory postsynaptic currents and inhibited LTD, presumably because the AMPARs that were internalized by the LTD induction protocol were not stabilized in an intracellular pool and therefore were able to cycle back to the synaptic membrane.
Some of our results differ from recent reports that also examined the effects of similar or identical mutations in GluR2 on its delivery to synapses. In one study, mutating the GRIP/ABP binding domain caused a decrease in the surface expression of a myc-tagged GluR2 construct expressed in hippocampal cultures by using Sindbis virus-mediated gene delivery (16). We find that interactions with GRIP/ABP or PICK1 are not necessary for the surface expression of AMPARs under our culture conditions, consistent with electrophysiological investigation in acute hippocampal slices (17). Another study, using Sindbis virus to express mutant forms of GluR2 in hippocampal slice cultures and electrophysiological methods to detect their expression at synapses, found that synaptic delivery of GluR2 appeared to be completely prevented by mutations that interfered with binding to NSF or GRIP/ABP (27). One possible explanation for this difference in results is that the construct used in this previous study required an electrophysiological tag to identify delivered receptors, making the receptors permeable to Ca2+, a key regulator of receptor cycling (2, 3). An increased Ca2+ influx may result in the endocytosis of delivered receptors, making their detection difficult. To directly test this possibility, we examined the surface expression of the Ca2+-permeable form of GluR2, FLAG-GFP-GluR2(Q), which also contained the ΔNSF mutation. To our surprise, this construct exhibited clear and robust punctate surface expression in dendrites (data not shown). Thus, we are unable to provide a definitive explanation for the difference between our and previous results except to note that, because surface expression of AMPARs is highly dependent on levels of activity (26, 28, 29), differences in culture conditions (e.g., age, density) and transfection techniques could have influenced the steady-state equilibrium between delivery and removal of the recombinant AMPARs. Our results clearly demonstrate that interactions with NSF and GRIP/ABP are not required for surface expression of GluR2 and that it is premature to reach definitive conclusions about the functions of these proteins in the synaptic delivery and/or synaptic stabilization of AMPARs.
Our results do not reveal a specific function for PICK1 in hippocampal neurons because we were unable to generate a construct that retains GRIP/ABP binding while eliminating the ability of PICK1 to bind. A recent study demonstrated that expression of PICK1 elicits the targeting of protein kinase C (PKC) to AMPARs, resulting in their phosphorylation and reduced surface expression (30). Another study found that loading cells with peptides that disrupt the GluR2–PICK1 interaction caused an increase in synaptic responses and inhibited LTD (20), suggesting that PICK1 may be important for AMPAR endocytosis. However, a similar electrophysiological study suggested a role of PKC phosphorylation and PICK1 interaction in the opposite movement of AMPARs from intracellular stores to the synaptic membrane (17). Thus, in the hippocampus, PICK1 may play a more generalized role in controlling AMPAR trafficking to and away from synapses.
Our results using mutant GluR2 constructs suggest an important role for the binding of NSF to GluR2-containing AMPARs in stabilizing these receptors in the synaptic plasma membrane and impeding their regulated endocytosis. On the other hand, the most straightforward explanation for the consequences of preventing GluR2 interactions with GRIP/ABP is that GRIP/ABP binding is important for stabilizing an intracellular pool of AMPARs after they have been internalized. Neither interaction appears to be critical for the control of the constitutive cycling of synaptic AMPARs, lending further support to the idea that there are two distinct pools of synaptic AMPARs: one that can be regulated by activity and one that serves a maintenance function (2, 3, 17, 22, 27).
Acknowledgments
We thank Tina Lizama for expert assistance in preparation of the hippocampal cultures and Jerron Fisher for assistance with molecular biology. This work was supported by grants from the National Institutes of Health (to R.C.M.) and a Wellcome Prize Traveling Research Fellowship (to S.P.B.).
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid
- NMDA
N-methyl-d-aspartate
- AMPAR
AMPA receptor
- APV
2-amino-5-phosphonovaleric acid
- NSF
N-ethylmaleimide-sensitive fusion
- ABP
AMPAR binding protein
- GRIP
glutamate receptor-interacting protein
- GFP
green fluorescent protein
- LTD
long-term depression
- NBQX
2,3-dihydroxy-6-nitro-7-sulfamoylbenzo[f]quinoxaline
- MS
mean square
- PICK1
protein interacting with C-kinase-1
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Malenka R C, Nicoll R A. Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- 2.Malinow R, Mainen Z F, Hayashi Y. Curr Opin Neurobiol. 2000;10:352–357. doi: 10.1016/s0959-4388(00)00099-4. [DOI] [PubMed] [Google Scholar]
- 3.Carroll R C, Beattie E C, Von Zastrow M, Malenka R C. Nat Rev Neurosci. 2001;2:315–324. doi: 10.1038/35072500. [DOI] [PubMed] [Google Scholar]
- 4.Geiger J R, Melcher T, Koh D S, Sakmann B, Seeburg P H, Jonas P, Monyer H. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 5.Wisden W, Seeburg P H. Curr Opin Neurobiol. 1993;3:291–298. doi: 10.1016/0959-4388(93)90120-n. [DOI] [PubMed] [Google Scholar]
- 6.Braithwaite S P, Meyer G, Henley J M. Neuropharmacology. 2000;39:919–930. doi: 10.1016/s0028-3908(99)00171-9. [DOI] [PubMed] [Google Scholar]
- 7.Garner C C, Nash J, Huganir R L. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- 8.Sheng M, Sala C. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Rothman J E. Nature (London) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 10.Nishimune A, Isaac J T, Molnar E, Noel J, Nash S R, Tagaya M, Collingridge G L, Nakanishi S, Henley J M. Neuron. 1998;21:87–97. doi: 10.1016/s0896-6273(00)80517-6. [DOI] [PubMed] [Google Scholar]
- 11.Song I, Kamboj S, Xia J, Dong H, Liao D, Huganir R L. Neuron. 1998;21:393–400. doi: 10.1016/s0896-6273(00)80548-6. [DOI] [PubMed] [Google Scholar]
- 12.Luscher C, Xia H, Beattie E C, Carroll R C, von Zastrow M, Malenka R C, Nicoll R A. Neuron. 1999;24:649–658. doi: 10.1016/s0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 13.Noel J, Ralph G S, Pickard L, Williams J, Molnar E, Uney J B, Collingridge G L, Henley J M. Neuron. 1999;23:365–376. doi: 10.1016/s0896-6273(00)80786-2. [DOI] [PubMed] [Google Scholar]
- 14.Dong H, O'Brien R J, Fung E T, Lanahan A A, Worley P F, Huganir R L. Nature (London) 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 15.Xia J, Zhang X, Staudinger J, Huganir R L. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 16.Osten P, Khatri L, Perez J L, Kohr G, Giese G, Daly C, Schulz T W, Wensky A, Lee L M, Ziff E B. Neuron. 2000;27:313–325. doi: 10.1016/s0896-6273(00)00039-8. [DOI] [PubMed] [Google Scholar]
- 17.Daw M I, Chittajallu R, Bortolotto Z A, Dev K K, Duprat F, Henley J M, Collingridge G L, Isaac J T. Neuron. 2000;28:873–886. doi: 10.1016/s0896-6273(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 18.Matsuda S, Launey T, Mikawa S, Hirai H. EMBO J. 2000;19:2765–2774. doi: 10.1093/emboj/19.12.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia J, Chung H J, Wihler C, Huganir R L, Linden D J. Neuron. 2000;28:499–510. doi: 10.1016/s0896-6273(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 20.Kim C-H, Chung H J, Lee H-K, Huganir R L. Proc Natl Acad Sci USA. 2001;98:11725–11730. doi: 10.1073/pnas.211132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lissin D V, Gomperts S N, Carroll R C, Christine C W, Kalman D, Kitamura M, Hardy S, Nicoll R A, Malenka R C, von Zastrow M. Proc Natl Acad Sci USA. 1998;95:7097–7102. doi: 10.1073/pnas.95.12.7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carroll R C, Lissin D V, von Zastrow M, Nicoll R A, Malenka R C. Nat Neurosci. 1999;2:454–460. doi: 10.1038/8123. [DOI] [PubMed] [Google Scholar]
- 23.Burnashev N, Monyer H, Seeburg P H, Sakmann B. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda S, Mikawa S, Hirai H. J Neurochem. 1999;73:1765–1768. doi: 10.1046/j.1471-4159.1999.731765.x. [DOI] [PubMed] [Google Scholar]
- 25.Chung H J, Scannevin R H, Zhang X, Huganir R L. J Neurosci. 2000;20:7258–7267. doi: 10.1523/JNEUROSCI.20-19-07258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beattie E C, Carroll R C, Yu X, Morishita W, Yasuda H, von Zastrow M, Malenka R C. Nat Neurosci. 2000;3:1291–1300. doi: 10.1038/81823. [DOI] [PubMed] [Google Scholar]
- 27.Shi S, Hayashi Y, Esteban J A, Malinow R. Cell. 2001;105:331–343. doi: 10.1016/s0092-8674(01)00321-x. [DOI] [PubMed] [Google Scholar]
- 28.Ehlers M D. Neuron. 2000;28:511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 29.Lin J W, Ju W, Foster K, Lee S H, Ahmadian G, Wyszynski M, Wang Y T, Sheng M. Nat Neurosci. 2000;3:1282–1290. doi: 10.1038/81814. [DOI] [PubMed] [Google Scholar]
- 30.Perez J L, Khatri L, Chang C, Srivastava S, Osten P, Ziff E B. J Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]