Abstract
The P2X7 receptor, an ATP-gated ion channel which belongs to the P2X receptor family, plays critical roles in recognizing extracellular adenosine 5’-triphosphate (ATP) and is widely expressed in most tumor cells as well as inflammatory cells. Previously, the P2X7 receptor has been demonstrated to modulate the progression of various malignancies, including glioblastoma, pancreatic cancer, lung cancer, leukemia, and lymphoma. However, the biological function and prognostic values of P2X7 receptor in hepatocellular carcinoma remain to be determined. Here, we investigated the expression level of P2X7 receptor in patients with hepatocellular carcinoma. Then MTS and EdU assays were carried out to study the role of P2X7 receptor blockade in the proliferation of hepatocellular carcinoma cells. In addition, the underlying mechanism was further elucidated by bulk RNAseq. Compared to the control group, the P2X7 receptor was significantly up-regulated in the hepatocellular carcinoma group. Interestingly, A740003 and A438079, two selective antagonists at P2X7 receptor, significantly blocked Ca2+ influx and decreased the proliferative rate of hepatocellular carcinoma cells. Furthermore, the expression level of chondroitin sulfate synthase 1 (CHSY1), an enzyme that mediates the polymerization step of chondroitin sulfate, was reduced by both A740003 and A438079. In conclusion, inhibition of the P2X7 receptor attenuated the proliferation of hepatocellular carcinoma cells, and this process was largely modulated by CHSY1. Thus, our findings reveal a previously unknown role for P2X7 receptor in the proliferation of hepatocellular carcinoma cells and imply that the P2X7 receptor may represent a new target for the treatment of hepatocellular carcinoma.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11302-024-10064-5.
Keywords: Hepatocellular carcinoma, Purinergic receptor, P2X7, Proliferation, CHSY1
Introduction
Hepatocellular carcinoma (HCC) is the most common form of liver cancer, accounting for ~ 90% cases and represents a major global health-care challenge [1]. Despite currently available treatment modalities, such as surgery, radiotherapy, chemotherapy, and immunotherapy, HCC remains a major medical challenge that has not been conquered [2, 3]. Thus, further research to find better methods for treating HCC is necessary. Nucleotides are extracellular messengers, and they play important roles in accelerating the progression of malignant by activating purinergic receptors on the cell membrane [4]. The P2 purinergic receptors (P2Rs), mainly responsible for recognizing extracellular purine or pyrimidine nucleotides, are classified into P2YR and P2XR family [5]. Accumulating evidence has revealed that P2YRs are involved in driving cancer cell proliferation by increasing the intracellular Ca2+ concentration and activating the PI3K-AKT and ERK-MAPK signaling pathways [6]. Furthermore, P2XRs have been reported to promote VEGF-dependent angiogenesis and PI3K-GSK3β-dependent proliferation of various malignancies [7–9]. However, little is known regarding the role of P2Rs in the occurrence and development of HCC. Understanding the cellular and molecular basis underlying P2R signaling in HCC progression can help to develop promising approaches for patients suffering from HCC.
The P2X7 receptor is a member of ATP-gated ionotropic P2X receptor family, which is expressed predominantly on immunocytes and some tumor cells [10]. P2X7 receptor activity is largely dependent on the concentration of extracellular ATP. Transient recognition of extracellular ATP (0.05–1 mM) induces K+ efflux while Na+ and Ca2+ influx to the cytoplasm rapidly, but long activation of the P2X7 receptor could form pores large enough to allow the non-selective movements of molecules up to 900 Da [11, 12]. Previously, we have reported that the P2X7 receptor could increase the proliferative rate of AML (acute myeloid leukemia) cells and amplify the amount of leukemia stem cells by up-regulating the expression level of Pre-B-Cell leukemia homeobox 3 (PBX3) [13]. Moreover, we also demonstrated that the P2X7 receptor can accelerate the progression of DLBCL (diffused large B-cell lymphoma) cells by up-regulated pyrimidine or purine metabolic activity mediated by carbamoyl-phosphate synthase 1 (CPS1) [14]. However, the biological function and molecular mechanism of P2X7 receptor in the progression of HCC are poorly understood. To date, only two original studies attempted to preliminarily elucidate the role of P2X7 receptor in HCC. A study published in 2015 reported that the expression level of P2X7 receptor in peritumor section was much higher than paired intratumor section, and there was a positive correlation between the increased expression of P2X7 receptor and the poor prognosis of HCC patients from China [15]. Additionally, another study from the Chinese Han population revealed that 3 polymorphisms of P2RX7 (946G > A, 1068G > A, and 1513A > C) were significantly associated with HCC susceptibility [16]. Taken together, these observations provide limited evidence that the P2X7 receptor might get involved in the progression of HCC. However, the potential role of P2X7 receptor signaling in the proliferation of HCC cells, particular the underlying molecular mechanism, needs further elucidation.
In this study, we assessed the expression level of P2RX7 in patients with HCC and confirmed that the expression level of P2RX7 was significantly up-regulated. Hence, we speculate that the P2X7 receptor signaling might modulate the proliferation of HCC cells. Recent studies have shown that the administration of the P2X7 receptor antagonists A740003 or A438079 could significantly decrease the proliferative rate of tumor cells. On the one hand, A740003 inhibited the proliferation of glioblastoma by down-regulating the activity of PI3K-GSK3β-VEGF signaling [17]. On the other hand, A438079 reduced the expression levels of caspase-1, caspase-3, CDC25C, and p21, leading to an inhibited growth of pancreatic cancer [18]. To test our hypothesis, we chose A740003 and A438079 to assess whether the P2X7 receptor signaling blockade could inhibit the proliferative rate of HCC cells. In addition, we set up a bulk RNAseq experiment to investigate the possible molecular mechanism involved in above process. Our findings indicated that inhibiting the P2X7 receptor signaling significantly decreased the proliferation rate of HCC cells. Moreover, chondroitin sulfate synthase 1 (CHSY1), an enzyme with dual glucuronyltransferase and galactosaminyltransferase activity, was essential for P2X7 receptor-mediated proliferation of HCC cells.
Materials and methods
Cell culture
The human HCC cell line HCC-LM3 was obtained from the type culture collection of the Chinese academy of sciences (Shanghai, China), and HepG2 was obtained from the American type culture collection (Rockville, MD, USA). HCC-LM3 and HepG2 cells were cultured in DMEM medium (Gibco, CA, USA) supplemented with 10% fetal bovine serum (Gibco, CA, USA), and maintained in a humidified cell incubator at 37 °C and 95% humidity with 5% CO2. After reaching 50% confluence, HCC cells were incubated with the P2X7 receptor antagonists A740003 or 438,079 (1, 5, 10, 20 to 50 μM) (Medchemexpress, NJ, USA). Treatments were performed at indicated timepoints (24 h, 48 h or 72 h) for each experimental purpose.
Data acquisition and processing
The public RNAseq datasets of patients with HCC were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Dataset GSE22058 contains 69 HCC patient samples and 20 normal samples [19], GSE36376 contains 240 HCC patient samples and 193 normal samples [20], GSE14520 contains 225 HCC patient samples and 220 normal samples [21], GSE25097 contains 268 HCC patient samples and 243 normal samples [22]. Additional RNAseq datasets of patients with HCC patients were downloaded from the International Cancer Genome Consortium (ICGC) database (https://dcc.icgc.org/). The dataset ICGC-LIRI-JP contains 212 HCC patient samples and 177 normal samples [23]. Above datasets were further analyzed to compare the gene expression level of P2RX7 between the normal group and the HCC group, by using unpaired student t test.
The Kaplan Meier Plotter online database (www.kmplot.com) was used for analyzing the survival of patients with HCC [24]. Online analysis of overall survival (OS) and relapse-free survival (RFS) was performed to evaluate the relationship between P2RX7 expression and patient prognosis. Patients from the liver hepatocellular carcinoma dataset (n = 371) in the Pan-cancer RNAseq unit were used for the above analysis.
The Human Protein Atlas (HPA) (https://www.proteinatlas.org/) is a worldwide public database for the comprehensive analysis of human protein, providing data from cells, tissues, and organs that are mainly used to assess the expression level of specific proteins [25]. The expression level of P2X7 receptor protein in HCC tissue and normal liver tissue was assessed by HPA databases. Typical immunohistochemical images of P2X7 receptor protein in normal liver tissues and HCC tissues were obtained by staining with the antibody HPA042013.
Moreover, the TCGA-LIHC dataset was downloaded and further analyzed via the Xena platform [26], to determine the correlation of P2RX7 expression and CHSY1 expression in patients with HCC (n = 424), using Spearman correlation.
Measurement of intracellular free Ca2+
Two categories of fluorescent Ca2+ indicators have been developed: single excitation wavelength indicators (Fluo‐4 and Fluo‐3) or dual‐wavelength dyes (Fura‐2 or Indo‐1) [27]. Through single wavelength indicators have a high quantum yield, the main limitation of single wavelength indicators is that they are unable to provide quantitative Ca2+ data because the emission intensities are largely influenced by dye concentration or photobleaching during imaging [28]. Fura‐2 is a ratiometric fluorescent Ca2+ indicator that was developed as an improved alternative to the Ca2+ indicator [29]. Compared to Indo-1, Fura-2 has a larger dynamic range between Ca2+ bound and free states and is more resistant to photobleaching [30, 31]. Different from the salt forms, the acetoxymethyl (AM) esters of Fura-2 can passively diffuse across cell membranes, enabling researchers to avoid the use of invasive loading techniques. In this study, we chose Fura-2AM (Medchemexpress, NJ, USA) as a fluorescent indicator to measure the intracellular free Ca2+ concentration. Firstly, HCC cells were treated with 10 μM Fura-2AM for 20 min at 37 °C, then cells were washed twice with Locke's solution (Gibco, CA, USA). Next, HCC cells were resuspended in Locke's solution at 1 × 106 cells/ml. The cell suspension was transferred into a 96-well plate and incubated with Locke's solution, A740003 (20 μM), or A438079 (20 μM) for 5 min. Finally, the value of fluorescence intensities at 510 nm which was separately excited by laser at 340 nm and 380 nm, were recorded by SpectraMax iD5 (Molecular Devices, CA, USA) every 12 s. Bz-ATP solution (50 μM) was injected at the indicated time point. The value of intracellular free Ca2+ was calculated as fluorescence intensities of Em 510—Ex 340 / fluorescence intensities of Em 510—Ex 380.
Cell proliferation assay
Cell proliferation was analyzed by MTS assay (Promega, WI, USA). This assay is used to quantify cellular proliferation and is based upon the cleavage of the yellow 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) to purple formazan by metabolic active cells. According to the manufactures’ instruction, HepG2 or HCC-LM3 cells were transferred into 96-well plates at an initial concentration of 5 × 103 cells/well (100 μl volume/well), then incubated for 4 h (indicated starting point). Next, the cells were treated with PBS, 20 μM A740003, or 20 μM A438079 respectively and cultured for 72 h. Every 24 h (at 24 h, 48 h and 72 h timepoints), MTS solution (20 μl/well) was supplied to culture systems and incubated for 2 h in a humidified cell incubator at 37 °C and 95% humidity with 5% CO2, then the OD490 value was measured by SpectraMax iD5. Background absorbance was first subtracted using a set of wells containing medium only. In dose-depend experiment, the HepG2 and HCC-LM3 cells were treated with PBS, A740003 or A438079 (1, 5, 10, 20 or 50 μM) and cultured for 48 h, then MTS assay was performed. In addition, HepG2 cells were incubated with A740003 or A438079 (20 μM), combined with or without recombinant SHH protein (0.4 mg/ml), or combined with or without 2 mM SHH inhibitor vismodegib (Medchemexpress, NJ, USA), and cultured for 48 h, then MTS assay was performed. All above experiments were performed in triplicate.
EdU incorporation assay
BeyoClick™ EdU cell proliferation kit (Beyotime, Shanghai, China) was used for performing the EdU incorporation assay. Firstly, after administration of A740003 or A438079, the HCC cells were bathed in EdU solution (50 μM) and incubated for 2 h. Next, HCC cells were fixed and permeabilized, then labeled with Alexa Fluor 647 azide. Last, the nuclear DNA of HCC cells were labeled by DAPI (10 μM). Fluorescent microscope Axio Observer3 (Zeiss, Oberkochen, Germany) was used to capture images. All the images were quantified by ImageJ (NIH) software 1.52a, and the percentage of EdU+ cell count / DAPI+ cell count was defined as the proliferation rate.
cDNA synthesis and real time PCR
RNAex Pro reagent (AGbio, Hunan, China) was used for cell lysis, and total RNA isolation was carried out by following the supplier’s instructions. PrimeScript™ 1st Strand Synthesis System (TAKARA, Tokyo, Japan) was used for reverse transcription. StepOne real-time PCR system (Applied Biosystems, CA, USA) was used for realtime PCR. To quantify the expression level of target genes, the ΔΔCt method [ΔΔCt = (CtTARGET − CtGAPDH) sample − (CtTARGET − CtGAPDH) calibrator] was applied. The sequences of primers used in this study were listed in Table 1.
Table 1.
Primers used in this study
| Gene | Sequence |
|---|---|
|
RN7SL1 (RNA component of signal recognition particle 7SL1) |
Forward 5’- GCTACTCGGGAGGCTGAGGCT -3’ Reverse 5’- TATTCACAGGCGCGATCC -3’ |
|
B3GALT6 (Beta-1,3-Galactosyltransferase 6) |
Forward 5’- GACGCCTACGAAAACCTCAC -3’ Reverse 5’- CCTTGAGCACGAACTCGAAG -3’ |
|
IGF2 (Insulin Like Growth Factor 2) FLNA (Filamin A) CHSY1 (Chondroitin Sulfate Synthase 1) GAPDH (glyceraldehyde-3-phosphate dehydrogenase) |
Forward 5’- CTTGGACTTTGAGTCAAATTGGCCT-3’ Reverse 5’- GAGGAGCCAGTCTGGGTTGTTGCTA-3’ Forward 5’- ACTGTTTCTAGCCTTCAGGAG -3’ Reverse 5’- GCACAGCATACTTATCTTGGTC -3’ Forward 5’- GGCACGCGTATTTACAGCAG-3’ Reverse 5’- TTGTGCTCACTCTTCGACCC-3’ Forward 5’- GAAGGTGAAGGTCGGAGTC-3’ Reverse 5’- GAAGATGGTGATGGGATTTC-3’ |
RNA sequencing and bioinformatic analysis
HepG2 cells were incubated with PBS, A740003 (20 μM), or A438079 (20 μM) and cells were harvested after 48 h. All samples were sent to Benagen (Hubei, China), and RNAseq was performed following standard protocols. Illumina NovaSeq platform was used for sequencing the sample libraries. Then Benagen performed the primary standard bioinformatics analysis. All the RNAseq raw data of HepG2-control, HepG2-A740003, and HepG2-A438079 were uploaded and available in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE268508. Different expression genes (DEGs) were defined as genes with fold change (FC) > 2, while false discovery rate (FDR) < 0.05. In addition, the gene set enrichment analysis (GSEA) and the KEGG pathway analysis were performed by Benagencloud tools, which was provided with a time-limited permission by Benagen. The p value was the main parameter for the infiltration of KEGG pathways.
Statistical analysis
Data are presented as the mean ± SD. Two-tailed unpaired Student’s t-tests were used to compare the differences between two groups. GraphPad 7 (San Diego, CA, USA) was mainly used to perform the statistical analyses. The results were defined as statistically significant when p value < 0.05.
Results
The P2X7 receptor is highly expressed in HCC patients
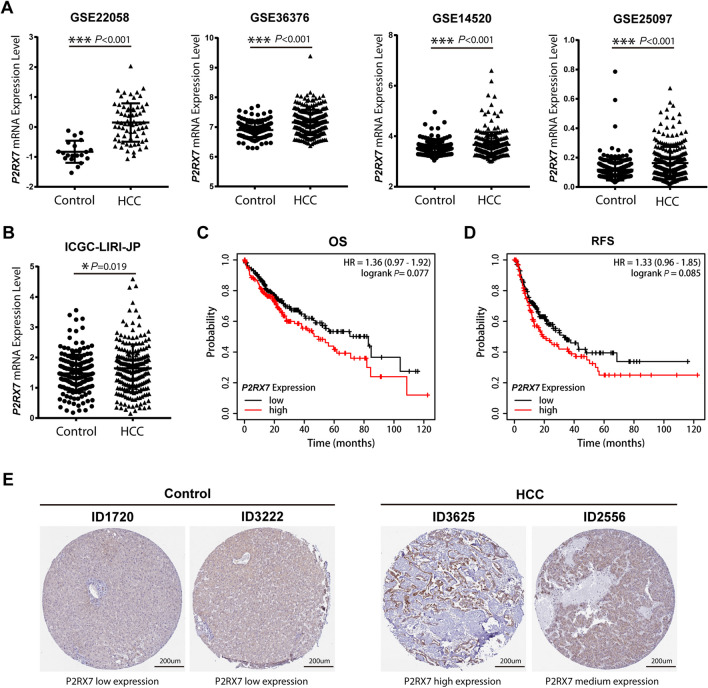
Highly expressed genes in HCC samples may predict what accelerates the progression of HCC. Firstly, the expression level of P2RX7 was analyzed with GEO datasets GSE22058, GSE36376, GSE14520, and GSE25097 [19–22]. Compared to normal liver tissues, the expression level of P2RX7 was significantly increased in HCC patient samples (Fig. 1A). Moreover, in the ICGC-LIRI-JP subset from the ICGC database [23], P2RX7 was significantly up-regulated in HCC samples when compared to normal liver tissues (Fig. 1B). Furthermore, we analyzed the OS and RFS of 470 patients with HCC from the Kaplan–Meier Plotter database [24]. The OS rate (HR = 1.36, 95% CI: 0.97–1.92, p = 0.077) and RFS rate (HR = 1.33, 95% CI: 0.96–1.85, p = 0.085) in the P2X7-high expressed group tended to be lower, but the differences were not significant (Fig. 1C, D). Furthermore, the typical immunohistochemical images of P2X7 receptor in normal liver tissue and HCC tissues which were downloaded from the HPA database were analyzed [25]. The P2X7 receptor was expressed at lower levels in the samples of normal liver tissue, while high and medium levels were expressed in the samples of patients with HCC (Fig. 1E). The above results revealed that the P2X7 receptor was highly expressed in HCC patients, and this may correlate with the worse prognosis of patients with HCC.
Fig. 1.
Expression level of the P2X7 receptor in patients with HCC. The expression level and prognostic value of P2X7 receptor in HCC patients were calculated with data which were downloaded from the GEO database, the ICGC database, the Kaplan–Meier Plotter database, and the Human Protein Atlas (HPA) database respectively. A The gene expression level of P2RX7 between normal liver tissue and HCC samples in datasets GSE22058 (normal n = 20, HCC n = 69), GSE36376 (normal n = 193, HCC n = 240), GSE14520 (normal n = 220, HCC n = 225), GSE25097 (normal n = 243, HCC n = 268) from the GEO database. B The gene expression level of P2RX7 between normal liver tissue (n = 177) and HCC samples (n = 212) in ICGC-LIRI-JP dataset from the ICGC database. C, D Prognostic value of P2RX7 for OS (C) and RFS (D) of patients with HCC in the Kaplan–Meier Plotter database. E Typical immunohistochemical staining of P2X7 receptor from the HPA database was shown, normal tissues were left panel (ID 1720 and ID 3222, P2X7 low expression), HCC samples were right panel (ID 3625 and ID 2556, P2X7 high expression and middle expression respectively). The data of gene expression level were calculated by unpaired Student's t-test. Each dot represents a sample from normal individual or patient with HCC
The proliferation rate of HCC cells was attenuated by P2X7 receptor inhibitors
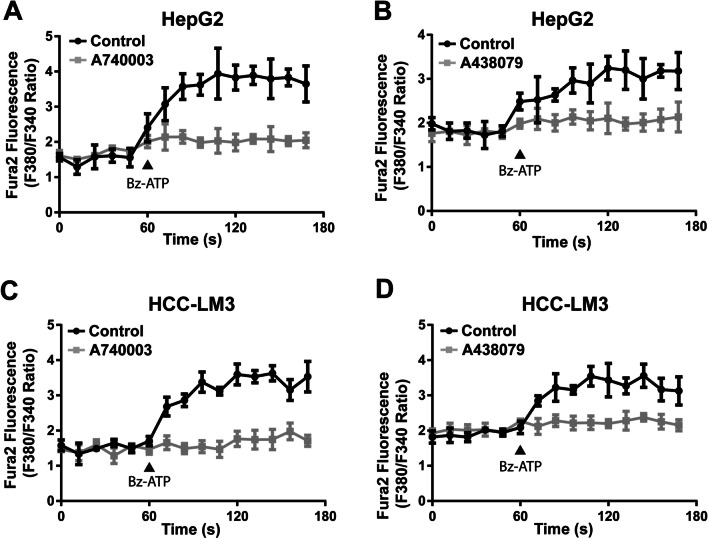
Selective P2X7 receptor antagonists, A740003 and A438079, have been demonstrated to inhibit P2X7 receptor function in previous studies [13, 32]. The HepG2 and HCC-LM3 cells were exposed to Bz-ATP, a well-recognized P2X7 receptor agonist, which increased the concentration of intracellular free Ca2+, while pretreatment with A740003 or A438079 completely blocked this effect (Fig. 2).
Fig. 2.
The P2X7 receptor mediated Ca2+ influx was blocked by antagonists in HCC cell lines. The Ca2+ influx function of P2X7 receptor was attenuated by A740003 in HepG2 cells (A) or HCC-LM3 cells (C), and was attenuated by A438079 in HepG2 cells (B) or HCC-LM3 cells (D). HepG2 cells and HCC-LM3 cells were pre-incubated with control Lock’s solution, 20 μΜ A740003, or 20 μΜ A438079 for 5 min before the supplement of Bz-ATP. The administration of 50 μΜ Bz-ATP was labeled with a triangle symbol in panels. At each timepoint, the fluorescence value of Fura-2AM was recorded as emission value in wavelength 510 nm which was excited by wavelength 340 nm or wavelength 380 nm separately. The level of Ca2+ influx was indicated as excitation ratio (340/380). Data were quantified and presented as the mean ± SD from three independent experiments
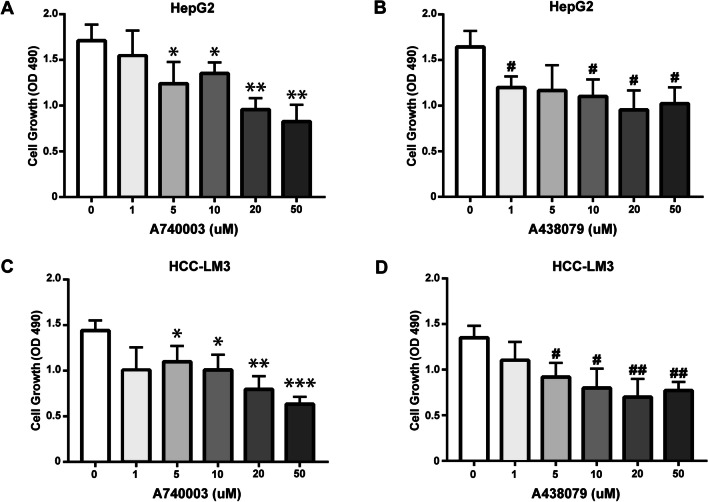
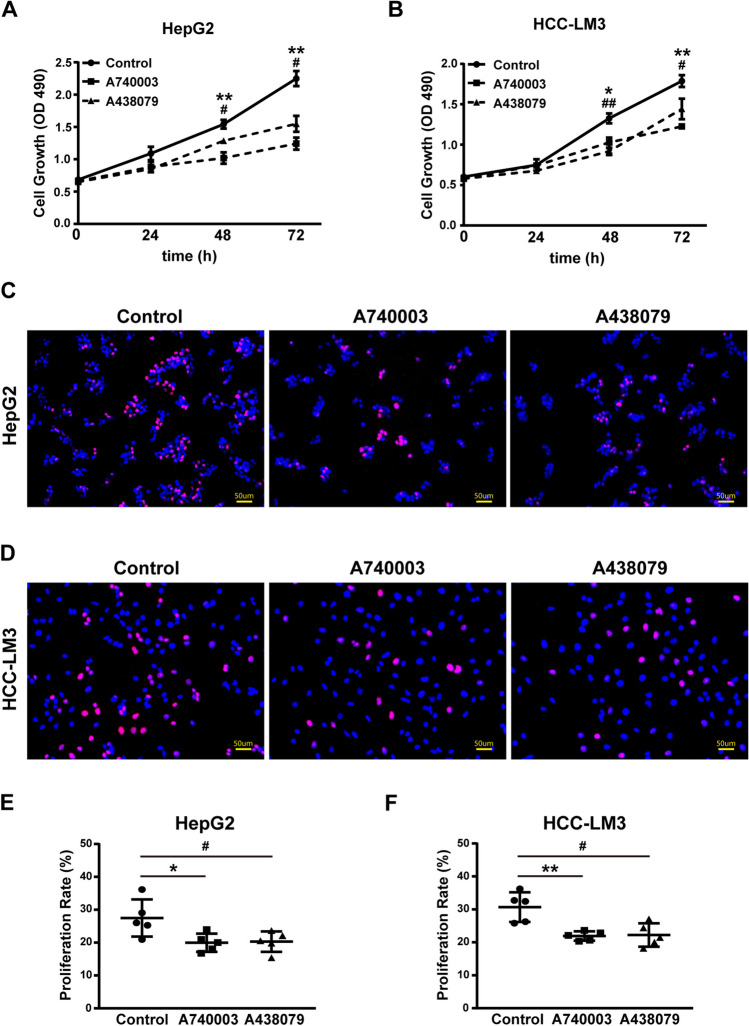
To further characterize the roles of P2X7 receptors in the proliferation of HCC cells, an MTS assay was carried out. A740003 and A438079 significantly inhibited the proliferation of HepG2 cells (Figs. 3A, B and 4A) and HCC-LM3 cells (Figs. 3C, D and 4B) in a concentration- and time-dependent manner. To confirm this effect, we performed the EdU incorporation assay for further validation. Administration of A740003 or A438079 significantly decreased the proliferative rate of HCC cells as evidenced by the lower incorporation level of EdU (Fig. 4C-F). Taken together, these findings show that the blockade of P2X7 receptor can effectively reduce the proliferative rate of HCC cells.
Fig. 3.
Dose-dependent effect of P2X7 receptor on the proliferation of HCC cells. Dose-dependent (48 h) effects of A740003 or 438,079 on the proliferation of HCC cells were analyzed by MTS assay. A, B Effects of gradient A740003 (A) or A438079 (B) on the proliferation of HepG2 cells. C, D Effects of gradient A740003 (C) or A438079 (D) on the proliferation of HCC-LM3 cells. Results are presented as the mean ± SD of three independent experiments. Data were analyzed by unpaired Student's t-test. A740003 vs Control: *p < 0.05, **p < 0.01, ***p < 0.001. A438079 vs Control: #p < 0.05, ##p < 0.01
Fig. 4.
The proliferative rate of HCC cells was attenuated by P2X7 receptor antagonist A740003 or A438079. Cell proliferation assays were conducted by the MTS method or the EdU incorporation method, HepG2 and HCC-LM3 cells were administrated with P2X7 receptor antagonists A740003 or A438079 respectively. A, B In the MTS assay, HepG2 and HCC-LM3 cells were administrated with 20 µM A740003, 20 µM A438079, or PBS for 72 h. The proliferative rate (indicated by OD490) of HepG2 (A) and HCC-LM3 (B) cells was detected at each time point. In panel A and panel B, data were presented as the mean ± SD from three independent experiments, and unpaired Student's t-test was conducted. Partial SDs in panel A and panel B appear absent, for the reason that they are too small to be seen. C-F In the EdU incorporation assay, HepG2 and HCC-LM3 cells were administrated with 20 µM A740003, 20 µM A438079, or PBS for 48 h. Then above cells were treated with EdU solution (50 µM) for 2 h, labeled by a click reaction and observed by fluorescence microscope. The representative image from three independent experiments was shown, the proliferating HepG2 (C) and HCC-LM3 (D) cells were indicated as the DAPI-blue+ and EdU-red+ cells. Scale bars: 50 µm. Percentage of proliferating HepG2 (E) and HCC-LM3 (F) cells were calculated within 5 randomly selected sections by software Image J, data were shown as the mean ± SD in panels and calculated by unpaired Student's t-test. A740003 vs. Control: *p < 0.05, **p < 0.01. A438079 vs. Control: #p < 0.05, ##p < 0.01
RNAseq of HCC cells with P2X7 receptor blockade
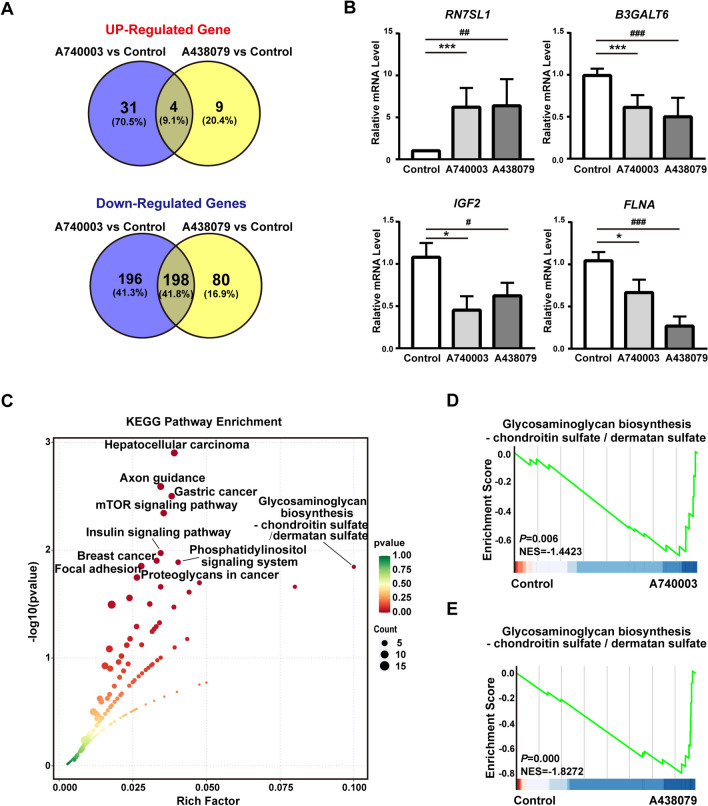
To characterize the potential mechanism of P2X7 receptor signaling in HCC, we performed bulk RNAseq to acquire the gene expression profiles, and data was uploaded to the GEO database and is publicly available with the accession number GSE268508. HepG2 cells were incubated with PBS, A740003, or A438079 for 48 h, then we isolated the total RNA and performed mRNA sequencing. DEGs were screened out with two parameters: FC > 2 while FDR < 0.05. Compared to the control group, the expression levels of 35 genes were increased and those of 394 genes were decreased in the A740003 group (Figs. 5A and S1A), while those of 13 genes were increased and 278 genes were decreased in the A438079 group (Figs. 5B and S1B). Then the expression level of RN7SL1 (selected from 4 genes which were both up-regulated in A740003 group and A438079 group), and the expression level of B3GALT6, IGF2, and FLNA (selected from 198 genes which were both down-regulated in A740003 group and A438079 group), were further validated by qRT-PCR (Fig. 5B). Moreover, KEGG analysis were performed with the 198 genes from gene sets which were both down-regulated in the A740003 and A438079 groups. This showed that Glycosaminoglycan biosynthesis-chondroitin sulfate/dermatan sulfate, Proteoglycans in cancer were enriched in these gene sets (Fig. 5C). Furthermore, GSEA results demonstrated that both the A740003 (NES = − 1.4423, P.adj = 0.006) and A438079 (NES = − 1.8272, P.adj < 0.001) groups were significantly associated with the Glycosaminoglycan biosynthesis-chondroitin sulfate/dermatan sulfate signaling pathway (Fig. 5D, E). These findings demonstrated that the inhibited P2X7 receptor signaling may block the proliferation of HCC cells by proteoglycans-associated signaling pathways.
Fig. 5.
Gene expression profiles of HCC cells with P2X7 receptor blockade. HepG2 cells were incubated with 20 µM A740003, 20 µM A438079, or PBS for 48 h respectively. Then cells were harvested and RNAseq experiment was conducted. A The number of overlapped DEGs (up-regulated DEGs in the upper panel and down-regulated DEGs in the lower panel) was illustrated by Venn diagram. B The expression level of DEGs in gene sets (4 genes) and gene sets (198 genes) were further validated by RT-PCR. The RQ value of control group treated with PBS was designated as 1.000. Data from panel B was shown as the mean ± SD of three independent experiments and calculated by unpaired Student's t-test. Partial SDs in control group appear absent, they are too small to be seen. C Bioinformatic analysis of KEGG pathway was carried out with DEGs from gene sets (198 genes), the abscissa is the rich factor, the ordinate is the p value, color indicates the p value (lower value stands for higher significance), and size indicates the enriched gene count (bigger size indicates more genes included). Top 10 pathways with minimum q value from the KEGG experiment were shown. D, E GSEA analysis of A740003 group vs control (D), and A438079 group vs control (E) in Glycosaminoglycan biosynthesis-chondroitin sulfate/dermatan sulfate signaling pathway. A740003 vs. Control: *p < 0.05, ***p < 0.001. A438079 vs. Control: #p < 0.05, ##p < 0.01, ###p < 0.001
CHSY1 was involved in the proliferation of HCC cells mediated by P2X7 receptor signaling
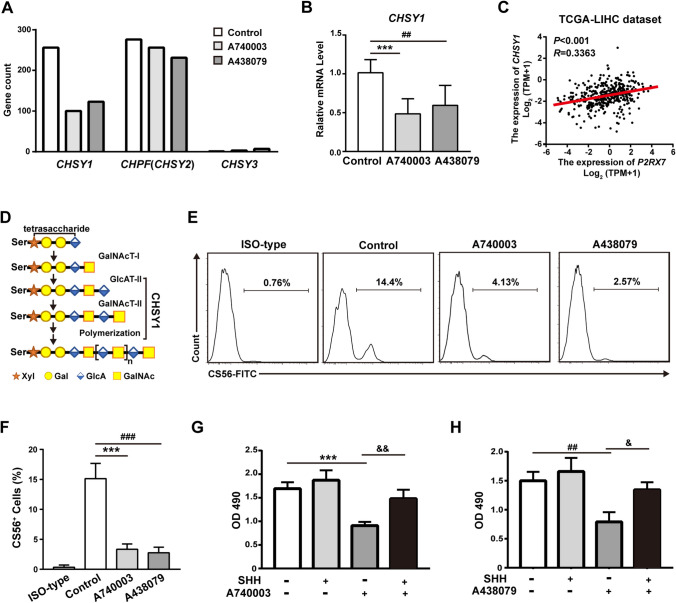
To identify the key molecules that are involved in the above process, we selected genes in the family of chondroitin sulfate synthases, which were associated with Glycosaminoglycan biosynthesis-chondroitin sulfate/dermatan sulfate signaling pathway in our RNAseq raw data. Interestingly, only CHSY1 was both down-regulated in the A740003 or A438079 groups (Fig. 6A). This result was further validated by qRT-PCR (Fig. 6B). Moreover, the correlation of P2RX7 expression and CHSY1 expression in the patient dataset with HCC from the TCGA database was analyzed (Fig. 6C) [26]. CHSY1 is a biosynthetic enzyme that is important in forming the repeating chondroitin sulfate (CS) backbone disaccharide structure [4GlcAβ1-3GalNAcβ1] and is also essential for polymerizing the chondroitin chain [33] (Fig. 6D). Chondroitin sulfate can be recognized by anti-chondroitin sulfate antibody (CS56) and detected by flow cytometry. Notably, CS56 did not recognize all types of chondroitin sulfate, and only the chondroitin sulfate expressed on cell surface expression can be labeled [34]. Administration of A740003 or A438079 significantly reduced the percentage of CS56+ cells (Fig. 6E, F).
Fig. 6.
P2X7 receptors regulating the proliferation of HCC cells by CHSY1. A The gene count of chondroitin sulfate synthase family members from our RNAseq raw data. B The gene expression level of CHSY1 in HepG2 cells (PBS group, A740003 group, or A438079 group) was further validated by RT-PCR. The RQ value of control group treated with PBS was designated as 1.000. C The correlation between P2RX7 expression and CHSY1 expression was assessed by the TCGA-LIHC dataset, Spearman correlation was performed. D CHSY1 functions in the biosynthesis of chondroitin sulfate. Abbreviations: Ser, serine; GalNAcT-I, N-acetylgalactosaminyltransferase I; GlcAT-II, β1,3-glucuronyltransferase II; CHSY1, chondroitin synthase 1; Xyl, xylose; Gal, galactose; GlcA, glucuron acid; GalNAc, N-acetylgalactosamine. E, F Administration with A740003 or A438079 reduced the expression of chondroitin sulfate on HCC cell surfaces. Representative images from the flow cytometry experiment were shown (E). Results from flow cytometry were quantified and presented as the mean ± SD from three independent experiments (F). G, H MTS assay of HepG2 cells were treated with A740003 (G) or A438079 (H) in combination with or without recombinant SHH protein (0.4 mg/ml). Results in panel B, F, G, or H are presented as the mean ± SD of three independent experiments. Data were analyzed by unpaired Student's t-test. A740003 vs. Control: ***p < 0.001. A438079 vs. Control: ##p < 0.01, ###p < 0.001. SHH vs. A740003 or A438079: &p < 0.05, &&p < 0.01
Recently, CHSY1 has been demonstrated to promote growth and metastasis of HCC cells by activating the Sonic Hedgehog (SHH) pathway [35]. To understand the role of the CHSY1-SHH signaling axis in P2X7 receptor antagonists mediating proliferative inhibition, we treated HepG2 cells with recombinant SHH protein. This significantly rescued A740003- and A438079-induced cell growth inhibition in HepG2 cells (Fig. 6G, H). Furthermore, to determine whether the blockade of the SHH pathway had a synergistic effect with A740003 and A438079, we used vismodegib, a U.S. Food and Drug Administration approved drug for blocking the SHH signaling pathway [36]. Vismodegib (2 mM) did not inhibit the growth of HepG2 cells, but significantly decreased the proliferation rate when combined with A740003 or A438079 compared to A740003 or A438079 alone (Fig. S2A, B). Collectively, these results demonstrated that CHSY1 was involved in P2X7 receptor signaling mediated proliferation of HCC cells.
Discussion
In this study, we have investigated the role of P2X7 receptor antagonism in the proliferation of human HCC cells. Our results show that the expression level of P2X7 receptor was up-regulated in the samples of HCC patients, and inhibiting P2X7 receptors signaling attenuated the Ca2+ influx and the proliferation of HCC cells. Previous studies have demonstrated that blocking P2X7 receptor can inhibit the tumor growth of melanoma, pancreatic cancer, and acute myeloid leukemia [13, 18, 37]. Consistent with these findings, our results suggest that targeting the P2X7 receptor could be a potential therapeutic approach for treating patients with HCC. In the molecular mechanism section of this study, bulk RNAseq experiment revealed the involvement of CHSY1 in this process. Antagonism of P2X7 receptor by A740003 and A438079 significantly reduced the expression level of CHSY1 and the chondroitin sulfate on the surface of HCC cells. Furthermore, supplementation with recombinant SHH protein, a key component of Sonic Hedgehog signaling pathway downstream activated by CHSY1 [35], rescued A740003- and A438079-induced cell growth inhibition in HCC cells. Overall, our data clearly demonstrate that the proliferative rate of HCC cells could be attenuated by P2X7 antagonists A740003 and A438079 in vitro, and suggest the preliminary mechanisms that this effect is largely modulated by CHSY1.
Liver damage induced by P2X7 receptor may play important roles in driving HCC carcinogenesis [38]. Previously, the P2X7 receptor has been reported to activate NLRP3 inflammasome in Kupffer cells and hepatocyte cell lines (HepG2 and L02), contributing to the inflammatory response and subsequently leading to the occurrence of liver damage, which often progresses to cirrhosis, fibrosis and HCC [39, 40]. These findings suggest that the P2X7 receptor may serve as a risk factor for HCC. However, the biological functions (i.e., proliferation, migration, invasion, and angiogenesis) of P2X7 receptor in HCC cells have not been fully elucidated. Interestingly, we observed higher expression of P2X7 receptor in the samples from HCC patients compared to normal control. To evaluate the involvement of P2X7 receptor in HCC cells proliferation, we chose antagonists A740003 and A438079 to inhibit its signaling in HCC cell lines HepG2 and HCC-LM3. In Ca2+ influx assay, we found that both 20 μΜ A740003 and A438079 could significantly block the Ca2+ influx into the cytoplasm of HCC cells. Importantly, this observation provided solid evidence that the P2X7 receptor is functional in HCC cell lines. Our results are consistent with those obtained for other tumor cells such as neuroblastoma cells, pancreatic ductal adenocarcinoma cells, glioma cells, etc. [41–43]. MTS assay and EdU incorporation assay demonstrated that A740003 and A438079 could inhibit the proliferation of HCC cells in a concentration- and time-dependent manner. Therefore, in addition to the proinflammatory action of P2X7 receptor on immune cells and hepatocytes in the early phase of liver damage [39, 40, 44], our data indicate that P2X7 receptor may serve as a key regulator in the proliferation of HCC cells. Moreover, this observation gives rise to an intriguing question regarding how P2X7 receptor blockade contributes to the proliferative inhibition of HCC cells.
In terms of the molecular mechanisms underlying P2X7 receptor blockade and its inhibitory effect on HCC cells proliferation, we have identified CHSY1 as an important mediator through RNAseq analysis. While our previous studies have demonstrated that the P2X7 receptor modulates AML cell proliferation through PBX3 and DLBCL cell proliferation through CPS1 [13, 14], our findings indicate that CHSY1, an enzyme critical for of chondroitin sulfate biosynthesis, is involved in regulating the proliferation of HCC cells. Despite high expression and significant signaling functionality of the P2X7 receptor in various human cancers [45], our results suggest that downstream mediators of P2X7 receptor signaling may be specific to cancer type. Furthermore, we elucidated that A740003 and A438079 significantly down-regulate mRNA expression level of CHSY1 in HepG2 cells, subsequently leading to the lower expression level of chondroitin sulfate on the cell surface. Interestingly, previous studies have revealed that knockdown of CHSY1 reduces the accumulation of chondroitin sulfate and inhibits the epithelial-mesenchymal transition process in HCC cells by suppressing the activity of hedgehog signaling pathway [46–48]. In support of this, our rescue experiment demonstrated that exogenous SHH protein effectively reversed the proliferation inhibition induced by A438079 and A740003 in HCC cells. Additionally, vismodegib (a widely used inhibitor for blocking the SHH signaling pathway) has been demonstrated to synergistically enhance the anti-proliferative effects of A740003 and A438079 in HCC cells. Collectively, these findings indicate that P2X7 receptor blockade may reduce the proliferative capacity of HCC cells through CHSY1 and its associated SHH signaling pathway. Future investigations are planned to elucidate the molecular mechanisms underlying the interaction between P2X7 receptor and CHSY1.
It should be noted that there are still some limitations and deficiencies in this study: I. Theoretically, we have primarily focused on the inhibitory effects and mechanisms of P2X7 receptor in the proliferation of HCC cells, however, further investigation is required to elucidate whether the activation of P2X7 receptor has a pro-tumor growth effect on HCC cells. Given that Bz-ATP effectively induced Ca2+ influx in HepG2 cells and HCC-LM3 cells, we have confirmed that the P2X7 receptor in HCC cells was activated with fully ion channel functionality. Nevertheless, the biological functions and underlying mechanisms of P2X7 receptor activation in HCC cells were still largely unknown. II. Methodologically, although we have provided compelling evidence that P2X7 receptor antagonists A740003 and A438079 significantly attenuated the proliferation of HepG2 and HCC-LM3 cells in vitro, future xenograft animal model (such as nude mice transplanted with human HCC cell lines or NSG mice transplanted with patient-derived HCC cells) should be employed to determine whether the P2X7 receptor antagonism can inhibit the HCC tumor growth in vivo. III. While the administration of antagonists is widely accepted as an effective approach to inhibit P2X7 receptor signaling, further validation of our findings will involve investigating knockout of the P2RX7 gene in HCC cells in our future studies. IV. The potential synergistic effect of P2X7 receptor antagonists with current first-line treatment for HCC patients on the proliferation of HCC cells remains to be determined.
In conclusion, aberrant up-regulated expression of P2X7 receptor is observed in patients with HCC. Blockade of the P2X7 receptor by antagonists A740003 and A438079 reduces the proliferative rate of HepG2 and HCC-LM3 cells. Most importantly, CHSY1 and its associated SHH pathway are involved in this process. These findings reveal the role of P2X7 receptor in the proliferation of HCC cells, and suggest its potential as a molecular target for treating patients with HCC.
Supplementary Information
Below is the link to the electronic supplementary material.
Figure S1. Volcano Plot of DEGs in HCC cells with P2X7 receptor inhibition. (A) Volcano plot of DEGs between HepG2 cells treated with A740003 and PBS control. (B) Volcano plot of DEGs between HepG2 cells treated with A438079 and PBS control. The orange dot represents the genes with adjust P<0.05 and log2 FC >1, and the green dot represents the genes with adjust P<0.05 and log2 FC <−1. (PNG 180 kb)
Figure S2. Synergistic effect of P2X7 receptor inhibitor A740003 or 438079 with SHH inhibitor vismodegib. (A, B) HepG2 cells were treated with 20 μm A740003 (A) or 20 μm A438079 (B) in combination with or without recombinant vismodegib protein (2 mM) for 48 hours, then MTS assay was performed. Results are presented as the mean ± SD of three independent experiments. Data were analyzed by unpaired Student's t-test. A740003 vs Control: **p < 0.01. A438079 vs Control: ###p < 0.001. A740003 or A438079 vs vismodegib: &p < 0.05. (PNG 60 kb)
Xinxing Tantai
is a Physician in the Department of Gastroenterology, the Second Affiliated Hospital of Xi’an Jiaotong University.

Author contributions
Xinxing Tantai: performed experiments, analyzed and interpreted data. Xin Yang: performed experiments, designed experiments, interpreted data. Xinyuan Liu: designed experiments, interpreted data, wrote the manuscript. Xiao Yang: acquired funding, designed and performed experiments, analyzed and interpreted data, wrote & review the manuscript.
Funding
This work was supported by grant 2024JC-YBMS-631 and grant 2020JQ-546 from the Natural Science Basic Research Program of Shaanxi; grant 81802862 from the National Natural Science Foundation of China (NSFC); the Personnel Training Specialized Research Foundation RC(XM)202002 from the Second Affiliated Hospital of Xi'an Jiaotong University.
Data availability
Data is provided within the manuscript or supplementary information files in Materials and Methods Scetion-Data Acquisition and Processing.
Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE268508.
Declarations
Ethical approval
There’s no animal experiment in this study, so no need to approve the experimental animal ethics. Moreover, all the clinical patients data were downloaded from public databases, and the ethical approval were stated in the references or cited websites.
Consent for publication
All authors approved to publish the study in this journal.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xinyuan Liu, Email: liuxinyuancsu@outlook.com.
Xiao Yang, Email: yang_x@xjtu.edu.cn.
References
- 1.Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A (2022) Hepatocellular carcinoma. Lancet 400:1345–1362. 10.1016/S0140-6736(22)01200-4 [DOI] [PubMed] [Google Scholar]
- 2.Li X, Bai X, Tang Y, Qiao C, Zhao R, Peng X (2023) Research progress on the P2X7 receptor in liver injury and hepatocellular carcinoma. Chem Biol Drug Des 101:794–808. 10.1111/cbdd.14182 [DOI] [PubMed] [Google Scholar]
- 3.Kwan SY, Sheel A, Song CQ, Zhang XO, Jiang T, Dang H et al (2020) Depletion of TRRAP induces p53-independent senescence in liver cancer by down-regulating mitotic genes. Hepatology 71:275–290. 10.1002/hep.30807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaur J, Dora S (2023) Purinergic signaling: diverse effects and therapeutic potential in cancer. Front Oncol 13:1058371. 10.3389/fonc.2023.1058371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S (2017) The P2X7 receptor in infection and inflammation. Immunity 47:15–31. 10.1016/j.immuni.2017.06.020 [DOI] [PubMed] [Google Scholar]
- 6.Burnstock G, Di Virgilio F (2013) Purinergic signalling and cancer. Purinergic Signal 9:491–540. 10.1007/s11302-013-9372-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mai Y, Guo Z, Yin W, Zhong N, Dicpinigaitis PV, Chen R (2021) P2X receptors: potential therapeutic targets for symptoms associated with lung cancer - a mini review. Front Oncol 11:691956. 10.3389/fonc.2021.691956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagel D, Vincendeau M, Eitelhuber AC, Krappmann D (2014) Mechanisms and consequences of constitutive NF-kappaB activation in B-cell lymphoid malignancies. Oncogene 33:5655–5665. 10.1038/onc.2013.565 [DOI] [PubMed] [Google Scholar]
- 9.Jelassi B, Chantome A, Alcaraz-Perez F, Baroja-Mazo A, Cayuela ML, Pelegrin P et al (2011) P2X(7) receptor activation enhances SK3 channels- and cystein cathepsin-dependent cancer cells invasiveness. Oncogene 30:2108–2122. 10.1038/onc.2010.593 [DOI] [PubMed] [Google Scholar]
- 10.Douguet L, JanhoDitHreich S, Benzaquen J, Seguin L, Juhel T, Dezitter X et al (2021) A small-molecule P2RX7 activator promotes anti-tumor immune responses and sensitizes lung tumor to immunotherapy. Nat Commun 12:653. 10.1038/s41467-021-20912-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnstock G, Kennedy C (2011) P2X receptors in health and disease. Adv Pharmacol 61:333–372. 10.1016/B978-0-12-385526-8.00011-4 [DOI] [PubMed] [Google Scholar]
- 12.Alves LA, de Melo Reis RA, de Souza CA, de Freitas MS, Teixeira PC, Neto Moreira Ferreira D et al (2014) The P2X7 receptor: shifting from a low- to a high-conductance channel - an enigmatic phenomenon? Biochim Biophys Acta 1838:2578–2587. 10.1016/j.bbamem.2014.05.015 [DOI] [PubMed] [Google Scholar]
- 13.Feng W, Yang X, Wang L, Wang R, Yang F, Wang H et al (2021) P2X7 promotes the progression of MLL-AF9 induced acute myeloid leukemia by upregulation of Pbx3. Haematologica 106:1278–1289. 10.3324/haematol.2019.243360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Ji Y, Mei L, Jing W, Yang X, Liu Q (2024) Potential role of the P2X7 receptor in the proliferation of human diffused large B-cell lymphoma. Purinergic Signal 20:273–284. 10.1007/s11302-023-09947-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Liu W, Liu Z, Liu Y, Zhang W, Xu L et al (2015) Prognostic value of purinergic P2X7 receptor expression in patients with hepatocellular carcinoma after curative resection. Tumour Biol 36:5039–5049. 10.1007/s13277-015-3155-2 [DOI] [PubMed] [Google Scholar]
- 16.Duan S, Yu J, Han Z, Cheng Z, Liang P (2016) Association between P2RX7 gene and hepatocellular carcinoma susceptibility: a case-control study in a Chinese Han Population. Med Sci Monit 22:1916–1923. 10.12659/msm.895763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amoroso F, Capece M, Rotondo A, Cangelosi D, Ferracin M, Franceschini A et al (2015) The P2X7 receptor is a key modulator of the PI3K/GSK3beta/VEGF signaling network: evidence in experimental neuroblastoma. Oncogene 34:5240–5251. 10.1038/onc.2014.444 [DOI] [PubMed] [Google Scholar]
- 18.Mohammed A, Janakiram NB, Madka V, Pathuri G, Li Q, Ritchie R et al (2017) Lack of chemopreventive effects of P2X7R inhibitors against pancreatic cancer. Oncotarget 8:97822–97834. 10.18632/oncotarget.22085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST et al (2012) Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open 2:e000825. 10.1136/bmjopen-2012-000825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho YA, Choi S, Park S, Park CK, Ha SY (2020) expression of pregnancy up-regulated non-ubiquitous calmodulin kinase (PNCK) in hepatocellular carcinoma. Cancer Genomics Proteomics 17:747–755. 10.21873/cgp.20229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu B, Liu DA, Guan L, Myint PK, Chin L, Dang H et al (2023) Stiff matrix induces exosome secretion to promote tumour growth. Nat Cell Biol 25:415–424. 10.1038/s41556-023-01092-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y et al (2012) Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 44:765–769. 10.1038/ng.2295 [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Bajari R, Andric D, Gerthoffert F, Lepsa A, Nahal-Bose H et al (2019) The international cancer genome consortium data portal. Nat Biotechnol 37:367–369. 10.1038/s41587-019-0055-9 [DOI] [PubMed] [Google Scholar]
- 24.Gyorffy B (2024) Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation (Camb) 5:100625. 10.1016/j.xinn.2024.100625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A et al (2015) Proteomics. Tissue-based map of the human proteome. Science 347:1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 26.Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A et al (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 38:675–678. 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi A, Camacho P, Lechleiter JD, Herman B (1999) Measurement of intracellular calcium. Physiol Rev 79:1089–1125. 10.1152/physrev.1999.79.4.1089 [DOI] [PubMed] [Google Scholar]
- 28.Maravall M, Mainen ZF, Sabatini BL, Svoboda K (2000) Estimating intracellular calcium concentrations and buffering without wavelength ratioing. Biophys J 78:2655–2667. 10.1016/S0006-3495(00)76809-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grynkiewicz G, Poenie M, Tsien RY (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem 260:3440–3450 [PubMed] [Google Scholar]
- 30.Paredes RM, Etzler JC, Watts LT, Zheng W, Lechleiter JD (2008) Chemical calcium indicators. Methods 46:143–151. 10.1016/j.ymeth.2008.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bootman MD, Rietdorf K, Collins T, Walker S, Sanderson M (2013) Ca2+-sensitive fluorescent dyes and intracellular Ca2+ imaging. Cold Spring Harb Protoc 2013:83–99. 10.1101/pdb.top066050 [DOI] [PubMed] [Google Scholar]
- 32.Martins JP, Silva RB, Coutinho-Silva R, Takiya CM, Battastini AM, Morrone FB et al (2012) The role of P2X7 purinergic receptors in inflammatory and nociceptive changes accompanying cyclophosphamide-induced haemorrhagic cystitis in mice. Br J Pharmacol 165:183–196. 10.1111/j.1476-5381.2011.01535.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikami T, Kitagawa H (2013) Biosynthesis and function of chondroitin sulfate. Biochim Biophys Acta 1830:4719–4733. 10.1016/j.bbagen.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 34.Ito Y, Hikino M, Yajima Y, Mikami T, Sirko S, von Holst A et al (2005) Structural characterization of the epitopes of the monoclonal antibodies 473HD, CS-56, and MO-225 specific for chondroitin sulfate D-type using the oligosaccharide library. Glycobiology 15:593–603. 10.1093/glycob/cwi036 [DOI] [PubMed] [Google Scholar]
- 35.Liu CH, Lan CT, Chou JF, Tseng TJ, Liao WC (2017) CHSY1 promotes aggressive phenotypes of hepatocellular carcinoma cells via activation of the hedgehog signaling pathway. Cancer Lett 403:280–288. 10.1016/j.canlet.2017.06.023 [DOI] [PubMed] [Google Scholar]
- 36.Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD et al (2012) Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med 366:2171–2179. 10.1056/NEJMoa1113713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Marchi E, Orioli E, Pegoraro A, Sangaletti S, Portararo P, Curti A et al (2019) The P2X7 receptor modulates immune cells infiltration, ectonucleotidases expression and extracellular ATP levels in the tumor microenvironment. Oncogene 38:3636–3650. 10.1038/s41388-019-0684-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Yu Q, Bai X, Li X, Sun Y, Peng X et al (2024) The role of the purinergic ligand-gated ion channel 7 receptor in common digestive system cancers. Eur J Cancer Prev 33:271–281. 10.1097/CEJ.0000000000000851 [DOI] [PubMed] [Google Scholar]
- 39.Saha M, Manna K, Das Saha K (2022) Melatonin suppresses NLRP3 inflammasome activation via TLR4/NF-kappaB and P2X7R signaling in high-fat diet-induced murine NASH model. J Inflamm Res 15:3235–3258. 10.2147/JIR.S343236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni J, Zhang Z, Luo X, Xiao L, Wang N (2016) Plasticizer DBP activates NLRP3 inflammasome through the P2X7 receptor in HepG2 and L02 cells. J Biochem Mol Toxicol 30:178–185. 10.1002/jbt.21776 [DOI] [PubMed] [Google Scholar]
- 41.Raffaghello L, Chiozzi P, Falzoni S, Di Virgilio F, Pistoia V (2006) The P2X7 receptor sustains the growth of human neuroblastoma cells through a substance P-dependent mechanism. Cancer Res 66:907–914. 10.1158/0008-5472.CAN-05-3185 [DOI] [PubMed] [Google Scholar]
- 42.Giannuzzo A, Pedersen SF, Novak I (2015) The P2X7 receptor regulates cell survival, migration and invasion of pancreatic ductal adenocarcinoma cells. Mol Cancer 14:203. 10.1186/s12943-015-0472-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei W, Ryu JK, Choi HB, McLarnon JG (2008) Expression and function of the P2X(7) receptor in rat C6 glioma cells. Cancer Lett 260:79–87. 10.1016/j.canlet.2007.10.025 [DOI] [PubMed] [Google Scholar]
- 44.Taylor JM, Han Z (2010) Purinergic receptor functionality is necessary for infection of human hepatocytes by hepatitis delta virus and hepatitis B virus. PLoS ONE 5:e15784. 10.1371/journal.pone.0015784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lara R, Adinolfi E, Harwood CA, Philpott M, Barden JA, Di Virgilio F et al (2020) P2X7 in cancer: from molecular mechanisms to therapeutics. Front Pharmacol 11:793. 10.3389/fphar.2020.00793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada S, Sugahara K (2008) Potential therapeutic application of chondroitin sulfate/dermatan sulfate. Curr Drug Discov Technol 5:289–301. 10.2174/157016308786733564 [DOI] [PubMed] [Google Scholar]
- 47.Whalen DM, Malinauskas T, Gilbert RJ, Siebold C (2013) Structural insights into proteoglycan-shaped Hedgehog signaling. Proc Natl Acad Sci U S A 110:16420–16425. 10.1073/pnas.1310097110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Lingala S, Khoobyari S, Nolta J, Zern MA, Wu J (2011) Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J Hepatol 55:838–845. 10.1016/j.jhep.2010.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Volcano Plot of DEGs in HCC cells with P2X7 receptor inhibition. (A) Volcano plot of DEGs between HepG2 cells treated with A740003 and PBS control. (B) Volcano plot of DEGs between HepG2 cells treated with A438079 and PBS control. The orange dot represents the genes with adjust P<0.05 and log2 FC >1, and the green dot represents the genes with adjust P<0.05 and log2 FC <−1. (PNG 180 kb)
Figure S2. Synergistic effect of P2X7 receptor inhibitor A740003 or 438079 with SHH inhibitor vismodegib. (A, B) HepG2 cells were treated with 20 μm A740003 (A) or 20 μm A438079 (B) in combination with or without recombinant vismodegib protein (2 mM) for 48 hours, then MTS assay was performed. Results are presented as the mean ± SD of three independent experiments. Data were analyzed by unpaired Student's t-test. A740003 vs Control: **p < 0.01. A438079 vs Control: ###p < 0.001. A740003 or A438079 vs vismodegib: &p < 0.05. (PNG 60 kb)
Data Availability Statement
Data is provided within the manuscript or supplementary information files in Materials and Methods Scetion-Data Acquisition and Processing.
Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE268508.