Abstract
The growing resistance to antifungal drugs and the limited number of antifungals currently available highlight the need for new antifungal strategies. In this context, there is significant potential for plant and insect antifungal defensins, which target fungal glucosylceramides (GlcCer), a growth and virulence determinant. The ETD151 peptide, optimized from the insect defensin Heliomicin, binds to fungal GlcCer as a crucial step in its activity. Nevertheless, further investigation is necessary to elucidate the mechanisms by which ETD151 targets fungal GlcCer at the molecular and atomic scales. The binding affinity was experimentally measured with isothermal titration calorimetry and microscale thermophoresis. The results collectively revealed affinity with a membrane-bound methylated GlcCer in the micromolar range. NMR has been employed to identify the area of ETD151 that would monopolize methylated GlcCer binding, including the two adjacent hydrophobic loops. Furthermore, our findings indicate that ETD151 specifically inserts into and disorders phosphatidylcholine lipid vesicles containing methylated GlcCer. Finally, it is worth noting that the C9-methyl in the sphingoid base plays a pivotal role in the ETD151–GlcCer interaction. Its presence increases the binding affinity between the two partners, resulting in stronger structural changes in ETD151 and deeper insertion into the hydrophobic core of model membranes. This study reveals key biochemical and structural elements of the ETD151–GlcCer interaction, provides a basis for elucidating the structure and dynamics of other GlcCer-targeting defensins in relation to their function, and facilitates the development of defensin-mimetic antifungals.
Keywords: antimicrobial peptide, defensin, sphingolipid, protein–lipid interaction, membrane biophysics, antifungal, fungal glucosylceramides
Fungal diseases have historically been underestimated and neglected. However, recent data indicate that fungal pathogens infect billions of people and cause at least 1.5 million deaths annually (1, 2). Despite the significant impact of fungal pathogens on global human health (3), only three frontline antifungal drug classes are currently employed in clinical practices to treat invasive infections: azoles, polyenes, and echinocandins (4). These drugs target either the cell membrane or the cell wall to inhibit their fungistatic/fungicidal activities (5). Azoles bind to ergosterol, the primary sterol in fungal membranes. Polyenes inhibit the ergosterol biosynthesis pathway, while echinocandins inhibit β-1,3-glucan synthase, a crucial enzyme for maintaining cell wall integrity (6, 7, 8). Despite their efficacy in the treatment of systemic fungal infections, their toxicity to the host and unfavorable pharmacokinetics limit their clinical use (9). Furthermore, there is an increased risk of drug-resistant fungal pathogens emerging due to the overuse of these limited antifungal classes (10, 11). This highlights the urgent need to identify new antifungal agents with innovative mechanisms of action (MoA) to treat systemic fungal infections. However, the development of new antifungals is challenging due to the close evolutionary relationship between fungi and Humans (12).
Antifungal defensins, disulfide-rich peptides present in the immune systems of plants and insects (13), show great promise for drug development as a potential alternative to conventional antifungals (14, 15). This is attributed to three factors: (i) in vivo efficacy against a wide variety of fungi, (ii) minimal toxicity to human cells, and (iii) high stability against proteases. Structurally, antifungal defensins share a common monomeric fold known as the cysteine-stabilized αβ motif (CSαβ). The latter comprises a three-stranded antiparallel β-sheet connected to an α-helix via two disulfide bridges. Despite their strong structural similarities, antifungal defensins act according to different MoAs, and are undoubtedly more complex than the membrane-lytic mechanism triggered by short linear antimicrobial peptides (AMPs). The MoAs of antifungal defensins involves interaction with one or more specific cell wall and/or plasma membrane compound(s), which can lead to membrane disruption and downstream events (16).
Among the promising cellular targets for antifungal defensins, glucosylceramides (GlcCer) have been identified as the most abundant neutral glycosphingolipid component in the fungal membranes. Their role as growth and virulence determinants has been established in various fungal species (17, 18). The presence of GlcCer in fungal membranes plays a pivotal role in the activity of certain plant and insect defensins, including RsAFP2 from the radish Raphanus sativus (19), Heliomicin from the tobacco budworm Heliothis virescens (19), Psd1 from the pea Pisum sativum (20), MsDef1 from the alfalfa/lucerne Medicago sativa (21) and PvD1 from the common bean Phaseolus vulgaris (22). On the one hand, direct in vitro molecular interaction with GlcCer has been demonstrated for RsAFP2, Heliomicin, Psd1, and Psd2 (19, 20, 23). Consequently, they are referred to as GlcCer-interacting defensins. On the other hand, the potential for GlcCer to serve as a target for some defensins (e.g., MsDef1 and PvD1) remains hypothetical, and further investigation is required to validate the direct molecular recognition with GlcCer.
We recently demonstrated that the recombinant form of ETD151 (produced in the yeast Saccharomyces cerevisiae), a 44-aa defensin derived from Heliomicin (24), requires the presence of GlcCer in fungal membranes to exert its full antifungal activity (25). The ETD151 peptide is active in vitro at micromolar concentrations against pathogenic fungi such as Cryptococcus, Candida, and Aspergillus (24), classified as a critical group in the World Health Organization list of priority fungal pathogens (https://www.who.int/publications/i/item/9789240060241). Furthermore, ETD151 was shown to be nontoxic after systemic administration and more effective than Amphotericin B, the current standard for the treatment of invasive infections, in a murine model infected with the yeast Candida albicans and the filamentous fungus Aspergillus fumigatus (26). Furthermore, ETD151 has demonstrated efficacy against the plant pathogenic fungus Botrytis cinerea (27). This filamentous fungus is a necrotrophic pathogen responsible for gray mold (28), which has developed resistance to current fungicides (29). The MoAs of ETD151 on B. cinerea have been partly elucidated, revealing a multifaceted mechanism (27) similar to that of certain plant defensins (30). To exert its anti-Botrytis activity, ETD151 binds directly to GlcCer located on the cell wall and/or plasma membrane, remaining mainly outside the cells (25) and causing membrane permeabilization (27). A proteomic study revealed that ETD151 disrupts proteins corresponding to six different pathways (spliceosome, ribosome, protein processing in endoplasmic reticulum endocytosis, mitogen-activated protein kinase signaling pathway, and oxidative phosphorylation) in B. cinerea without directly affecting the respiratory chain (27). These findings collectively indicate that ETD151 represents a promising alternative to current therapies against fungal infections by targeting GlcCer. It is therefore crucial to elucidate the mechanisms by which ETD151 targets fungal GlcCer, at the molecular and atomic scale.
The typical structure of fungal GlcCer consists of a methylated sphingoid base (C9-methyl-4,8-sphingadienine; d19:2) linked to a 2-hydroxylated C16-C18 fatty acid and attached to glucose by an ester bond (31). Interestingly, fungal GlcCer exhibit distinct structural features, including a double bond at C8 (Δ8) and a methyl group at C9 on the sphingoid base, modifications that are absent in mammalian GlcCer which typically lack both the Δ8 double bond and C9-methylation. Although plant GlcCer may contain a Δ8 double bond, they generally do not possess the C9-methylation. The methyl group at C9, added by the fungal-specific enzyme sphingolipid C9-methyltransferase (SMT), thus represents a unique structural hallmark of fungal GlcCer, distinguishing them from their mammalian and plant counterparts (32). Similarly to GlcCer synthesis, C9-methylation in the sphingoid base is essential for fungal growth, differentiation, and pathogenesis (33). Disruption of the gene encoding SMT in the encapsulated yeast Cryptococcus neoformans (Δsmt), which does not produce methylated GlcCer, resulted in a significant reduction in virulence in a murine model of cryptococcosis (34). The filamentous fungus Neurospora crassa Δsmt mutant cells were entirely devoid of methylated GlcCer production, exhibiting significant growth defects compared to their respective WT strains (35). In the case of the filamentous fungi Fusarium gramineurum and Aspergillus nidulans, the two SMTs encoded by respectively the smt1 and smt2 genes are suggested to exhibit redundant enzymatic functions (33, 36). F. gramineurum Δsmt1 has been found to produce methylated GlcCer like the WT strains. However, F. gramineurum Δsmt2 produces 75% of nonmethylated GlcCer and exhibits significant growth abnormalities and decreased pathogenicity in wheat (36). In addition to its critical role in fungal virulence, C9-methylation is a key factor influencing the activity of certain antifungal defensins. Indeed, the plant defensin AFP1 from the mustard plant Brassica juncea induces membrane permeabilization and reactive oxygen species production in WT C. albicans strains. However, this effect is not observed in C. albicans Δmts1 mutants, which lack the methylated GlcCer (37). These observations demonstrate that targeting methylated GlcCer provides a selective approach to combat fungal infections while minimizing host toxicity. It is therefore crucial to investigate whether ETD151 is selective for methylated GlcCer.
This work presents evidence of the in vitro recognition of fungal methylated GlcCer by ETD151 from the perspective of peptide structure and membrane lipid order. In light of these considerations, we conducted a molecular-scale investigation employing biophysical approaches to (i) estimate the binding affinity between ETD151 and fungal methylated GlcCer using isothermal titration calorimetry (ITC) and microscale thermophoresis (MST) techniques; (ii) determine which part of ETD151 is impacted by the interaction with fungal methylated GlcCer using solution NMR; and (iii) assess the impact of ETD151 presence on membrane lipid dynamics using solid-state NMR (ss-NMR). We also examine the role of the methyl group in the molecular recognition between GlcCer and ETD151, by comparing methylated GlcCer and nonmethylated GlcCer. The biochemical and structural characterization of the interaction between ETD151 and its fungal molecular target could facilitate the development of novel antifungals strategies that target pathogenic fungal cells through methylated GlcCer.
Results
To study the interaction between ETD151, a mutant derived from the Heliomicin defensin (Fig. 1A), and GlcCer in a membrane context, 100 nm large unilamellar vesicles (LUVs) were used for all experiments, except for ss-NMR, which required the preparation of multilamellar vesicles (MLVs). While MLVs are not as biologically relevant as LUVs due to their lack of a defined interior, their small curvature imposes local lipid constraints that closely resemble those found in cellular membranes (38). These membrane models are primarily composed of phosphatidylcholine (PC), the main phospholipid present in the fungal plasma membrane (39). PC is a zwitterionic lipid, which prevents nonspecific electrostatic attraction with the positively charged ETD151 (net charge +4 at physiological pH). Considering that sphingolipids account for approximately 7 to 16% of the fungal membrane lipid composition (40), GlcCer were incorporated into LUVs with 10% mol of total lipid. According to the literature, incorporating 10% GlcCer into lipid vesicles does not induce observable phase separation, thus supporting the formation of a homogeneous liquid-disordered phase, with GlcCer well mixed within the PC bilayer (23, 41). Two commercially available GlcCer were considered: a typical fungal GlcCer referred to as “methylated GlcCer,” and a plant GlcCer lacking the methyl group at C9 in the sphingoid base, referred to as “nonmethylated GlcCer.” The sole structural difference between these two GlcCer is the presence of the methyl group (Fig. 1, B and C).
Figure 1.
Sequences of Heliomicin and ETD151 defensins, chemical structures of glucosylceramide species, and thermodynamic (ITC), and thermophoretic (MST) analysis of the binding affinity of ETD151 with GlcCer-containing LUVs.A, sequence alignment of Heliomicin and ETD151 defensins. Conserved residues are highlighted in blue. Disulfide bridges between corresponding cysteine residues are indicated by black brackets (top). B and C, chemical structures of plant nonmethylated GlcCer (B) and fungal methylated GlcCer (C). D–F, ITC titrations of 3.5 mM LUVs suspension into a solution of 250 μM ETD151. Top: ITC thermograms show the differential power (DP) over time. Bottom: binding isotherms present enthalpy (ΔH) in different molar ratios. Pure PC-based LUVs were used as a control (D). ITC results shown are representative of triplicate titrations for each condition. Kd is the dissociation constant of the ETD151–LUVs complex with PC-nonmethylated GlcCer LUVs (E) and PC-methylated GlcCer LUVs (F). G and H, MST binding curves of fluorescently labeled ETD151 (Atto647-ETD151) to PC-nonmethylated GlcCer LUVs (G) and PC-methylated GlcCer LUVs (H). Kd values estimated by MST assay are presented as the mean of three independent experiments ± SD. Peptide and liposome solutions were prepared in phosphate buffer 10 mM, pH 5.8 for all ITC and MST experiments. ITC, isothermal titration calorimetry; LUV, large unilamellar vesicle; MST, microscale thermophoresis; PC, phosphatidylcholine.
The affinity of ETD151 for GlcCer is in the micromolar range, with a greater affinity observed when GlcCer is methylated
To investigate the binding affinity of ETD151 to GlcCer and to determine to what extent the C9-methyl structural feature in the sphingoid base may be involved in this binding affinity, ITC experiments were carried out using PC-based LUVs as control vesicles, PC-nonmethylated GlcCer LUVs, and PC-methylated GlcCer LUVs. As illustrated in Figure 1D, the titration of 3.5 mM pure PC-based vesicles into a ETD151 solution (250 μM, final concentration) did not generate any heat signal, confirming the absence of a strong interaction between PC and ETD151. The addition of 10% mol of nonmethylated or methylated GlcCer incorporated into PC-based vesicles resulted in an exothermic binding isotherm (Fig. 1, E and F). This heat signal is associated with the specific interaction between ETD151 and GlcCer (methylated or not), as no such exothermic signal results were observed in the titration of these GlcCer-containing vesicles into 10 mM phosphate buffer at pH 5.8 (see supp info Fig. S1). Using the "one-site binding model," the dissociation constant (Kd) values were determined of 1.9 mM and 100 μM were obtained with nonmethylated GlcCer (Fig. 1E) and methylated GlcCer (Fig. 1F), respectively. The thermodynamic results demonstrated that the strength of the interaction between ETD151 and methylated GlcCer-containing vesicles (determined by a free energy of binding ΔG of −5.45 kcal/mol) appears to be primarily governed by an entropic contribution (−TΔS = −5.03 kcal/mol) (see supp info Table S1), under our experimental conditions. The ITC results demonstrated that the incorporation of GlcCer enhances the binding affinity of ETD151 to the PC membrane but also that the mere presence of the C9-methyl group significantly affects this binding (19-fold higher affinity).
As we have determined the binding affinity of ETD151 to GlcCer extracted from B. cinerea using the MST assays (25), the affinity of ETD151 was compared with nonmethylated GlcCer and methylated GlcCer, using the LUVs compositions defined in the ITC experiments. The reported values result from two distinct effects: a fast effect (e.g., the local responses of the fluorophore to the temperature jump, which depends on the environment), and a slow effect (e.g., the thermophoresis diffusion fluorescence changes). The MST results indicated that the Kd values of 50 μM fluorescently labeled ETD151 (Atto647-ETD151) for both GlcCer-containing PC vesicles are in the micromolar range: Kd 5.3 μM for nonmethylated GlcCer (Fig. 1G) and Kd 0.7 μM for methylated GlcCer (Fig. 1H). In line with the ITC results, the Kd estimated values using the single-site binding model from MST experiments showed that ETD151 binds to GlcCer-containing PC vesicles with a 7.5-fold higher affinity in the presence of C9-methyl.
The presence of GlcCer primarily affects the hydrophobic L1 and L3 loops of ETD151, with a greater alteration observed with methylated GlcCer
To determine which part of ETD151 governs its interaction with the lipid target, a series of 1H-15N heteronuclear single quantum coherence spectroscopy NMR experiments were conducted with varying compositions and concentrations of LUVs. The reference 15N-ETD151 spectrum displays a high degree of peak dispersion, typical of a well-structured peptide (see Fig. S2). Peaks corresponding to 40 residues were observed, that is, 44 residues minus the N-terminal D1 residue, N13 which is masked under the intense C22 peak, G33 peak which is not in the range of the recorded chemical shift (6–10 ppm) and V38 located in the L3 loop. The latter residue is not visible, most likely due to intense peak broadening, a characteristic of an intermediate exchange regime.
Upon addition of 5 mM either PC-based LUVs or PC-nonmethylated GlcCer LUVs or PC-methylated GlcCer to a solution of 15N-ETD151 (50 μM), no clear chemical shift perturbations (CSPs) were detected (see Fig. S3). However, an overall decrease in peak intensity ratios due to the change in effective correlation time upon binding was observed for both GlcCer-containing LUVs (Fig. 2, nonmethyalted GlcCer in orange and methylated GlcCer in cyan), in contrast to control PC-based LUVs (Fig. 2, black). A similar phenomenon, that is, linewidth broadening and attenuation of signal intensities without significant CSPs, has been described in other NMR studies (42, 43). For example, Mehdi Mobli’s team recently reported NMR titrations of membrane-active peptides with model membranes (43).
Figure 2.
Relative 1H-15N SOFAST-HMQC signal intensities of 15N-ETD151 in the presence of different compositions of LUVs. All liposomes were used at the same concentration (5 mM): PC-based LUVs (IPC/I0, in black), PC-nonmethylated GlcCer LUVs (InonmethylatedGlcCer/IPC, in orange), and PC-methylated GlcCer (Imethylated GlcCer/IPC, in cyan), with I0 corresponding to the peak intensities of free-state 15N-ETD151. The peptide 15N-ETD151 was used at the same concentration (50 μM) for all samples. The horizontal lines indicate the mean ratio intensity for each sample. Residues of ETD151 are shown as consecutive numbers on the x-axis. Asterisks indicate residues where the resonance is absent in the free and bound states of ETD151. GlcCer, glucosylceramide; LUV, large unilamellar vesicle; PC, phosphatidylcholine; SOFAST-HMQC, band-selective optimized flip angle short transient heteronuclear multiple quantum coherence.
As the peak ratio intensities for LUVs containing GlcCer were calculated relative to IPC (intensity of peak in the ETD151-PC LUVs spectrum), the decrease in intensity is linked to the presence of GlcCer. Interestingly, the presence of methylated GlcCer results in a greater overall loss of mean intensity, decreasing by approximately 75%, than that of nonmethylated GlcCer, decreasing by around 60% (Fig. 2). It is worth noting that the peaks corresponding to G10, C18, and N37 are undetectable when GlcCer, methylated or not, is added. However, the peaks corresponding to V8, N39, C40, and C42 are undetectable only when methylated GlcCer is added, suggesting that these four residues are specifically affected, directly or indirectly, by the presence of the C9-methyl (Fig. 2).
To go further, NMR spectra of ETD151 (50 μM, final concentration) in the presence of PC-methylated GlcCer LUVs at different molar ratios (ETD151: methylated GlcCer; 10:1, 1:1, and 1:10) were performed to identify which residues were most affected in the binding, and thus define which part(s) of the peptide chain would play a key role in ETD151’s affinity for the target, namely methylated GlcCer.
As seen in Figure 3A, the increased concentrations of methylated GlcCer-containing PC LUVs did not result in a discernible shift in the resonance positions of ETD151 residues. However, this resulted in a significant and progressive loss of the overall signal intensity of 15N-ETD151 (Fig. 3B). The progressive attenuation of ETD151 signal intensity is indicative of its binding to PC-methylated LUVs due to the slower tumbling of the large complex formed. Interestingly, differential perturbations in the intensity of the cross-peaks of certain residues were obtained, indicating a differential contribution to the interaction. To quantitatively discriminate the most perturbed residues in the presence of methylated GlcCer, we calculated the mean ratio intensity (Imethylated GlcCer/I0, where I0 is the intensity of ETD151 peak in free state), determined the SD (σ), and set a threshold at −1 SD from the mean (mean −1σ). Any residue with a ratio intensity below this threshold indicated a potential perturbation due to the presence of methylated GlcCer (Fig. 3C). The absence of resonance in some residues was attributed to linewidth broadening, indicating complete partitioning of the peptide into the lipid bilayers or strong binding to the liposome surface. These residues, namely V8, G10, C18, N37, N39, C40, and C42 (Fig. 3C, color blue), are likely to be the most affected by the presence of the methylated GlcCer. The intensity of several peaks decreased significantly (intensity below the threshold defined by mean −1σ), specifically the peaks of residues L3, I4, W9, A11, A20, G30, C32, F35, and A36 (Fig. 3C, color cyan). The ETD151 residues highlighted in Figure 3C show that the area comprising the L1 and L3 loops is likely to be the most involved in lipid binding, involving mainly hydrophobic residues (W9, F35, V8, A11, F35, A36), and some polar residues (N37, N39).
Figure 3.
Solution NMR analysis for ETD151 structure in the presence of methylated GlcCer-containing PC-based LUVs. A, superimposed 15N-ETD151 (50 μM) 2D SOFAST-HMQC spectra in the presence of PC-methylated GlcCer LUVs at different ETD151: methylated GlcCer molar ratios; 10:1 (black), 1:1 (pink), and 1:10 (cyan). B, peak intensity ratios of 15N-ETD151 (50 μM) in the presence of PC-methylated GlcCer LUVs at different ETD151: methylated GlcCer molar ratios; 10:1 (black), 1:1 (pink), and 1:10 (cyan) over the I0 intensity of the corresponding peak in the reference spectrum (15N-ETD151 free-state at 50 μM). Residues of ETD151 are shown as consecutive numbers on the x-axis. The horizontal line marks the mean of the intensity ratio in the presence of ETD151: methylated GlcCer at a molar ratio of 1:10, −1 SD (−1σ). Asterisks indicate residues for which peaks are absent in the free and bound states of ETD151. In ETD151 sequence, residues with the most dramatic decrease in intensity due to significant exchange broadening are reported in cyan (below the threshold) or in blue (missing residues at ETD151: PC-methylated GlcCer LUV, 1:10 ratio). The secondary structure elements are displayed above the sequence, with black arrows indicating the three β-strands (β1, β2, and β3), and a black rectangle for the α-helix, connected by three loops (L1, L2, and L3). C, 3D structure of ETD151 showing the most affected amino acids by the addition of PC: methylated GlcCer LUVs (intensity below the threshold defined in (B). Residues with the most dramatic decrease in intensity due to significant exchange broadening are reported in cyan (below the threshold) or in blue (missing residues at ETD151: PC-methylated GlcCer LUV 1:10 ratio). GlcCer, glucosylceramide; LUV, large unilamellar vesicle; PC, phosphatidylcholine; SOFAST-HMQC, band-selective optimized flip angle short transient heteronuclear multiple quantum coherence.
ETD151 increases the lipid dynamics of GlcCer-containing membranes, with a more pronounced effect with methylated GlcCer
Methylation of GlcCer plays a pivotal role in regulating the physical properties of the plasma membrane in terms of lipid organization and membrane fluidity. Since GlcCer are the membrane target of ETD151, we suggest a biophysical link between GlcCer-containing vesicles and ETD151 interaction, raising the possibility of modification of lipid order and dynamics. Due to their high sensitivity to changes in lipid dynamics, 31P and 2H ss-NMR experiments have been widely used to investigate the impact of peptide presence on membrane models (44).
In this study, static 31P ss-NMR experiments were conducted to gain insight into the molecular mobility of the polar head group of PC lipid and static 2H ss-NMR provided additional information on the dynamics of the hydrophobic core using a deuterated 1-palmitoyl-d31-2-oleoyl-sn-glycero-3-phosphocholine 16:0-d31-18:1 (d31-POPC) chain. MLVs containing GlcCer without ETD151 were used as a control and compared to MLVs with the presence of a ratio of 1 ETD151 to 20 lipids. It is worth noting that although the lipids are initially well mixed, the preparation of MLVs may induce phase separation. Moreover, the presence or absence of ETD151 could differentially affect this separation, leading to lipid segregation that we cannot assess.
All 31P spectra (Fig. 4, A and C), recorded with methylated or nonmethylated GlcCer-containing MLVs, in the absence or presence of ETD151, are characteristic of a lamellar phase with axial symmetry. Also characteristic of a lamellar fluid phase, all 2H spectra (Fig. 4, B and D) exhibited a symmetric spectrum. For nonmethylated GlcCer-containing MLVs, ETD151 increased the dynamic of the phosphate group of PC lipids (chemical shift anisotropy [CSA] decreased from approximately 31–29.5 ppm), with a slight decrease in the downfield edge (//edge) intensity of the 31P spectra (Fig. 4A, circled gray area). This observation indicates the deformation of spherical liposomes into “prolate-like” liposomes in the magnetic field, following an increase in membrane elastic properties (45). However, no significant differences were observed in static 2H spectra with or without ETD151 for these vesicles (Fig. 4B), suggesting that ETD151 did not disrupt the dynamics of the d31-POPC in this membrane model. This also suggests that the effect on nonmethylated GlcCer-containing MLVs order is very subtle, with a 31P spectral shape modification indicating a change of orientation in the phosphate group.
Figure 4.
Solid state-NMR study of the effect of ETD151 presence on the dynamics of GlcCer-containing PC-based vesicles at a peptide/lipid ratio 1:20. A, static 31P spectra and (B) static 2H spectra of nonmethylated GlcCer-containing vesicles in the absence (black) or presence (orange) of ETD151.//edge and ⟂ edge indicate the downfield edge and perpendicular edge, respectively. C, static 31P spectra and (D) static 2H spectra of methylated GlcCer-containing vesicles in the absence (black) or presence (blue) of ETD151. Chemical shift anisotropy (CSA) values are indicated in ppm. GlcCer, glucosylceramide; PC, phosphatidylcholine.
Regarding methylated GlcCer-containing MLVs, a decrease in the CSA from 31.5 to approximately 29 ppm is also observed for lipid systems upon the addition of ETD151 (Fig. 4C), indicating increased dynamics in the headgroup part of the membrane. The addition of ETD151 has a clear impact on static 2H spectra obtained with the methylated GlcCer model, leading to an overall decrease in spectral width (Fig. 4D), corresponding to an increase in the dynamics of the hydrophobic core of the membrane, where the deuterons are located. While order parameters at each position could theoretically be measured, an overall quantification could be assessed easily by measuring the decrease of 2H second spectral moment M2, reflecting a more disordered membrane when containing ETD151 (see Table S2). This disordering is often interpreted as an increase in membrane fluidity, whereas increased order is associated with augmented rigidity or stiffening (46).
Overall, the ss-NMR results obtained in our experimental conditions suggest that ETD151 was not deeply inserted into the hydrophobic part of the nonmethylated GlcCer-containing model membranes and is primarily located near the phosphate group of phospholipids, at the vesicle surface. However, ETD151 insertion in POPC-containing methylated GlcCer is more profound, with a greater impact on overall membrane dynamics and a notable disordering primarily observed of the hydrophobic part.
Discussion
ETD151 was optimized from the defensin Heliomicin isolated from the tobacco budworm H. virescens (24). It is highly effective against most fungal pathogens (e.g., A. fumigatus, C. albicans, C. neoformans) (24), which have developed multiresistance to available antifungal drugs (47). Moreover, ETD151 showed low toxicity in mouse models (26). ETD151 therefore warrants further investigation as a promising antifungal peptide. While some steps of its MoAs (27) and the crucial role of fungal GlcCer for ETD151 activity have been revealed (25), the mechanisms by which ETD151 targets fungal GlcCer, at the molecular and atomic scale require further investigation.
GlcCer are well-conserved glycosphingolipids in fungal membranes and play essential structural and signaling roles in cell survival and pathogenicity (17, 48). As their presence in the fungal membrane is important for the activity of certain CSαβ defensins (20, 21, 22, 37), there is a strong case for GlcCer to be a putative binding target for these defensins, such as Psd1, MtDef1, and AFP1. GlcCer interaction is a key step for antifungal defensin activity and the cascade of events leading to the attack on fungi. It is therefore essential to understand the structural and biochemical bases of this interaction to fully exploit the potential of GlcCer-interacting defensins as antifungal agents. In this study, we initially assessed the binding affinity using MST and ITC techniques. Then, we identified the disruptions at the atomic scale from the perspective of either the protein or the membrane, by NMR studies. Finally, given that methylated GlcCer are found only in fungi, we investigated the impact of the C9-methyl on the ETD151–GlcCer interaction. To characterize the molecular recognition between ETD151-fungal GlcCer and determine whether the methyl impacts this recognition, we used two commercially available GlcCer differing only by the presence of the C9-methyl: a fungal methylated GlcCer from the golden oyster mushroom Pleurotus citrinopileatus (d19:2/h16:0) and a plant nonmethylated GlcCer (d18:2/h16:0) from soybean. Each GlcCer was incorporated into PC-based LUVs at the same ratio to create simplified biomimetic membranes.
ETD151 demonstrates a greater affinity for methylated GlcCer
For the characterization of drug–target interactions, the determination of the binding (i.e., binding affinity) between the two partners is a key issue. This study combined ITC and MST assays, which indicated that ETD151 binds to PC-based vesicles when containing GlcCer. Unlike other GlcCer-interacting defensins, including Psd1, Psd2, and Sd5 defensins which have shown affinity for PC-based vesicles (20, 23, 49, 50), a nonsignificant interaction was observable between PC and ETD151 in PC-LUVs used as control. It is noteworthy that with both techniques, the measured affinity of ETD151 is higher when the incorporated GlcCer is methylated, clearly indicating the influence of the C9-methyl. Previous in vitro studies have demonstrated the following fungus-specific feature: C9-methylation plays a role in the binding of GlcCer by the pea defensin Psd1 (33) and AFP1 from the brown Eastern mustard B. juncea (37). Indeed, its absence in the sphingoid base prevents RsAFP2 from interacting with GlcCer (19) and reduces the binding affinity of Psd1 to GlcCer-containing lipid vesicles by 2.5-fold (20). It is worth noting that these previous studies (19, 20) used fungal and plant GlcCer, which, differ in fatty acid length and unsaturation state, in addition to the difference in C9-methyl in the sphingoid base. Therefore, the role of these structural variations in the binding affinity of antifungal CSαβ defensins to GlcCer cannot be ruled out. To our knowledge, this study demonstrates the in vitro impact of C9-methyl on ETD151-GlcCer binding.
There are limited data available in the literature regarding the estimation of defensin-GlcCer binding, data acquired with different biophysical techniques, such as reverse ELISA (19), surface plasmon resonance (SPR) (20, 23), MST, and ITC (this work). This makes affinity comparisons quite challenging (51). In a reverse ELISA assay, glycolipids are immobilized on the hydrophobic surface of the wells of microtiter plates and the bound defensin is detected immunologically. This technique has been used to characterize the binding of RsAFP2 and Heliomicin to fungal methylated GlcCer and showed a dose-dependent interaction (20). SPR experiments were conducted to examine the interaction of mimetic fungal membranes with the pea defensins Psd1 and Psd2. In these experiments, liposomes were immobilized on the surface of the chip before peptide injection at a given concentration under constant flow. The SPR assay provides binding affinity and kinetic parameters, association and dissociation rates (Ka and Kd). However, in all studies of defensin–GlcCer interaction by SPR, the defensin-membrane dissociation phase was insignificant (Ka >>> Kd). Therefore kinetic data (Ka and Kd values) could not be obtained (20, 23). Binding affinity was therefore assessed by the equilibrium response before dissociation (Req) and/or by the speed of initial association indicated by the initial association rate (α-value). Higher Req and α values correlate with greater peptide binding affinity to membranes. The incorporation of 30% mol GlcCerFusarium solani into a PC-based vesicles increased binding 5-fold for Psd1 compared to a PC-based vesicles, based on an α-value (20). However, Psd2, which shares approximately 45% sequence identity with Psd1, showed a 2-fold stronger binding response intensity with pure POPC-based vesicles than with POPC: GlcCerF. solani vesicles (70:30), based on Req value (23).
To examine ETD151 interaction with methylated GlcCer, we employed both MST and ITC techniques to estimate the Kd values, each method having its own advantages. The Kd value estimated by MST with the commercial fungal GlcCer (d19:2/h16:0, corresponding to one of the two main GlcCer directly extracted from B. cinerea, and named methylated GlcCer in this work) is in the range of 0.7 μM. This value is consistent with that obtained in our previous work for ETD151 binding to liposomes containing GlcCer directly extracted from B. cinerea (mixture of d19:2/h16:0 and d19:2/h16:1, Kd is in the range of 0.5 μM) (25). With the commercial fungal GlcCer (d19:2/h16:0), we were able to evaluate the Kd value by ITC too. ITC-derived Kd values were consistent with those obtained from MST experiments in that both methods indicated micromolar affinities and a stronger binding of ETD151 to GlcCer when the latter is methylated. However, the absolute Kd values measured by ITC were higher than those obtained by MST. This discrepancy between ITC and MST measurements is well-documented in the literature and largely attributed to differences in experimental setup (52).
Among these variations, the choice of appropriate concentration ranges for both macromolecule and ligand is a crucial parameter. In MST experiments, this choice is simpler; the concentration of the fluorescent molecule is generally kept below the expected Kd, minimizing the risk of aggregation. On the other hand, the choice of appropriate concentrations for ITC is more complex due to multiple influencing factors. In our study, MST was performed using more diluted concentrations of both ETD151 peptide and LUVs compared with ITC. In the ITC setup, we cannot rule out the formation of (nano)-aggregates of either ETD151 and/or liposomes during titration, which may have contributed to the higher apparent affinities measured by ITC. A relevant example is shown by Winiewska et al., in which the binding of halogenated benzotriazoles to human CK2 resulted in ITC-derived Kd values that were approximately 10-fold higher than those from MST (53). This discrepancy was attributed to ligand nano-aggregation upon injection into the ITC cell, the aggregates that dissociate slowly distorting, therefore, the affinity measurement. Similar artifacts could occur even below the solubility limit, potentially biasing ITC-derived Kd values.
Timing of data acquisition is another important consideration. ITC records heat changes in real time during titration, capturing transient events and making it useful for studying reaction kinetics (54). However, this also means ITC may yield biased affinity values in systems with intermediate or slow binding kinetics, such as the interaction between ETD151 and GlcCer-containing LUVs as indicated by our NMR data, or when time-dependent phenomena such as conformational changes, oligomerization, or dissolution of (nano)-aggregates occur postinjection. In contrast, MST measures interactions after the system has reached equilibrium and is therefore less susceptible to such effects. Nevertheless, each method has its own limitations. While ITC is a label-free technique, MST requires fluorescent labeling. Although our labeling strategy selectively targeted the N-terminal amino group which, based on NMR data, is located far from the binding interface, we cannot entirely exclude the possibility of minor effects on ligand interaction.
The hydrophobic adjacent loops of ETD151 are impacted by the presence of GlcCer in model membranes
Identifying which parts of the ligand contribute to binding with the target is essential for understanding the ligand–target interaction at the atomic level. The positive charge of several antifungal defensins has been reported to determine membrane surface binding activity, particularly with the negatively charged phosphate head group of phospholipids (55). In solution NMR, signals of free-moving peptides in solution are typically sharp and intense. In contrast, peptides bound to liposomes are expected to show significant line broadening and intensity loss due to the slow tumbling motion of liposomes. Thus, if the addition of the liposomes does not induce a notable change in chemical shift or relative intensity to the peptide alone in the solution, it is considered a nonstrong interaction. By contrast, when the addition of the liposomes results in a significant loss of intensity and/or clear CSP, the residues affected by the interaction can be identified. Moreover, at identical liposomes and peptide concentrations, greater attenuation of peptide signal intensity can be interpreted as higher peptide/liposomes affinity, allowing peptides to be classified according to their liposome affinity.
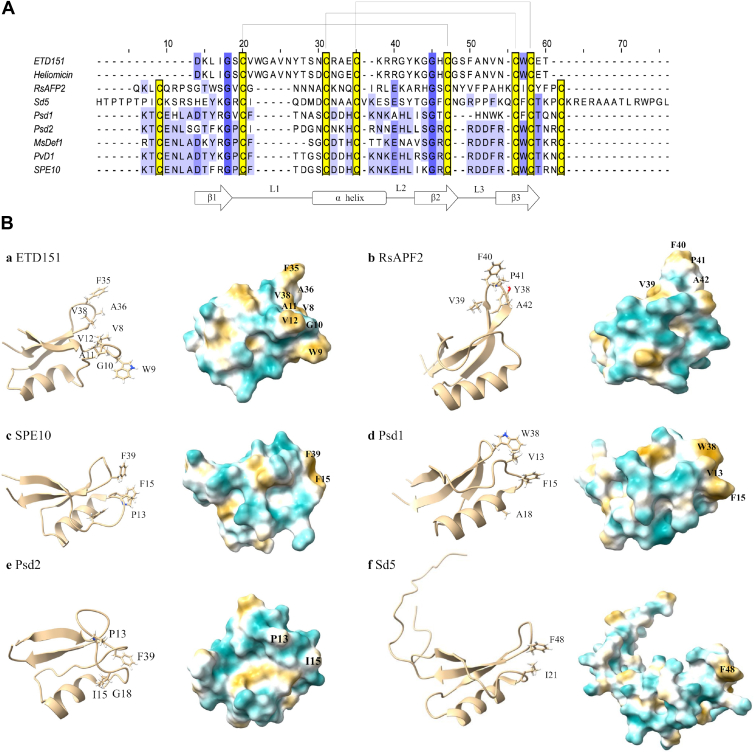
The NMR results suggest that the basic residues of ETD151 play a minor role in recognizing fungal GlcCer-containing vesicles. However, they demonstrate that the two L1 and L3 hydrophobic loops are seemingly those by which ETD151 interacts with or inserts into liposomes containing GlcCer (Fig. 3B). This predominant hydrophobic role correlates with ITC results indicating an entropy-driven interaction. Except the conservation of the disulfide bridges array (in yellow, Fig. 5A), sequence conservation is generally low in the antifungal defensin family. In particular, the L1 loop of ETD151 is highly atypical and longer than usual (Fig. 5A). The hydrophobic residues V8/W9/G10/A11/V12 of L1 are close to F35/A36/V38 of the L3 loop, forming a significant hydrophobic patch (Fig. 5, B-a). Note that V8, W9, G10, A11, F35 and A36 are among the most disturbed residues when ETD151 is in interaction with GlcCer-containing membranes (Fig. 3B). Such hydrophobic patches involving L1 and L3 loops are found in some antifungal plant defensins, with hydrophobic residues playing important biological roles, including activity, pathogen target recognition, and/or dimer formation (56, 57). For RsAFP2, mutagenesis of key hydrophobic residues in L3 loop, namely Y38, V39, F40, P41, and A42 (Fig. 5, B-b), resulted in a sharp decrease in antifungal activity compared to the WT (56). Note that V39, F40 and A42 correspond to F35, A36 and V38 in ETD151 (Fig. 5, A and B-a and B-b). In SPE10 from Pachyrhizus erosus jicama seeds, residues P13, F15 located in the L1 loop, and F39 located in the L3 loop forming the hydrophobic patch (Fig. 5B-c) are required for antifungal activity and dimer formation (with F15 and F39 corresponding to V8 and V38 in ETD151, respectively). In particular, residue F39 has been identified as essential for antifungal activity (57). Solution NMR analysis of defensin Psd1 showed that hydrophobic residues V13, F15, and A18 in the L1 loop and residue W38 in the L3 loop (Fig. 5B-d) (with F15 and W38 corresponding to V8 and V38 in ETD151, respectively) are disrupted by GlcCerF. solani in PC lipid vesicles (58). Note that these four residues correspond to less hydrophobic residues in Psd2 (P13, I15, G18, and F39, respectively), for which no clear hydrophobic area is visible (Fig. 5B-e) (59). In addition to the dominant hydrophobic interactions, the involvement of other types of interactions between ETD151 and methylated GlcCer, such as hydrogen bonds, remains a possibility. Furthermore, a conformational adaptation of the ETD151 structure to recognize GlcCer may be suggested, since two cysteines (C18, C40) forming one of the three disulfide bridges were disrupted. The multiple effects induced by the interaction between defensin and GlcCer have already been postulated for Psd1 (58).
Figure 5.
Sequences, 3D structures, and hydrophobic surface properties of some antifungal defensins.A, sequence alignment for a selected list of antifungal defensins. Cysteines are shown in yellow, while conserved residues are highlighted in blue. Top: black brackets represent disulfide bridges between corresponding cysteine residues. Bottom: the secondary structure elements are displayed above the sequence, with black arrows indicating the three β-strands (β1, β2, and β3), and a black rectangle for the α-helix, connected by three loops (L1, L2, and L3). B, structures of some of the discussed antifungal defensins and their hydrophobic surface properties. Hydrophobic and hydrophilic potential areas are displayed in yellow and blue, respectively. Residues that are discussed are shown in 3D structures and surfaces. B-a, ETD151 (PDB code: 1P00); B-b, RsAFP2 (PDB code: 2n2r); B-c, SPE10 (PDB code: 3psm); B-d, Psd1 (PDB code: 1jkz); B-e, Psd2 (PDB code: 6nom); and B-f, Sd5 (PDB code: 2ksk).
Structural changes in defensins recognizing GlcCer in the bound state have rarely been studied. However, there are indications that ETD151 shares structural changes in common with Psd1 when binding to fungal GlcCer. Indeed, for Psd1, the L1/L3 region exhibits reduced conformational exchange, likely due to the stabilization of a specific membrane-bound conformation of the peptide (58). In contrast, different structural regions of the sugar cane Saccharum officinarum defensin Sd5 were highlighted upon recognition of fungal GlcCer. The L1/L3 hydrophobic patch is small, involving I21 in the L1 loop and F48 in the L3 (corresponding to V8 and V38 in ETD151) (Fig. 5B-f). The Sd5 region considered specific for Sd5–GlcCer interaction comprises part of the α-helix and the L2 loop (49).
Dynamic NMR studies for Psd1 and Sd5, two peptides with completely different dynamic properties, showed molecular recognition of fungal GlcCer by conformational selection. Upon binding to GlcCer, a modification of ETD151 dynamics could not be excluded. The internal dynamic of ETD151 in the free state, studied using NMR relaxation experiments, is in favor of a compact peptide (longitudinal relaxation rate R1, Fig. S4A), without large amplitude internal motions on the ps∼ns timescale (heteronuclear nuclear overhauser effect, Fig. S4C). Interestingly, transverse relaxation rate values (R2) and dispersion experiments (Carr-Purcell-Meiboom-Gill [CPMG]) of ETD151 suggest that exchange processes occur on the μs∼ms timescale (see Fig. S4, B and D), particularly for residues I4, S34, F35, N37, N39, and C40 (see Fig. S4E). These residues are identified as potentially involved in the target-binding event (solution NMR results). Importantly, the signal of V38 is undetectable due to an increase of its linewidth (via “exchange broadening”) also reflecting the intermediate exchange regime. Dynamic processes in this time window include side chain reorientation, loop motion, secondary structure changes, and hinged domain movements. Such motions may affect processes including ligand binding and release, folding and unfolding events, allostery, and the rate of catalytic turnover. Therefore, despite sharing a common CSαβ structural fold, there seems to be no consensus among antifungal defensins regarding interaction with lipid targets, including fungal GlcCer. For ETD151, the two adjacent L1 and L3 loops constitute an essential region that would monopolize methylated GlcCer binding.
ETD151 specifically inserts into and disorders PC lipid vesicles containing methylated GlcCer
The lipid-binding activity of AMPs is thought to modify several structural and dynamic properties of the membrane (60, 61, 62). While the effects of several linear AMPs with pore-forming activity (e.g., melittin, magainin, gramicidin, aurein, caerin) on lipids have been reported via ss-NMR (63, 64), the impact of disulfide-rich peptides has rarely been investigated (e.g., human β-defensin-3 analogs (65)). In this study, we employed 31P and 2H ss-NMR experiments to examine the disruption of lipid order caused by the interaction with ETD151. In the case of MLVs containing nonmethylated-GlcCer, ETD151 seems to remain on the lipid surface near the phosphate head group, deforming MLVs into nonspherical vesicles without disrupting the dynamics of lipid hydrophobic chains. In the case of MLVs containing methylated-GlcCer, ETD151 binding disrupts the order in the polar and hydrophobic parts of the lipid membranes, suggesting peptide insertion into the membrane. This suggests that the presence of the C9-methyl in GlcCer plays a role for ETD151-induced membrane disordering and is a key factor for ETD151 to insert deeper into fungal membrane models. Moreover, our results show that ETD151 binding induces a slight increase in membrane disorder. Under similar conditions, we have observed the broadening of ETD151 15N-1H heteronuclear single quantum coherence spectroscopy NMR signals, indicating reduced dynamics and a stiffening of the peptide as it penetrates the membrane. Interestingly, analogous observations have been reported for rhamnolipids, potential biocontrol agents for crop protection with antifungal activities, using representative fungal membrane models (66). This differs from the human β-defensin-3 analog, which rigidified negatively charged lipids mimicking bacterial membranes in addition to membrane morphological disruption (65). This indicates a possible different mechanism, at least in the membrane binding step of pore formation.
The C9-methyl in the sphingoid base plays a major role in the ETD151–GlcCer interaction
In a membrane context, the presence of C9-methyl in the GlcCer markedly enhances affinity with ETD151, influences the two adjacent loops of the ETD151 structure, and reinforces the destabilization of lipid dynamics caused by the peptide. Changes in the biophysical properties of membranes due to the presence of C9-methyl and/or the direct involvement of C9-methyl in the binding sites with ETD151 may explain the selectivity of ETD151 for membranes containing methylated GlcCer rather than nonmethylated GlcCer. Indeed, C9-methylation in GlcCer has been reported to influence membrane topography by affecting lipid organization, which directly impacts membrane permeabilization (35, 67, 68). Structural features of fungal GlcCer, for example, C9-methylation and the Δ8 double bond, could increase the physical distance between the hydrophobic core of GlcCer and other membrane lipids (34). Such effects on the biophysical properties of membranes may facilitate ETD151 access to the hydrophobic core of fungal membranes. It is worth noting that a correlation between the structural role of methylated GlcCer in membranes and virulence in fungal cells has been documented. For example, Δsmt C. neoformans mutants, unable to infect a mouse model, showed a defect in cell membrane structures, as confirmed by fluorescence spectroscopy and atomic force microscopy studies (34). The second hypothesis is that C9-methyl is directly involved in the binding sites with ETD151. Our findings demonstrate that ETD151 binds primarily by hydrophobic effects to GlcCer-containing membranes and that increased hydrophobicity by C9-methyl correlates with enhanced affinity of ETD151 for GlcCer. The dependence of defensin activity on the presence of methylated GlcCer in fungal cells is controversial. Although RsAFP2, MsDef1, and Psd1 showed similar activity against Δsmt fungal mutants and their WT strains (23, 36), Psd2 showed reduced activity against Δsmt fungal mutants compared to WT strains (33). The C9-methyl in the sphingoid base plays a major role in the defensin–GlcCer interactions. However, the precise mechanism by which the C9-methyl is involved in GlcCer recognition by antifungal defensins has yet to be determined.
Finally, using biophysical and spectroscopic methods, this study provides molecular-level insights into how the antifungal insect defensin ETD151 interacts with its fungal membrane target, GlcCer. Furthermore, the presence of the methyl in GlcCer plays a role in ETD151-induced membrane disordering and is a key factor in ETD151's deeper insertion into fungal membrane models, explaining why ETD151's binding affinity for GlcCer is greater when GlcCer is methylated. These new findings pave the way for the optimization and development of defensin mimetics to selectively combat pathogenic fungi by targeting methylated GlcCer while minimizing toxicity to the host.
Experimental procedures
Peptides and lipids
The unlabeled recombinant ETD151 was provided by Dr Philippe Bulet. Recombinant expression of the uniformly isotopically 15N-labeled ETD151 peptide (15N-ETD151) in Saccharomyces cerevisiae was entrusted to the Promise company.
GlcCer from the golden oyster mushroom P. citrinopileatus was purchased from Funakoshi and referred to here as “methylated GlcCer.” Soy GlcCer referred to here as “nonmethylated GlcCer” and egg L-α-PC and d31-POPC were purchased from Avanti Polar Lipids.
Preparation of LUVs
The lipid stock solutions (PC, nonmethylated GlcCer, and methylated GlcCer) were prepared in pure chloroform (purity higher than 99.8%, Sigma-Aldrich) and mixed with appropriate volumes to obtain the desired concentrations. The organic solvent was dried under a stream of nitrogen and lyophilized overnight. The resulting dry lipid films were hydrated with phosphate buffer 10 mM, pH 5.8 (buffer A), and homogenized by five cycles of freezing (liquid nitrogen) and thawing (40 °C) to produce vesicle suspensions. To calibrate and homogenize the size of the vesicles, the suspensions were passed 11 times using a mini-extruder through 0.1 μm pore-size polycarbonate membrane filters (Avanti Polar Lipids). The polydispersity index and the hydrodynamic diameter of the generated LUVs were assessed by dynamic light scattering (DLS) spectroscopy at 20 °C. DLS experiments were conducted using a Zetasizer (Malvern Panalytical, Instruments version 7.11, https://www.malvernpanalytical.com/en/support/product-support/software/zetasizer-family-software-update-v7-11) with low-volume cuvettes (VWR International). DLS data were acquired using the Zetasizer software version 7.11. All LUV suspensions were freshly prepared and presented a hydrodynamic diameter ∼100 nm and a polydispersity index < 0.01, and used for all experiments, except for ss-NMR.
Isothermal titration calorimetry
Prior to the ITC experiment, all solutions (buffer A, peptide solution, liposome solutions) were degassed under vacuum to remove air bubbles. LUV suspensions of 3.5 mM PC or PC: nonmethylated GlcCer (9:1, molar ratio) or PC: methylated GlcCer (9:1, molar ratio) in buffer A were titrated into a 250 μM ETD151 solution in the same buffer A. The freshly prepared LUVs suspensions were flushed with argon and kept at +4 °C for a maximum of 2 days. Blank titrations were conducted by titrations of LUVs with varying compositions into the buffer A.
All binding ITC experiments were performed using a MicroCal PEAQ-ITC microcalorimeter (Malvern Panalytical Ltd). Samples were equilibrated to 25 °C before measurement. Titrations were performed at 25 °C under constant stirring (750 rpm). Each experiment consisted of an initial injection of 0.3 μl followed by 16 separate injections of 2.5 μl into the sample calorimeter cell of 200 μl. The time between each injection was 180 s, and the measurements were performed with the reference power set to 5 μcal/s and the feedback mode set to “high.”
The acquired calorimetric data were analyzed using MicroCal PEAQ-ITC Analysis Software version 1.20 (Malvern Panalytical Ltd, https://www.malvernpanalytical.com/en/support/product-support/software/microcal-peaq-dsc-software-update-v1-20) and the graphs generated by the same software. To achieve the titrations with GlcCer-containing LUVs, the binding data were fit based on the “one set of sites” fitting model of the software, using the titrant concentration corresponding to that of GlcCer (10% mol of the total lipid concentration). Heat associated with the first injection was not included in the data analysis. Binding curves fitting generate a Kd, stoichiometries (n), enthalpy of binding (ΔH) within 95% confidence intervals, from which the apparent binding free energy (ΔG) and entropy (−TΔS) are calculated. Thermodynamic parameters are reported as the average of triplicates with SD. Blank titrations and dilution data were compared to and not subtracted from the binding data.
Microscale thermophoresis
MST experiments were performed using a Monolith NT.115 (red) instrument (NanoTemper Technologies). A serial dilution with liposome solution (½ dilution, 16 concentrations from 1 mM to 30.5 nM) were performed in buffer A. ETD151 was labeled with Atto647 (NHS ester reactive) from Sigma-Aldrich (07376) according to (25). The fluorescently labeled peptide (Atto647-ETD151) was diluted in buffer A and added to each liposome to a final concentration of 50 nM. All samples were measured after 5 min of incubation and centrifugation (2 min, 5000g). The experiments were performed at 40% MST power and between 20 and 80% LED power at 22 °C. MST traces were recorded using the following parameters: 5 s MST power off, 20 s MST power on, and 5 s MST power off. The Kd between peptide–liposome interaction was calculated based on the protocol provided by NanoTemper Technologies using the MO.Affinity Analysis 2.3 software (https://support.nanotempertech.com/hc/en-us/articles/19204575620241-Monolith-software-download-instructions).
Solution NMR
All NMR experiments were performed at 298 K on a 700 MHz Bruker Avance III HD spectrometer equipped with a triple resonance (1H, 13C, 15N) cryoprobe. All data were processed using the Topspin 3.2 Bruker software (https://www.bruker.com/en/products-and-solutions/mr/nmr-software/topspin.html), cross peaks assignment and intensities (height extraction) were performed with CcpNmr (69). Sensitive fast-pulse experiments 2D 1H-15N band-selective optimized flip angle short transient heteronuclear multiple quantum coherence (SOFAST-HMQC) (70) were recorded using Bruker pulse sequence “fhmqcf3gpph” with spectral widths of 1024 × 128 complex points in the 1H and 15N dimensions, respectively.
Lyophilized 15N-ETD151 was dissolved in buffer A to 100 μM. 1H-15N SOFAST-HMQC spectra were recorded for free ETD151 in solution (50 μM as final concentration), and cross-peak assignment was based on PDB 1P00 deposited data. Peak intensities of free-state 15N-ETD151 are referred to as I0. Next, 1H-15N SOFAST-HMQC spectra were recorded for 15N-ETD151 at 50 μM in the presence of 5 mM PC LUVs or PC-GlcCer LUVs (nonmethylated) or PC-methylated GlcCer LUVs, and the chemical shift and peak intensities were extracted. The peak intensities of 15N-ETD151 in the presence of each LUVs composition are referred to as IPC, Inonmethylated GlcCer, and Imethylated GlcCer, respectively. The effects of adding PC-methylated GlcCer LUVs to ETD151 at different molar ratios (ETD151: methylated GlcCer; 10:1, 1:1, and 1:10) on the 1H-15N SOFAST-HMQC spectrum of 15N-ETD151 at constant concentration (50 μM) were assessed. For each tube, 10% (v/v) of D2O was added as a final concentration. The effect of the addition of PC LUVs, PC-GlcCer LUVs, and PC-methylated GlcCer LUVs on the ETD151 structure was evaluated by the intensity ratio, namely (IPC/I0), (Inonmethylated GlcCer/IPC), and (Imethylated GlcCer/IPC), respectively. ETD151 residues showing a decrease in the intensity ratio of less than −1 SD (−1σ) were considered to be significantly impacted by the presence of lipids contained in LUVs. Figures 3C and 5B were generated using PyMOL (71) and ChimeraX 1.4.
NMR relaxation experiments
The recombinant 15N-ETD151 peptide was dissolved in phosphate buffer (40 mM, pH 5.5) to obtain a final concentration of 0.4 mM. NMR relaxation experiments were performed at 298 K on an Avance III HD Bruker 700 MHz spectrometer equipped with a cryoprobe. 15N R1 relaxation delays of 10, 20, 30, 50, 75, 100, 150, 200, 300, 500, 1000, 2000, and 3000 ms and R2 relaxation delays of 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 125, 150, 200, 250, and 300 ms were used for data collection. The above time series were fitted by a single exponential decay.
The 15N-NOE spectra were collected with a 3 s relaxation delay. The 1H-15N heteronuclear NOE was calculated from the equation NOE = Isat ⁄ Ieq, where Isat and Ieq represent the volumes of a cross-peak in the spectra collected with and without proton saturation, respectively. All experiments were performed three times under identical conditions. Volumes for the amide 15N-1H cross peaks were measured using Topspin TM 3.2 Bruker software. Uncertainties in the volumes were determined from the duplicate spectra.
1H-15N CPMG experiments were recorded with νCPMG = 0, 50, 75, 100, 125, 150, 175, 200, 250, 300, 350, 400, 500, 600, 700, 800, and 900 Hz, and a CPMG constant time delay of 40 ms. Peak intensities were plotted as effective relaxation rates and relaxation dispersion curves were fitted using the ShereKhan web application (72). The data were fit using a global two-state exchange process with the Carver–Richards model for slow exchange (73).
Solid-state NMR
For ss-NMR experiments, MLVs were prepared using the film method as previously described (74). Appropriate volumes of lipid stock solutions in chloroform were mixed to obtain the desired lipid compositions (i) PC: d31-POPC: nonmethylated GlcCer and (ii) PC: d31-POPC: methylated GlcCer, both with a molar ratio of 7:2:1. The organic solvent was dried under a nitrogen stream. The residual organic solvent in lipid film was removed by high vacuum for at least 4 h. The resulting dried lipid film was hydrated with 50 mM HEPES, 10 mM NaCl, pH 7.4 (buffer B) in deuterium-depleted water from Sigma-Aldrich (7732-18-5). Lipid dispersion was vortexed and subjected to three cycles of freeze-thawing (10 min at −20 °C followed by 10 min at 40 °C) and transferred directly to an insert, which was then fitted into a 4 mm rotor. For samples containing ETD151, hydration of the lipid film was performed with buffer B containing ETD151 at a molar ratio equal to ½ that of GlcCer.
The ss-NMR experiments were performed at 298K on a Bruker Avance III 500 wide bore spectrometer, with resonance frequencies of 500.5 MHz for 1H, 202.6 MHz for 31P spectra and 76.8 MHz for 2H spectra, equipped with a 4-mm magic-angle spinning triple-resonance probe. Static 31P NMR spectra for the MLVs control sample and the ETD151-containing sample were acquired using a phase-cycled Hahn echo pulse sequence (75) with high-power (50 kHz) 1H decoupling during acquisition. Using a 90° pulse length of 3 μs, an interpulse delay of 35 μs, data were collected using 4 k data points with a recycle delay of 3 s and a total of 1 k scans per spectrum, amounting to 1 h of acquisition. The static 2H NMR experiments were carried out using a quadrupolar echo sequence (76) with 60 k data points, acquired with a 90° pulse length of 5 μs, an interpulse delay of 96 μs, and a recycle time set to 500 ms. A total of 100 k scans per spectrum were acquired over 16 h. Line broadening of 50 and 100 Hz was applied to the 31P and 2H spectra, respectively. The 31P CSA was determined by line fitting using the solid lineshape analysis (SOLA) module included in the Bruker Topspin 3.2 software. Magic-angle spinning 2H ss-NMR experiments were carried out using a 10 kHz spinning frequency and a phase-cycled quadrupolar echo sequence (77), with 100 k data points, acquired with a 90° pulse length of 4 μs, a rotor-synchronized interpulse delay of 96 μs and a recycle time of 500 ms. A total of 1 k scans per spectra were acquired for 10 min. 2H spectral moment analysis was performed using MestRenova software V6.0 (Mestrelab Research). Second spectral moments (M2) values, provide a quantitative description of the membrane lipid ordering, were calculated as previously described (77).
Data availability
All the relevant data are contained within this article and supporting information. Source data and additional data can be obtained from the corresponding author upon reasonable request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the MO2VING/NMR facility (CBM, Orléans, France), particularly Hervé Meudal for his technical advice in acquiring solution NMR experiments. For their technical assistance and helpful guidance in conducting and analyzing the ITC experiments, we also want to thank Caroline Mas (Grenoble Instruct-ERIC center, France), Franck Coste, and Marcin Suskiewicz (Center for Molecular Biophysics, Orléans, France).
Author contributions
O. K., F. P., R. N., R. N., V. A., P. B., D. W., and C. L. writing–review and editing; O. K. and C. L. writing–original draft; O. K. and C. L. visualization; O. K., D. W., and C. L. methodology; O. K., F. P., R. N., J.-B. M., V. A., D. W., and C. L. investigation; R. N., R. N., V. A., P. B., D. W., and C. L. validation; P. B. resources; P. B. and C. L. funding acquisition; P. B., D. W., and C. L. conceptualization; C. L. supervision; C. L. project administration.
Funding and additional information
This work used the platforms of the Grenoble Instruct-ERIC center (ISBG; UAR 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by INSTRUCT project (PID 23320, VID 39727), FRISBI (ANR-10-INBS-000502) and GRAL and financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003).
Reviewed by members of the JBC Editorial Board. Edited by Karen Fleming
Supporting information
References
- 1.Bongomin F., Gago S., Oladele R.O., Denning D.W. Global and multi-national prevalence of fungal Diseases—estimate precision. J. Fungi. 2017;3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denning D.W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 2024;24:428–438. doi: 10.1016/S1473-3099(23)00692-8. [DOI] [PubMed] [Google Scholar]
- 3.Brown G.D., Ballou E.R., Bates S., Bignell E.M., Borman A.M., Brand A.C., et al. The pathobiology of human fungal infections. Nat. Rev. Microbiol. 2024;22:687–704. doi: 10.1038/s41579-024-01062-w. [DOI] [PubMed] [Google Scholar]
- 4.Roemer T., Krysan D.J. Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb Perspect. Med. 2014;4 doi: 10.1101/cshperspect.a019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odds F.C., Brown A.J.P., Gow N.A.R. Antifungal agents: mechanisms of action. Trends Microbiol. 2003;11:272–279. doi: 10.1016/s0966-842x(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 6.Francois I., Cammue B., Borgers M., Ausma J., Dispersyn G., Thevissen K. Azoles: mode of antifungal action and resistance development. Effect of miconazole on endogenous reactive oxygen species production in Candida albicans. AIAMC. 2006;5:3–13. [Google Scholar]
- 7.Carolus H., Pierson S., Lagrou K., Van Dijck P. Amphotericin B and other polyenes—Discovery, clinical use, mode of action and drug resistance. J. Fungi. 2020;6:321. doi: 10.3390/jof6040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perlin D.S. Mechanisms of echinocandin antifungal drug resistance. Ann. N. Y Acad. Sci. 2015;1354:1–11. doi: 10.1111/nyas.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houšť J., Spížek J., Havlíček V. Antifungal drugs. Metabolites. 2020;10:106. doi: 10.3390/metabo10030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockhart S.R., Chowdhary A., Gold J.A.W. The rapid emergence of antifungal-resistant human-pathogenic fungi. Nat. Rev. Microbiol. 2023;21:818–832. doi: 10.1038/s41579-023-00960-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y., Robbins N., Cowen L.E. Molecular mechanisms governing antifungal drug resistance. NPJ Antimicrob. Resist. 2023;1:5. doi: 10.1038/s44259-023-00007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins N., Wright G.D., Cowen L.E. Antifungal drugs: the Current armamentarium and development of new agents. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.FUNK-0002-2016. [DOI] [PubMed] [Google Scholar]
- 13.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 14.Thevissen K., Kristensen H.-H., Thomma B.P.H.J., Cammue B.P.A., François I.E.J.A. Therapeutic potential of antifungal plant and insect defensins. Drug Discov. Today. 2007;12:966–971. doi: 10.1016/j.drudis.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Silva P.M., Gonçalves S., Santos N.C. Defensins: antifungal lessons from eukaryotes. Front. Microbiol. 2014;5:97. doi: 10.3389/fmicb.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vriens K., Cammue B.P.A., Thevissen K. Antifungal plant defensins: mechanisms of action and production. Molecules. 2014;19:12280–12303. doi: 10.3390/molecules190812280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandes C.M., Goldman G.H., Del Poeta M. Biological roles played by sphingolipids in dimorphic and filamentous fungi. mBio. 2018;9 doi: 10.1128/mBio.00642-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rollin-Pinheiro R., Singh A., Barreto-Bergter E., Del Poeta M. Sphingolipids as targets for treatment of fungal infections. Future Med. Chem. 2016;8:1469–1484. doi: 10.4155/fmc-2016-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thevissen K., Warnecke D., François I., Leipelt M., Heinz E., Ott C., et al. Defensins from insects and plants interact with fungal glucosylceramides. J. Biol. Chem. 2004;279:3900–3905. doi: 10.1074/jbc.M311165200. [DOI] [PubMed] [Google Scholar]
- 20.Neves de Medeiros L., Domitrovic T., Cavalcante de Andrade P., Faria J., Bergter E.B., Weissmüller G., et al. Psd1 binding affinity toward fungal membrane components as assessed by SPR: the role of glucosylceramide in fungal recognition and entry. Biopolymers. 2014;102:456–464. doi: 10.1002/bip.22570. [DOI] [PubMed] [Google Scholar]
- 21.Ramamoorthy V., Cahoon E.B., Li J., Thokala M., Minto R.E., Shah D.M. Glucosylceramide synthase is essential for alfalfa defensin-mediated growth inhibition but not for pathogenicity of Fusarium graminearum. Mol. Microbiol. 2007;66:771–786. doi: 10.1111/j.1365-2958.2007.05955.x. [DOI] [PubMed] [Google Scholar]
- 22.Mello Ede O., dos Santos I.S., Carvalho Ade O., de Souza L.S., de Souza-Filho G.A., do Nascimento V.V., et al. Functional expression and activity of the recombinant antifungal defensin PvD1r from Phaseolus vulgaris L. (common bean) seeds. BMC Biochem. 2014;15:7. doi: 10.1186/1471-2091-15-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amaral V.S.G., Fernandes C.M., Felício M.R., Valle A.S., Quintana P.G., Almeida C.C., et al. Psd2 pea defensin shows a preference for mimetic membrane rafts enriched with glucosylceramide and ergosterol. Biochim. Biophys. Acta Biomembr. 2019;1861:713–728. doi: 10.1016/j.bbamem.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 24.Landon C., Barbault F., Legrain M., Menin L., Guenneugues M., Schott V., et al. Lead optimization of antifungal peptides with 3D NMR structures analysis. Protein Sci. 2004;13:703–713. doi: 10.1110/ps.03404404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kharrat O., Yamaryo-Botté Y., Nasreddine R., Voisin S., Aumer T., Cammue B.P.A., et al. The antimicrobial activity of ETD151 defensin is dictated by the presence of glycosphingolipids in the targeted organisms. Proc. Natl. Acad. Sci. U. S. A. 2025;122 doi: 10.1073/pnas.2415524122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrès E. Cationic antimicrobial peptides in clinical development, with special focus on thanatin and heliomicin. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:881–888. doi: 10.1007/s10096-011-1430-8. [DOI] [PubMed] [Google Scholar]
- 27.Aumer T., Voisin S.N., Knobloch T., Landon C., Bulet P. Impact of an antifungal insect defensin on the proteome of the phytopathogenic fungus Botrytis cinerea. J. Proteome Res. 2020;19:1131–1146. doi: 10.1021/acs.jproteome.9b00638. [DOI] [PubMed] [Google Scholar]
- 28.Williamson B., Tudzynski B., Tudzynski P., van Kan J.A.L. Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 2007;8:561–580. doi: 10.1111/j.1364-3703.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 29.Hahn M. The rising threat of fungicide resistance in plant pathogenic fungi: botrytis as a case study. J. Chem. Biol. 2014;7:133–141. doi: 10.1007/s12154-014-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cools T.L., Struyfs C., Cammue B.P., Thevissen K. Antifungal plant defensins: increased insight in their mode of action as a basis for their use to combat fungal infections. Future Microbiol. 2017;12:441–454. doi: 10.2217/fmb-2016-0181. [DOI] [PubMed] [Google Scholar]
- 31.Barreto-Bergter E., Pinto M.R., Rodrigues M.L. Structure and biological functions of fungal cerebrosides. Acad. Bras Cienc. 2004;76:67–84. doi: 10.1590/s0001-37652004000100007. [DOI] [PubMed] [Google Scholar]
- 32.Ternes P., Sperling P., Albrecht S., Franke S., Cregg J.M., Warnecke D., Heinz E. Identification of fungal sphingolipid C9-methyltransferases by phylogenetic profiling. J. Biol. Chem. 2006;281:5582–5592. doi: 10.1074/jbc.M512864200. [DOI] [PubMed] [Google Scholar]
- 33.Fernandes C.M., de Castro P.A., Singh A., Fonseca F.L., Pereira M.D., Vila T.V.M., et al. Functional characterization of the Aspergillus nidulans glucosylceramide pathway reveals that LCB Δ8-desaturation and C9-methylation are relevant to filamentous growth, lipid raft localization and Psd1 defensin activity. Mol. Microbiol. 2016;102:488–505. doi: 10.1111/mmi.13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh A., Wang H., Silva L.C., Na C., Prieto M., Futerman A.H., et al. Methylation of glycosylated sphingolipid modulates membrane lipid topography and pathogenicity of Cryptococcus neoformans. Cell Microbiol. 2012;14:500–516. doi: 10.1111/j.1462-5822.2011.01735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huber A., Oemer G., Malanovic N., Lohner K., Kovács L., Salvenmoser W., et al. Membrane sphingolipids regulate the fitness and antifungal protein susceptibility of Neurospora crassa. Front. Microbiol. 2019;10:605. doi: 10.3389/fmicb.2019.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramamoorthy V., Cahoon E.B., Thokala M., Kaur J., Li J., Shah D.M. Sphingolipid C-9 methyltransferases are important for growth and virulence but not for sensitivity to antifungal plant defensins in Fusarium graminearum. Eukaryot. Cell. 2009;8:217–229. doi: 10.1128/EC.00255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oguro Y., Yamazaki H., Takagi M., Takaku H. Antifungal activity of plant defensin AFP1 in Brassica juncea involves the recognition of the methyl residue in glucosylceramide of target pathogen Candida albicans. Curr. Genet. 2014;60:89–97. doi: 10.1007/s00294-013-0416-8. [DOI] [PubMed] [Google Scholar]
- 38.Warschawski D.E., Arnold A.A., Beaugrand M., Gravel A., Chartrand É., Marcotte I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta. 2011;1808:1957–1974. doi: 10.1016/j.bbamem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 39.Deleu M., Crowet J.-M., Nasir M.N., Lins L. Complementary biophysical tools to investigate lipid specificity in the interaction between bioactive molecules and the plasma membrane: a review. Biochim. Biophys. Acta. 2014;1838:3171–3190. doi: 10.1016/j.bbamem.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 40.Rautenbach M., Troskie A.M., Vosloo J.A. Antifungal peptides: to be or not to be membrane active. Biochimie. 2016;130:132–145. doi: 10.1016/j.biochi.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 41.Zajchowski L.D., Robbins S.M. Lipid rafts and little caves. Eur. J. Biochem. 2002;269:737–752. doi: 10.1046/j.0014-2956.2001.02715.x. [DOI] [PubMed] [Google Scholar]
- 42.Bodner C.R., Dobson C.M., Bax A. Multiple tight phospholipid-binding modes of alpha-synuclein revealed by solution NMR spectroscopy. J. Mol. Biol. 2009;390:775–790. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang A.H., Edwards I.A., Mishra B.P., Sharma G., Healy M.D., Elliott A.G., et al. Elucidating the lipid binding properties of membrane-active peptides using cyclised nanodiscs. Front. Chem. 2019;7:238. doi: 10.3389/fchem.2019.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcotte I., Ouellet M., Auger M. Insights on the interaction of met-enkephalin with negatively charged membranes--an infrared and solid-state NMR spectroscopic study. Chem. Phys. Lipids. 2004;127:175–187. doi: 10.1016/j.chemphyslip.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Picard F., Paquet M.-J., Levesque J., Bélanger A., Auger M. 31P NMR first spectral moment Study of the partial magnetic orientation of phospholipid membranes. Biophys. J. 1999;77:888–902. doi: 10.1016/S0006-3495(99)76940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakraborty S., Doktorova M., Molugu T.R., Heberle F.A., Scott H.L., Dzikovski B., et al. How cholesterol stiffens unsaturated lipid membranes. Proc. Natl. Acad. Sci. U. S. A. 2020;117:21896–21905. doi: 10.1073/pnas.2004807117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wold Health Organization . 2022. WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action. [Google Scholar]
- 48.Jiang C., Ge J., He B., Zeng B. Glycosphingolipids in filamentous fungi: biological roles and potential applications in cosmetics and health foods. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.690211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva de Paula V., Razzera G., Barreto-Bergter E., Almeida F.C.L., Valente A.P. Portrayal of complex dynamic properties of sugarcane defensin 5 by NMR: multiple motions associated with membrane interaction. Structure. 2011;19:26–36. doi: 10.1016/j.str.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 50.Gonçalves S., Teixeira A., Abade J., de Medeiros L.N., Kurtenbach E., Santos N.C. Evaluation of the membrane lipid selectivity of the pea defensin Psd1. Biochim. Biophys. Acta. 2012;1818:1420–1426. doi: 10.1016/j.bbamem.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 51.Luo Y., Chen Y. Comparative analysis of the techniques for the determination of binding affinity between a small molecule inhibitor and a protein target. bioRxiv. 2024 doi: 10.1101/2024.05.16.594462. [preprint] [DOI] [Google Scholar]
- 52.Winiewska-Szajewska M., Poznański J. Differential scanning fluorimetry followed by microscale thermophoresis and/or isothermal titration calorimetry as an efficient tool for ligand screening. Biophys. Rev. 2025;17:199–223. doi: 10.1007/s12551-025-01280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winiewska M., Bugajska E., Poznański J. ITC-derived binding affinity may be biased due to titrant (nano)-aggregation. Binding of halogenated benzotriazoles to the catalytic domain of human protein kinase CK2. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Wang G., Moitessier N., Mittermaier A.K. Enzyme kinetics by isothermal titration calorimetry: allostery, inhibition, and dynamics. Front. Mol. Biosci. 2020 doi: 10.3389/fmolb.2020.583826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alvares D.S., Viegas T.G., Ruggiero Neto J. Lipid-packing perturbation of model membranes by pH-responsive antimicrobial peptides. Biophys. Rev. 2017;9:669–682. doi: 10.1007/s12551-017-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Samblanx G.W., Goderis I.J., Thevissen K., Raemaekers R., Fant F., Borremans F., et al. Mutational analysis of a plant defensin from radish (Raphanus sativus L.) reveals two adjacent sites important for antifungal activity. J. Biol. Chem. 1997;272:1171–1179. doi: 10.1074/jbc.272.2.1171. [DOI] [PubMed] [Google Scholar]
- 57.Song X., Zhang M., Zhou Z., Gong W. Ultra-high resolution crystal structure of a dimeric defensin SPE10. FEBS Lett. 2011;585:300–306. doi: 10.1016/j.febslet.2010.12.039. [DOI] [PubMed] [Google Scholar]
- 58.Neves de Medeiros L., Angeli R., Sarzedas C.G., Barreto-Bergter E., Valente A.P., Kurtenbach E., Almeida F.C.L. Backbone dynamics of the antifungal Psd1 pea defensin and its correlation with membrane interaction by NMR spectroscopy. Biochim. Biophys. Acta. 2010;1798:105–113. doi: 10.1016/j.bbamem.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 59.Pinheiro-Aguiar R., do Amaral V.S.G., Pereira I.B., Kurtenbach E., Almeida F.C.L. Nuclear magnetic resonance solution structure of Pisum sativum defensin 2 provides evidence for the presence of hydrophobic surface-clusters. Proteins. 2020;88:242–246. doi: 10.1002/prot.25783. [DOI] [PubMed] [Google Scholar]
- 60.Bechinger B. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta. 1999;1462:157–183. doi: 10.1016/s0005-2736(99)00205-9. [DOI] [PubMed] [Google Scholar]
- 61.Khandelia H., Ipsen J.H., Mouritsen O.G. The impact of peptides on lipid membranes. Biochim. Biophys. Acta. 2008;1778:1528–1536. doi: 10.1016/j.bbamem.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 62.Iversen A., Utterström J., Khare L.P., Aili D. Influence of lipid vesicle properties on the function of conjugation dependent membrane active peptides. J. Mater. Chem. B. 2024;12:10320–10331. doi: 10.1039/d4tb01107d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ludtke S.J., He K., Heller W.T., Harroun T.A., Yang L., Huang H.W. Membrane pores induced by Magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 64.Laadhari M., Arnold A.A., Gravel A.E., Separovic F., Marcotte I. Interaction of the antimicrobial peptides caerin 1.1 and aurein 1.2 with intact bacteria by 2H solid-state NMR. Biochim. Biophys. Acta. 2016;1858:2959–2964. doi: 10.1016/j.bbamem.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Kang X., Elson C., Penfield J., Kirui A., Chen A., Zhang L., Wang T. Integrated solid-state NMR and molecular dynamics modeling determines membrane insertion of human β-defensin analog. Commun. Biol. 2019;2:402. doi: 10.1038/s42003-019-0653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monnier N., Furlan A.L., Buchoux S., Deleu M., Dauchez M., Rippa S., Sarazin C. Exploring the dual interaction of natural rhamnolipids with plant and fungal biomimetic plasma membranes through biophysical studies. Int. J. Mol. Sci. 2019;20:1009. doi: 10.3390/ijms20051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santos F.C., Marquês J.T., Bento-Oliveira A., de Almeida R.F.M. Sphingolipid-enriched domains in fungi. FEBS Lett. 2020;594:3698–3718. doi: 10.1002/1873-3468.13986. [DOI] [PubMed] [Google Scholar]
- 68.Raj S., Nazemidashtarjandi S., Kim J., Joffe L., Zhang X., Singh A., et al. Changes in glucosylceramide structure affect virulence and membrane biophysical properties of Cryptococcus neoformans. Biochim. Biophys. Acta. 2017;1859:2224–2233. doi: 10.1016/j.bbamem.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vranken W., Boucher W., Stevens T., Fogh R.H., Pajon A., Llinas M., et al. The CCPN Data Model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 70.Schanda P., Kupce E., Brutscher B. SOFAST-HMQC experiments for recording two-dimensional deteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR. 2006;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 71.Delano W. Delano Scientific; San Carlos: 2002. The Pymol Molecular Graphics System. [Google Scholar]
- 72.Mazur A., Hammesfahr B., Griesinger C., Lee D., Kollmar M. ShereKhan - calculating exchange parameters in relaxation dispersion data from CPMG experiments. Bioinformatics. 2013;29:1819–1820. doi: 10.1093/bioinformatics/btt286. [DOI] [PubMed] [Google Scholar]
- 73.Carver J.P., Richards R.E. A general two-site solution for the chemical exchange produced dependence of T2 upon the carr-Purcell pulse separation. J. Magn. Reson. 1972;6:89–105. [Google Scholar]
- 74.Warschawski D.E., Arnold A., Marcotte I. A new method of assessing lipid mixtures by 31P magic-angle spinning NMR. Biophys. J. 2018;114:1368–1376. doi: 10.1016/j.bpj.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rance M., Byrd R.A. Obtaining high-fidelity powder spectra in anisotropic media: Phase-cycled Hahn echo spectroscopy. J. Magn. Reson. 1983;52:221–240. [Google Scholar]
- 76.Davis J.H., Jeffrey K.R., Bloom M., Valic M., Higgs T. Quadrupolar echo deuteron magnetic resonance spectroscopy in ordered hydrocarbon chains. Chem. Phys. Lett. 1976;42:390–394. [Google Scholar]
- 77.Warnet X., Laadhari M., Arnold A., Marcotte I., Warschawski D.E. A (2)H magic-angle spinning solid-state NMR characterisation of lipid membranes in intact bacteria. Biochim. Biophys. acta. 2015;1858:146–152. doi: 10.1016/j.bbamem.2015.10.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the relevant data are contained within this article and supporting information. Source data and additional data can be obtained from the corresponding author upon reasonable request.