The template for paramyxovirus RNA synthesis is not naked RNA but the helical nucleocapsid core of the virus, in which each nucleocapsid protein (N protein) is predicted to be associated with precisely 6 nucleotides (nt) (11). Presumably as a consequence of this association, paramyxovirus genomes are replicated efficiently only when they are a multiple of 6 nt in length, and this has been dubbed the “rule of six” (4). The structure of paramyxovirus nucleocapsids is thus central to understanding how this rule might operate.
ONE-DIMENSIONAL PROTEIN ASSEMBLIES
The information for forming many of the larger assemblies of macromolecules in cells is contained in the subunits themselves, as under appropriate conditions the isolated subunits can spontaneously assemble into the final structure. The first large macromolecular aggregate found to self-assemble from its component parts was tobacco mosaic virus (TMV) (15). TMV consists of a cylinder of ca. 2,200 coat protein subunits arranged around a helical RNA core of ca. 6,600 nt. The helical path of TMV RNA is imposed by the arrangement of the subunits themselves, which can be viewed as a one-dimensional assembly of a single subunit. One-dimensional assemblies form when the subunits contain a binding site which is complementary to a region of its own surface which does not include the binding site itself (Fig. 1) (1). Under certain special orientations of the two binding sites, the chain will run into itself and form a closed ring of subunits, as is found for purified vesicular stomatitis virus (VSV) N protein (2). More commonly an extended polymer of subunits will result, and provided that each of the subunits is bound to its neighbor in an identical way, the subunits in the polymer will be arranged in a helix that can be extended indefinitely. These structures are also found independently of RNA, e.g., the helical actin filament is composed only of actin. However, when they are associated with RNAs, the length of the assembly is determined by that of the RNA. For TMV, e.g., each coat protein subunit is associated with precisely 3 nt. For many paramyxovirus nucleocapsids, each N protein subunit is predicted to be associated with precisely 6 nt.
FIG. 1.
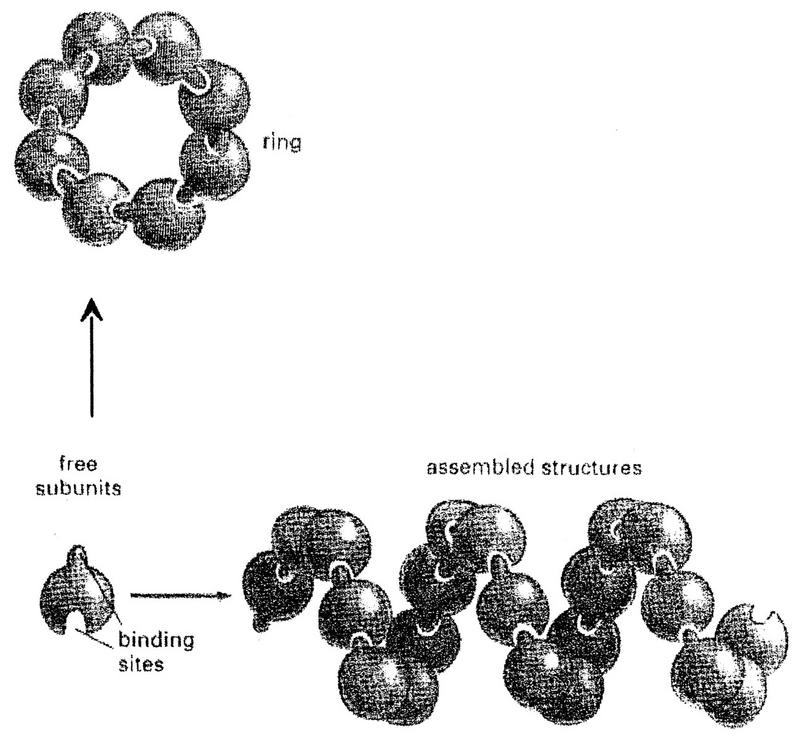
Single subunit assemblies. Large macromolecular assemblies can be formed from a single protein subunit, if the subunit interacts with itself repeatedly. This is possible if the binding site is complementary to a region of its own surface that does not include the binding site itself. Adapted from Alberts et al. (1).
PARAMYXOVIRUS NUCLEOCAPSIDS
Paramyxoviruses are enveloped animal viruses with nonsegmented negative-strand RNA genomes which are found almost exclusively in nucleocapsid structures. In contrast to the plus-strand TMV RNA genome which disassembles to function as mRNA and as a template for genome amplification, mononegavirus nucleocapsids never disassemble as far as we know; all RNA synthesis occurs here without reversing the structure of the N-RNA template. Transcriptionally active Sendai virus (SeV) nucleocapsids also contain ca. 50 L and 300 P proteins, which together form the viral polymerase. The P protein is found as a homotrimer (9), both together with the catalytic L subunit and independently of L. Both P complexes (P3-L and P3) together with the N-RNA template are required for reconstituting mRNA synthesis in cell-free extracts (9, 10, 16, 20).
Paramyxovirus nucleocapsids are seen in remarkable detail in negatively stained electron micrographs, in which they appear as regular left-handed helical coils of ca. 200 Å in diameter with a central hole of ca. 50 Å, ca. 1 μm in length, and with a pitch of ca. 60 Å. For SeV and VSV, nucleocapsids composed only of their N-RNA have also been examined and they are indistinguishable from holonucleocapsids containing P and L. That these structures were one-dimensional assemblies was evident from their characteristic herringbone or chevron-shaped appearance (when viewed perpendicularly to the helix axis) in the earliest negatively stained electron micrographs (14). In 1989, by analyzing electronically straightened images of negatively stained SeV nucleocapsids, Egelman et al. (11) found that they existed in discrete pitch states. More importantly, by reconstructing images of these nucleocapsids, they also concluded that each N subunit was associated with an integral number of nucleotides and predicted this to be 6 nt. Little attention was paid to this number then; in part because the VSV N protein was estimated to be associated with 9 nt (35).
Whereas nonenveloped RNA viruses such as TMV are tightly packed rigid rods designed for maximum protection of the RNA, mononegavirus nucleocapsids are generally more flexible and open structures, as befits their function in RNA synthesis. The RNA within the nucleocapsid is, however, insensitive to RNase attack at any salt concentration (19). Although the nucleocapsids can be very flexible when viewed in the electron microscope, they must be very stable as the assembly survives the high salt conditions of CsCl density gradient centrifugation, at which they band at ca. 1.3 g/ml (or 2.5 M CsCl). This stability suggests that hydrophobic forces are important in maintaining the N-N and N-RNA interactions, and consistent with this, the N protein can be separated from the RNA only with the aid of ionic detergents like sodium dodecyl sulfate or 3 M guanidine-HCl. If hydrophobic forces are important in maintaining the interactions between N and the highly charged RNA, the RNA would seem to be intimately associated with the N subunits. It is then unclear how this RNA within the assembly can act as a template for RNA synthesis, and two suggestions have been made in this regard. (i) Structural transitions in the assembly bring the RNA to the surface for the polymerase (11), and (ii) the N subunits might be displaced locally from the RNA by the action of the viral polymerase, much like the separation of the two strands of DNA which occurs in the template during DNA-directed RNA synthesis (23).
THE UNEXPECTED REQUIREMENT OF HEXAMER GENOME LENGTH FOR PARAMYXOVIRUSES: THE RULE OF SIX
Paramyxovirus genomes were first expressed from DNA as artificial defective interfering (DI) genomes in which the entire protein coding region of the nondefective genome was replaced with a chloramphenicol acetyltransferase (CAT) reporter gene (pSV-CAT) (25). Minus-strand T7 RNA polymerase transcripts of these DNAs were transfected into cells infected with helper virus, and CAT activity could be found both in these cells and in those infected with the progeny viruses. Although not all the constructs prepared by Park et al. were active (22a), there appeared to be no special requirements for the construction of active replicons, beyond that they contain the ends of the genome needed for replication and mRNA synthesis. The replication of pSV-CAT was, however, very limited; only the CAT activity could be detected, whereas genomes or mRNAs were below the level of detection. The next paramyxovirus genome expressed from DNA was that of an SeV DI RNA (H4) generated upon repeated passage in eggs, which was known to strongly interfere with the nondefective genome in cell culture infections and to accumulate to high levels (3). An exact copy of this RNA was then transcribed from cDNA in a transfection/infection assay, and it was indeed found to replicate to levels similar to those of natural infections (4). However, internal deletions of the H4 RNA, including those which removed only a few nucleotides, failed to replicate to a detectable level, for no apparent reason.
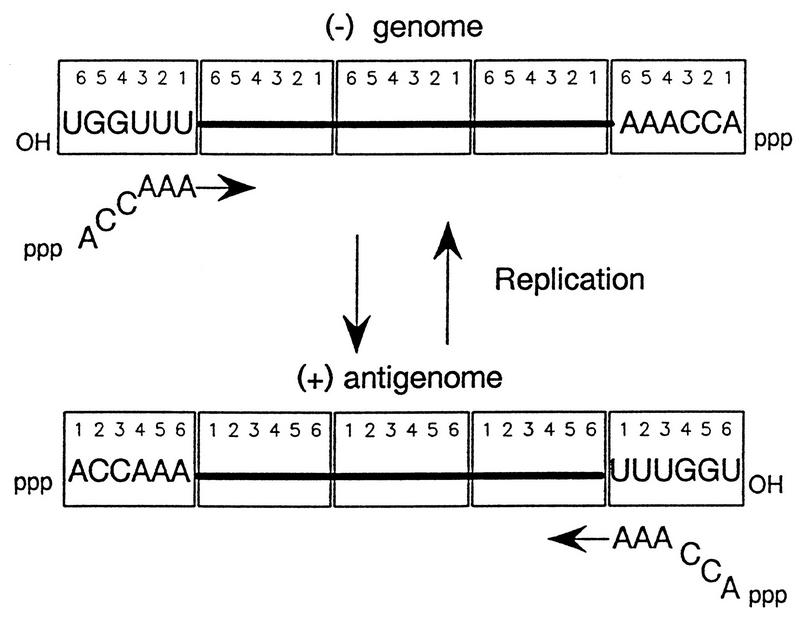
While puzzling over these unexpected results, we were visited by Tatsuo Shioda (University of Tokyo), who had become convinced that hexamer length was important for paramyxovirus RNA replication based on his extensive sequencing of the SeV and bovine parainfluenzavirus type 3 (bPIV3) genomes. This conviction was more intuitive than anything else, but as DI H4 was indeed of hexamer length (1,410 or 235 × 6 nt) and since internal deletions in the H4 clone routinely led to inactive constructs, P. Calain and L. Roux decided to examine Shioda’s intuition. Using five unique restriction sites in the H4 DNA, 17 derivatives were generated which produced RNAs of 6n+0, 6n+1, 6n+2, etc. Of the 17 derivatives, 5 replicated at high efficiency; their Northern blot signals were ca. 100-fold higher than those of the remaining 12. These five were 1,410 and 1,416 nt long, both hexamer lengths. The 12 nonreplicating RNAs, on the other hand, included every possibility but 6n+0. Since (i) each N subunit was predicted to be associated with precisely 6 nt, (ii) nucleocapsid assembly (at least for VSV) occurred from the very 5′ end of the nascent RNA chain (2), and (iii) assembly of the nonhexamer-length T7 transcripts in the transfected cells also appeared to be taking place, Calain and Roux (4) proposed that the 3′ ends of the nucleocapsid RNAs were efficient templates for replication only when they were precisely covered with N subunits. Having one more or one less nucleotide anywhere in the chain would result in the 3′ end of the template not being precisely covered by the subunits (Fig. 2).
FIG. 2.
Paramyxovirus genome replication and the requirement for hexamer genome length. Genome and antigenome nucleocapsids are shown as a linear array of N subunits (open rectangles), each with six sites for binding nucleotides (numbered 1 to 6, 5′ to 3′ on the antigenome). Only the sequences of the six 5′ (ppp) and 3′ (OH) conserved bases are shown. Nucleocapsid assembly occurs concomitantly with synthesis and is proposed to initiate flush with the 5′ pppACCAAA hexanucleotide. Chains which are not 6n+0 long alter the position of the 3′ OHUGGUUU promoter element relative to the terminal N subunit and are therefore inefficiently initiated.
In further support for this thesis, replication of the internal deletion DI RNAs generated in cell culture, and that of pSV-CAT itself, was found to depend on their being of hexamer length (13, 17). This rule of six appears to apply to morbilliviruses as well, because measles virus minireplicons expressing CAT genes do not replicate well unless they are of hexamer length (33) and infectious measles virus was not recovered from DNA until its genome was also of hexamer length (29). Furthermore, three measles virus DI RNAs were characterized in cell culture-grown virus and were found to be of hexamer length (12), as well as most copyback DI RNAs cloned from human (subacute sclerosing panencephalitis) brains (32). Direct biochemical evidence that N protein binds 6 nt, however, is lacking.
THE DOMINANT ROLE OF THE RULE OF SIX FOR SEV GENOME REPLICATION
Paramyxovirus genomes contain inverted terminal repeats of 12 nt specific to each genera, hence the 5′- and 3′-terminal 12 nt of the genome and antigenome of each virus are identical. These duodecamers are undoubtedly important cis-acting determinants for genome replication, and they would be precisely covered by the terminal two N subunits in hexamer-length genomes (Fig. 2). The rule of six implies that the viral polymerase is interacting with the template bases for RNA synthesis in the context of the N subunits, since it is difficult to imagine how such a rule could operate if the N subunit were completely displaced from the template RNA for the polymerase to interact with the bases. Genome replication would then begin opposite the first base of the first subunit, at genome map coordinate 1, or nt 1 (Fig. 2). The 15,384-nt-long SeV genome sequence can be represented as a linear array of 2,564 hexamers, and we were interested in the relationship of the internal cis-acting sequences important for mRNA synthesis. Was their position relative to the phase created by the N subunits conserved, and was this important?
The start site of the first mRNA is strictly conserved at nt 56 in the Paramyxovirinae (Table 1), so the N mRNAs would initiate opposite the 2nd base of the 10th N protein for all these viruses. To examine the effect changing the position of the N mRNA start site would have on the efficiency of its synthesis, a natural internal deletion SeV DI (E307) which expresses a single N/L fusion mRNA (13) was modified to contain unique BglII (at nt 47) and NsiI sites (at nt 67) at either side of the leader/N junction. E307A, which contained these four-base substitutions replicated normally in transfected cells (in the absence of C protein coexpression, see below), i.e., it was amplified to the same levels as E307, the wild-type (wt) control. Six nucleotides were then inserted at either nt 47 or nt 67, and these inserted E307 genomes also accumulated to wt levels in transfected cells. mRNA synthesis from the inserted templates was examined in cell extracts, and mRNA was found to be made as efficiently as from wt DI genomes. For the +6@nt47 construct, this mRNA synthesis could also be shown to have started opposite the 2nd base of the 11th subunit (at nt 62), so the start site can be moved to the same subunit position of the next subunit without much effect. Two nucleotides were then added at nt 47, and two were removed at nt 67. E307A+2/−2 also replicated normally and made normal amounts of mRNA by initiating opposite the 4th base of the 10th subunit (at nt 58). The conserved N mRNA start site can then also be moved within the subunit without much effect, at least as measured in this cell-free system (27).
TABLE 1.
Subunit hexamer phasing at gene junctions
| Virus (size in nt) | Protein | Sequencea | mRNA start position (nt) | 6n + x (phase)b |
|---|---|---|---|---|
| Paramyxoviruses | ||||
| bPIV3 (15,480) | ||||
| N | CTT AGGATTAAAG | 56 | 2 | |
| N/P | AGTAAGAAAAA CTT AGGATTAACG | 1,705 | 1 | |
| P/M | ATTAAGAAAAA CTT AGGATAAAAG | 3,703 | 1 | |
| M/F | AAAATCAAAAA CTT AGGATCAAAG | 4,855 | 1 | |
| F/H | AGTACAAAAAA CTT AGGAACAAAG | 6,751 | 1 | |
| H/L | ATTACAAAAAA CTT AGGAGGAAAG | 8,642 | 2 | |
| L | AGTAAGAAAAA CAT | 15,440 | ||
| Human PIV3 (15,462) | ||||
| N | CTT AGGATTAAAG | 56 | 2 | |
| N/P | AATAAGAAAAA CTT AGGATTAAAG | 1,705 | 1 | |
| P/M | AATAAGAAAAA CTT AGGATTAAAG | 3,721 | 1 | |
| M/F | ATAATCAAAAA CTT AGGACAAAAG | 4,879 | 1 | |
| F/H | ATTATAAAAAA CTT AGGAGTAAAG | 6,733 | 1 | |
| H/L | AATATAAAAAA CTT AGGAGCAAAG | 8,624 | 2 | |
| L | AGTAAGAAAAA CAT | 15,422 | ||
| SeV (15,384) | ||||
| N | TTT AGGGTCAAAG | 56 | 2 | |
| N/P | AGTAAGAAAAA CTT AGGGTGAAAG | 1,741 | 1 | |
| P/M | ATTAAGAAAAA CTT AGGGTGAAAG | 3,637 | 1 | |
| M/F | AATAAGAAAAA CTT AGGGATAAAG | 4,813 | 1 | |
| F/H | AATAAGAAAAA CTT AGGGTGAAAG | 6,637 | 1 | |
| H/L | ATTAAGAAAAA CCC AGGGTGAATG | 8,528 | 2 | |
| L | AGTAAGAAAAA CTT | 15,331 | ||
| Consensus | ADWAHVAAAAA CYY AGGRNNAAHG | |||
| Morbilliviruses | ||||
| Measles virus (15,894) | ||||
| N | CTT AGGATTCAAG | 56 | 2 | |
| N/P | GTTATAAAAAA CTT AGGAACCAGG | 1,748 | 2 | |
| P/M | CATTATAAAAA CTT AGGAGCAAAG | 3,406 | 4 | |
| M/F | ACTAAACAAAA CTT AGGGCCAAGG | 4,875 | 3 | |
| F/H | GTTAATTAAAA CTT AGGGTGCAAG | 7,251 | 3 | |
| H/L | ATTAAGAAAAA CGT AGGGTCCAAG | 9,212 | 2 | |
| L | ATTAAAGAAAA CTT | 15,858 | ||
| Rinderpest virus (15,882) | ||||
| N | CTT AGGATTCAAG | 56 | 2 | |
| N/P | ATTATAAAAAA CTT AGGACCCAGG | 1,748 | 2 | |
| P/M | ATTATAAAAAA CTT AGGAGCAAAG | 3,406 | 4 | |
| M/F | ACCAAACAAAA CTT AGGGTCAAAG | 4,869 | 3 | |
| F/HN | ATAAAGAAAAA CTT AGGATGCAAG | 7,239 | 3 | |
| HN/L | ATTATAAAAAA CGT AGGGTCCAAG | 9,200 | 2 | |
| L | ACTAAAGAAAA CTT | 15,846 | ||
| Dolphin morbillivirus (15,702) | ||||
| N | CTT AGGATTAATG | 56 | 2 | |
| N/P | ATTACAAAAAA CTT AGGACCAAAG | 1,742 | 2 | |
| P/M | ATTATAAAAAA CTT AGGATTCAAG | 3,400 | 4 | |
| M/F | ATTAAATAAAA CTT AGGAGTAAAG | 4,856 | 2 | |
| F/HN | ATTAAAGAAAA CTT AGGGTGCAAG | 7,071 | 3 | |
| HN/L | ATTAAGAAAAA CTT AGGGACCAGG | 9,020 | 2 | |
| L | ATTAAGAAAAA CAA | 15,666 | ||
| Consensus | VHHWHDnAAAA CKT AGGRnBMARG | |||
| Rubulavirusesc | ||||
| SV5 (15,246) | ||||
| N | AGGTCCGGAA | 56 | 2 | |
| N/P | AAAGAAAAAAA T AGGCCCGGAC | 1,789 | 1 | |
| P/M | TTTAGAAAAAA C (13) T AAGCCCGAAC | 3,108 | 6 | |
| M/F | TTCAAAGAAAA C (20) T AAGCACGAAC | 4,501 | 1 | |
| F/SH | TAAGAAAAAAA C (2) T AGGACCGAAC | 6,224 | 2 | |
| SH/HN | TAAAGAAAAAA T AGGCCCGAAC | 6,517 | 1 | |
| HN/L | TTTAAGAAAAA C (11) T AGGCCAGAAT | 8,406 | 6 | |
| L | TTAAGAAAAAA | |||
| Mumps virus (15,384) | ||||
| N | AAGCCAGGAA | 56 | 2 | |
| N/P | TTAAGAAAAAA T T AGGCCCGGAA | 1,909 | 1 | |
| P/M | TAAATAAAAAA T AAGCACGAAC | 3,228 | 6 | |
| M/F | TATAGAAAAAA T AAGCCTAGAA | 4,483 | 1 | |
| F/SH | TTAGAAAAAAA C (5) T AAGAATGAAT | 6,217 | 1 | |
| SH/HN | TAAAGAAAAAA G C AAGCCAGAAC | 6,535 | 1 | |
| HN/L | TTAAGAAAAAA C AGGCCAGAAT | 8,430 | 6 | |
| L | TTAAGAAAAAA | |||
| SV41 (15,450) | ||||
| N | AGGCCCGGAA | 56 | 2 | |
| N/P | TTTAAAGAAAAA C T T AGGCCCGGAC | 1,903 | 1 | |
| P/M | TTTAAGAAAAAA C (20) T AGGCACGAAC | 3,331 | 1 | |
| M/F | TTTAT AAA T (13) T AGGTCCGAAC | 4,663 | 1 | |
| F/HN | TTTAAGAAAAAA T (2) T AGGCACGAAC | 6,541 | 1 | |
| HN/L | TTTAAAGAAAAA T (5) T AGGCCAGAAT | 8,568 | 6 | |
| L | TTTAAGAAAAAA | |||
| Consensus | TWHRDRRAAAA B (N) Y ARGHMHRRAH |
The nucleotide sequences of the entire paramyxovirus genomes were obtained as follows: SeV, accession no. M30202; bPIV3, accession no. D84095; human PIV3, accession no. Z11575; measles virus, accession no. K01711; rinderpest virus, Tom Barrett, Pirbright, United Kingdom; dolphin morbillivirus and mumps virus, Bert Rima, Belfast, Northern Ireland; SV5, Reay Paterson and Bob Lamb, Evanston, Ill.; SV41, accession no. X64275; PIV2, Yasuhito Ito, Mie, Japan. The sequences provided by the various laboratories are considered to be most accurate to date and sometimes supersede previous information. The single-letter nucleotide codes used in the consensus sequences are as follows: R, purine; Y, pyrimidine; M (mino), A and C; K (keto), G and T; B, D, H, and V, anything but A, C, G, and U, respectively; W (weak), A and T.
Numbers in bold indicate conservation within each genus.
Numbers in parentheses refer to additional nucleotides in the intergenic region.
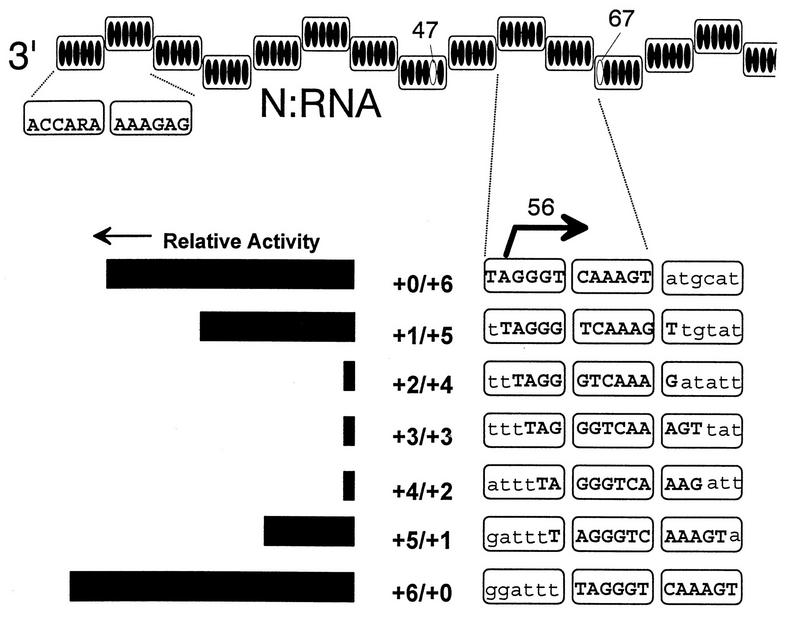
The above experiments were carried out to determine whether the subunit phase of the TAGGGT60 mRNA start site (initiating A residue is bold) was important for mRNA synthesis. No evidence was provided for this, but evidence was found for a sequence between nt 47 and 67 whose N subunit phase was clearly important for genome replication, albeit in a conditional fashion, as follows. Several of the constructs variously modified at nt 47 and 67 simply did not replicate, even though they could be shown to be of hexamer length. For example, whereas E307A+2/−2 replicated normally, we were unable to prepare active E307A+2/+4. This did not make sense, as E307A0/+6 replicated normally and so insertions at nt 67 were tolerated. When the entire insertion series (+0/+6, +1/+5, … +5/+1, +6/+0) was examined, the reasons for this became clearer (Fig. 3). Mutations which alter the subunit phase between nt 47 and 67 alone are well tolerated, and those which introduce 6 nt at either site without altering the subunit phase in between are well tolerated, but the simultaneous introduction of 6 nt and the altering of the subunit phase are not. A possible explanation for this “synthetic lethality” is that the genomic replication promoter has been found by mutational analysis to be composed of at least two elements; the first half of the leader sequence (nt 1 to 30) and part of the 5′ untranslated region of the N gene previously identified as the BB box (nt 79 to 96 for SeV) (34a). The existence of this downstream promoter element presumably accounts for why the insertion of 12 or 18 nt is not tolerated at sites where the insertion of 6 nt is well tolerated, i.e., the spacing of these two elements is important (27). The above results suggest that the phase of the nt 47 to 67 sequence is a third element of the genomic replication promoter (Fig. 3).
FIG. 3.
cis-acting promoter sequences and the rule of six. The N-RNA nucleocapsid template is shown above as a helical assembly of N subunits, each containing 6-nt binding sites (dark ovals). The SeV sequence within the 9th, 10th, and 11th subunits, containing the N mRNA start site at nt 56, is shown below; the YARRGT repeats at nt 55 to 66 are highlighted in bold capitals. The effect of adding a total of 6 nt at nt 47 and 67 on the hexamer phase of these repeats is shown on the right, and their effect on the amplification activity of a DI genome in the absence of C protein expression (27) is shown on the left. The polymerase is postulated to recognize these ARRG repeats equally well when they are contained entirely within the 9th and 10th or 10th and 11th subunits, but these repeats are less well recognized when their phase relative to the N subunits is altered. Adapted from reference 27.
The manner in which this phase might operate is suggested by inspection of the sequence, which includes the leader/N junction. N mRNAs, for example, start opposite the 2nd base of the 10th subunit, within TAGGGT60 (initiating A residue in bold) which is partially repeated in the 11th subunit as YARRGT66 (Fig. 3). When 6 nt are inserted, replication remains wt as long as the tetrapurine run remains in the center of the subunit. Moving the ARRG one position in either direction results in a ca. 2-fold reduction in replication efficiency, but moving it by two, three, or four positions results in a >10-fold reduction, when the ARRGs would no longer be contained within single N subunits (Fig. 3).
HEXAMER RULES FOR THE INITIATION OF MRNA SYNTHESIS?
Although we were unable to show that the phase of the N mRNA start site is important for mRNA synthesis in vitro, there is nevertheless remarkable circumstantial evidence that this is so. We have used the duodecamer TAGGGT60YAGGGT66 (phase indicated by spacing) as an example of how subunit phase could operate as an element of the replication promoter, because this duodecamer is so strongly conserved at the paramyxovirus mRNA start sites (Table 1). The subunit phase of this sequence must also be important, because this too has been conserved, but more variously, as detailed below.
The Paramyxovirinae are currently organized into three genera, the paramyxovirus genus (including SeV and parainfluenza virus types 1 and 3 [PIV1 and PIV3]), morbilliviruses (e.g., measles virus and the distemper viruses), and rubulaviruses (e.g., mumps virus and simian virus 5 [SV5]). The entire genome sequences of 11 of the Paramyxovirinae are now available, and 9 of these are in fact multiples of 6 nt long and have been analyzed for evidence of a hexamer phase. There is considerable conservation of the gene start and gene end sequences, and these are aligned in Table 1 around the intergenic regions, i.e., those nucleotides not present in mRNA. When the hexamer positions (or phases) of the initiating A residues of the various mRNAs are determined for each genus of the subfamily, these phases are clearly conserved. The strongest conservation is found among the paramyxoviruses SeV (2,564 × 6 nt), bPIV3 (2,580 × 6 nt), and human PIV3 (2,577 × 6 nt), in which the initiating A residue of the successive N, P, M, F, HN, and L mRNAs of each virus is found at hexamer positions 2, 1, 1, 1, 1, and 2, respectively. Here, for example, the ARRG of each of these 18 mRNA start sites is found within the same N subunit. Whether the initiating A is at hexamer position 1 or 2 also appears to matter, as the same position is strictly conserved for each particular cistron. Of the four morbilliviruses sequences available, measles virus (2,649 × 6 nt), dolphin morbillivirus (2,617 × 6 nt), and rinderpest virus (2,647 × 6 nt) were multiples of 6 nt long (only canine distemper virus contained 15,685 nt, or 6n+1 nt). For these morbilliviruses, the subunit phase of the initiating ARRG was somewhat less well conserved, over three hexamer positions (2–4) rather than two for the paramyxoviruses: the second tetrapurine run AARG is often present as MARG, and the phases of each cistron are strictly conserved except for that of the F mRNAs (Table 1).
A remarkable degree of conservation is also found for the remaining rubulavirus genus, for which the start sites are also clustered over three subunit positions—6, 1, 2—the two tetrapurine runs are reduced to ARGH and HRRA, and the start sites of the individual cistrons are again clearly conserved. Two of the rubulaviruses (SV5 and mumps virus) contain a seventh (SH) gene between F and HN, yet in spite of this variability, the hexamer phases of the N, P, F, HN, and L start sites are strictly conserved. At the time of this sequence gazing, a reverse genetic system for rubulaviruses, in which to test the rule of six, was not available. However, the fact that three of the four rubulavirus sequences reported are of hexamer length (only PIV2 is 15,653 nt, or 6n+5 nt) and the hexamer positions of its individual mRNA start sites are conserved is evidence that this rule applies as well to the rubulaviruses. This latter conservation is of particular significance, because rubulavirus intergenic regions are quite variable in length (1 to 20 nt) and composition, rather than the conserved CTT of the two other genera. Because paramyxoviruses and morbilliviruses contain this almost invariant CTT, the hexamer phases of both the gene start and end sequences are conserved and this phase may well be important for both mRNA initiation and polyadenylation/termination for these viruses. For the rubulaviruses in which the gene start and gene end sequences are no longer separated by a fixed number of nucleotides, it is clear that the hexamer phase has been conserved primarily for mRNA initiation. During review of this article, Murphy and Parks (23a) reported a reverse genetic system for the rubulavirus SV5 in which genome lengths that are divisible by six are not essential but enhance replication of their DI analogs by 5- to 20-fold. Their results are similar to those of SeV systems in which C protein expression is suppressed (reference 34 and below). A histogram of the phases of the 56 mRNA start sites of the nine paramyxoviruses listed in Table 1 is shown in Fig. 4. There is an overall preference for starting mRNAs at hexamer positions 1 and 2, but almost any position (except position 5) is used. In summary, different genera prefer different hexamer phases for the initiation of each particular cistron and these phases are clustered for each genus.
FIG. 4.
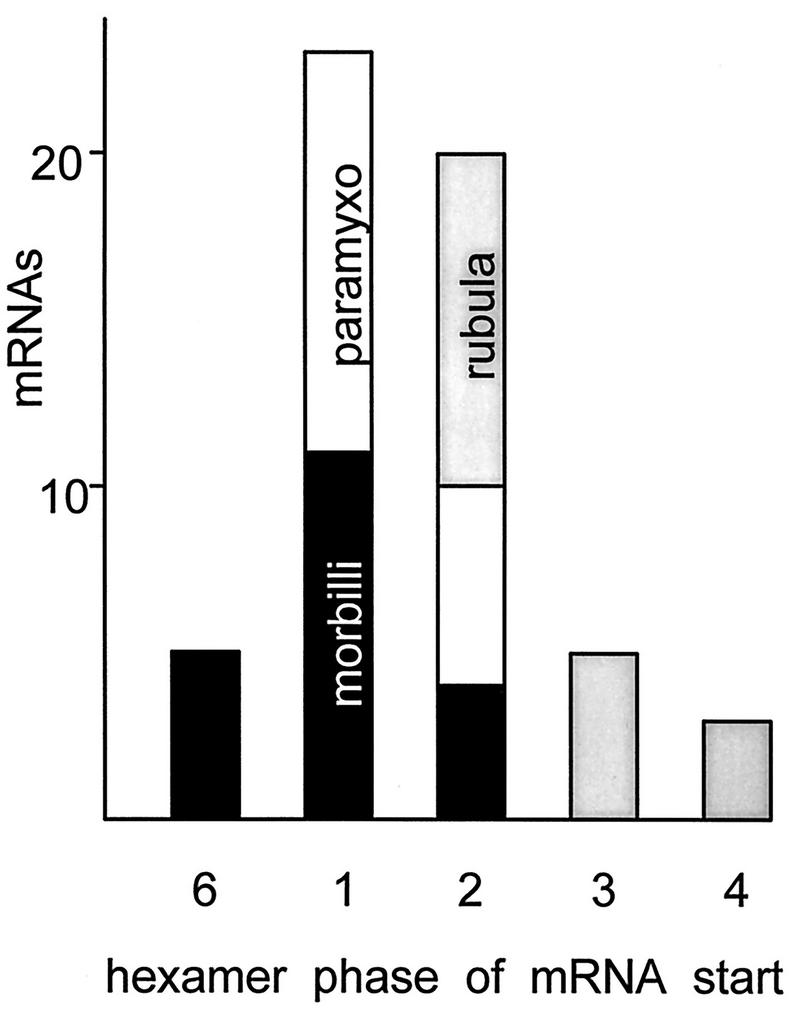
Distribution of mRNA start site subunit phases. The initiating adenosines of the 56 mRNA start sites listed in Table 1 are plotted in this histogram according to their N subunit positions (hexamer phase). The clustering of these phases for each genus of the subfamily is indicated by shading.
PARAMYXOVIRUS P GENE MRNA EDITING
Most of the P genes of the Paramyxovirinae contain an AnGn purine run at the start of the internal, overlapping V open reading frame (ORF). mRNAs with expanded G runs are transcribed from these genes in addition to those which are faithful copies of their templates (5, 36), and the number of G insertions which occur for each virus group mirrors their requirements to switch between the in-frame and out-of-frame ORFs (reviewed in reference 22). For the morbilliviruses and SeV, which require a +1 frameshift to access the V ORF from the genome-encoded P ORF, a single G is added as the predominant insertional event. For the rubulaviruses which require a +2 frameshift to access the remainder of the P ORF from the genome-encoded V ORF, two Gs are added at high frequency when insertions occur. For bPIV3, in which both V and another ORF (called D) overlap the middle of the genome-encoded P ORF, one to six Gs are added at roughly equal frequencies, so that mRNAs encoding all three overlapping ORFs are expressed.
When the hexamer phase of the start of the short G run expanded in mRNA editing (underlined in Table 2) is determined, it is clear that this position as well has been conserved according to virus group. The strongest conservation is found among the rubulaviruses, for which the start of the G run is found at hexamer position 3 in the four viruses for which we have this information, even though each site is at a different genome map coordinate. Similarly, the start of the G run is found at hexamer position 6 in the four morbilliviruses for which we have this information. Only in the paramyxovirus genus is the hexamer position of the start of the G run not strictly conserved, consistent with the fact that SeV and the two PIV3 viruses edit their P gene mRNAs quite differently. The hexamer phase of the AnGn run could also play a role in controlling the pattern of G insertions distinct to each virus group.
TABLE 2.
Subunit hexamer phasing at the P gene mRNA editing site
| Virus | nt | 6n+x |
|---|---|---|
| Paramyxovirusa | ||
| SeV CTCAAC AAAAAA GGGCAT AGGAGA | 2791 | 1 |
| bPIV3 GGGAAT TAAAAA AGGGGT TGGAAA | 2504 | 2 |
| hPIV3 AAGAAT TAAAAA AGGGGG AAAAGG | 2504 | 2 |
| Morbillivirusb | ||
| MeV CCCATT AAAAAG GGCACA GACGCG | 2496 | 6 |
| CDV TCCATT AAAAAG GGCACA GAAGAG | 2490 | 6 |
| RPV CCCATT AAAAAG GGCACA GACGTG | 2496 | 6 |
| DMV TCCATT AAAAAG GGCACA GGAGAG | 2490 | 6 |
| Rubulavirusc | ||
| MuV GACAGA ATTTAA GAGGGG GGCCGG | 2439 | 3 |
| SV5 CATCGA TTTTAA GAGGGG CAGGGA | 2337 | 3 |
| SV41 CCCCAA CTTTAA GAGGGG GGGAGA | 2457 | 3 |
| PIV2 CCCCAA CTTTAA GAGGGG GGGAGC | 2481 | 3 |
hPIV3, human PIV3.
MeV, measles virus; CDV, canine distemper virus; RPV, rinderpest virus; DMV, dolphin morbillivirus.
MuV, mumps virus.
TMV AS A MODEL FOR PARAMYXOVIRUS N PROTEIN-RNA INTERACTIONS
TMV and SeV nucleocapsids are single-subunit assemblies of ca. 96% protein and 4% RNA by weight. Their structures appear similar in negatively stained electron micrographs, except that TMV is a rigid, compact rod, whereas paramyxovirus nucleocapsids are more flexible, open coils. TMV is rigid because the coat subunits interact not only laterally along the helical path of the RNA (i.e., perpendicular to the helix axis) but also axially, making contact with the subunits on either side of the ribbon that forms the coil. The final structure forms both a protective shell for the viral genome and one that can be translationally disassembled within the plant cytoplasm for this positive-stranded RNA genome to function. TMV is a somewhat curious model for paramyxovirus nucleocapsids, as these two structures are clearly designed for different biochemical functions. However, both are helical assemblies in which each subunit is associated with a precise number (three or six) of nucleotides, and the manner in which the coat protein subunits of TMV interact with the cis-acting sequences of its RNA is clearly of interest.
The structure of TMV, as well as the location of its RNA, is known at atomic resolution, and this detail has provided insight into the mechanism of its assembly and disassembly (24). The RNA follows a path near the inside of the coat protein cylinder, within a groove formed by the axial coat protein-coat protein contacts of the stacked protein coils. The RNA makes contact only with the surfaces of the coat subunits, and the nucleotide binding sites are formed by the juxtaposition of opposite subunit surfaces of the stacked coils. The RNA in VSV nucleocapsids may also be present near the surfaces of their N subunits, as the Watson-Crick positions of their bases are accessible to the solvent (via chemical attack [1a, 21]). TMV RNA structure is strongly influenced by its interaction with the protein, leading to unusual RNA conformations. The first and third base of each trinucleotide point in the same direction as the helix axis and stack with the third and first base, respectively, of the neighboring trinucleotides. The middle base of each trinucleotide points toward the outside of the cylinder. While any base can be accommodated at the three binding sites, when a G is present in the first position it can form two additional hydrogen bonds to the protein, and this makes the binding of G at this position particularly favorable. The additional stability of this base-specific interaction at the first (5′) binding site is thought to explain why (i) the site for the initiation of virus assembly is composed of single-stranded RNA with the trinucleotide GNN repeated six times (37) and (ii) why the 5′ untranslated region of this capped mRNA is almost devoid of G residues, which could impede translational disassembly (38). Although different TMV strains are not necessarily of trimer length, their origin of assembly sequence is almost always positioned such that the initiating (5′) G residue is found at the first nucleotide binding site (39).
The base-specific interactions of the TMV coat protein with its RNA provide a precedent for how cis-acting RNA sequences can be recognized during paramyxovirus nucleocapsid assembly. The additional stability of simultaneously occupying all six phosphodiester backbone binding sites would also help ensure that nucleocapsid assembly started at the first base of the nascent chain and continued in register. Base-specific interactions in the nucleocapsid might also play a role in viral RNA synthesis, especially if structural transitions in the N subunits occur during this process (11). In this respect, we note that the strain-specific frequencies with which certain VSV and SeV polymerases read through the leader/N junction in virion reactions map to the N-RNA template rather than to the P-L polymerase (8, 28). Moreover, revertants of the high readthrough VSV phenotype were found to arise by extragenic suppression and presumably map to the L protein (6). Variations in the N protein sequences of these viruses can clearly affect the functioning of the viral polymerase at this particular junction, and in this respect, the N subunits are as much a part of the polymerase as the P and L proteins.
Although each base can be accommodated at each subunit binding site, additional interactions (via hydrogen bonds or stacking with aromatic side chains) of particular bases with particular sites might also be operating not only at junctions and other control regions but throughout the RNA chain. We have systematically examined whether there might be a preference for certain bases at each of the six hexamer positions throughout the entire sequence. However, we were able to detect only a trimer phase, which, as for TMV (39), appeared to be due to that imposed by codon usage.
RULES MAY BE RULES, BUT…
The variable conservation of the hexamer phase of the mRNA start sites among the Paramyxovirinae and the fact that different clusters of phases are preferred in the different genera (Table 1), suggest that the mRNA start site phase plays a role, but not a paramount one, in mRNA synthesis. This notion is also supported by experimental evidence. Schneider et al. (31) have prepared a recombinant measles virus in which 3 nt were inserted in the P gene mRNA editing site, and this was compensated for by the deletion of 3 nt at the last (H/L) junction. Even though the start site phases of the M, F, and H mRNAs were displaced by three hexamer positions downstream, this measles virus replicated normally in cell culture. The fact that the mRNA start site phase is more strongly conserved in the paramyxovirus genus than in the two others is of some interest, because there appears to be no rule at all for those viruses most closely related to this subfamily, namely, pneumoviruses like respiratory syncytial virus (RSV) (the other subfamily of the Paramyxoviridae) and rhabdoviruses such as VSV. Natural RSV genomes are in fact of hexamer length, but its infectious DNA clone contains one more nucleotide (7) and no evidence at all for the importance of hexamer length could be found using RSV CAT minireplicons (30). Similarly, the precise length of VSV DI genomes does not appear to be important (26).
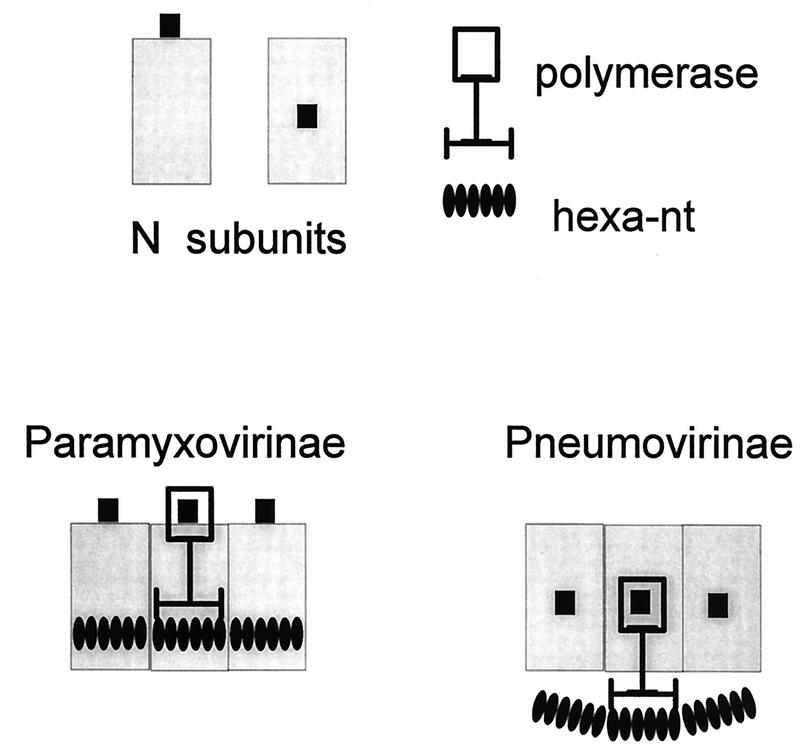
All these mononegavirus genomes are found as helical assemblies of N and RNA whose structures appear roughly similar in electron microscopy, and it seems unlikely that they would have chosen very different mechanisms of RNA synthesis. Nevertheless, if the rule of six for the Paramyxovirinae implies that their template bases are seen in the context of the N subunits for RNA synthesis, the absence of an integer rule must mean that the bases of VSV and RSV genomes are deciphered independently of the N protein backbone. It is possible that the degree to which a particular virus is governed by an integer rule depends on the extent that its N subunits are displaced from the bases during RNA synthesis. We assume that in all cases the P-L polymerase interacts with both the N subunits and the bases simultaneously, but one can imagine that for VSV and RSV the distance between these two interaction sites is greater. In the cartoon of Fig. 5, this occurs even though the relationships of the N subunit and RNA template binding sites of the polymerase are unchanged among the different viruses. However, the polymerase binding site on the pneumovirus N subunit is postulated to be much closer to the RNA template, hence the N subunits are separated from the template during RNA synthesis. If the separation is great enough, the spatial relationship of the RNA template to N subunits is determined only through the P-L polymerase complex and the N subunit phase is lost.
FIG. 5.
Paramyxoviridae and the rule of six. The N subunits are shown as open grey rectangles, with their six nucleotides shown as dark ovals. The polymerase binding site is shown as a small dark square, which is found at a different location relative to the RNA for each of the two subfamilies. The polymerase is shown as being composed of the N subunit binding site (open thick-lined rectangle) and the RNA interaction site (horizontal thick line) and is postulated to be similar for both subfamilies. The P and L components of the polymerase are not indicated. The simultaneous interaction of the polymerase N protein and RNA binding sites with their targets displaces the RNA from the N subunits for the pneumoviruses but does not disturb the RNA-N interactions for the paramyxoviruses.
MRNA EDITING, GENOME LENGTH CORRECTION, AND THE RULE OF SIX
The above model for how an integer rule might apply to some viruses but not to others, of course, begs the question of why the N subunit phase is required for some but apparently not required for other viruses. The requirement for an N subunit phase correlates with whether these viruses edit their P gene mRNA, as paramyxoviruses, morbilliviruses, and rubulaviruses all edit their P gene mRNAs, whereas pneumoviruses and rhabdoviruses apparently do not. Paramyxovirus P gene mRNA editing occurs by the programmed expansion of a short G run within an AnGn polypurine run during mRNA synthesis. As these G insertions are not found in the viral genomes, it was simply assumed that stuttering did not occur during genome replication. We now know that this is not strictly so, and this may offer one explanation of why the N subunit phase is required for viruses which edit their mRNAs.
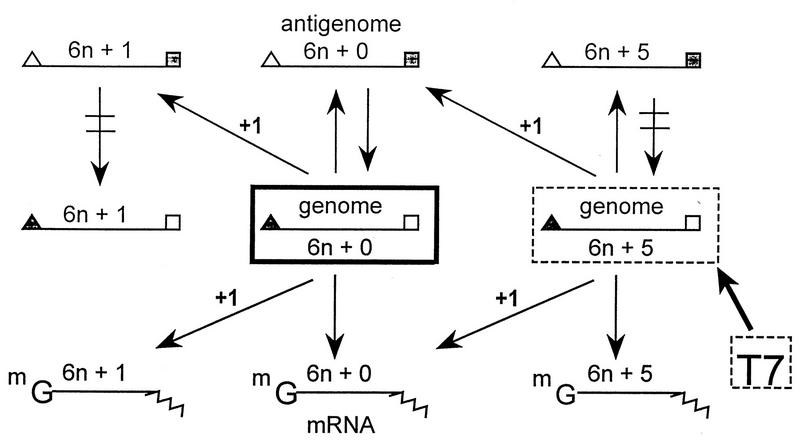
The lengths of all natural SeV genomes, which can vary by 20-fold in size, are in fact multiples of six. Changing their lengths to ones that are not multiples of six was found to result in very inefficient (50- to 100-fold less) genome amplification in transfected cells, especially when the overlapping C gene of P mRNA is expressed. However, when C protein expression is suppressed, nonhexamer-length DI genomes (especially those that contain 1 nt more or 1 nt less) replicate only 10-fold less well than hexamer-length genomes (reference 34 and unpublished data). Similar results have recently been reported for the rubulavirus SV5 (whose P gene does not contain an overlapping C protein ORF) (23a) and for human PIV3 DI analogs in a system in which C protein expression was suppressed (10a). It will be of interest to determine whether reexpression of the human PIV3 C protein increases the requirement for hexamer genome length for this paramyxovirus, as it does for SeV. In addition, if the SeV DI genomes also contain the P gene mRNA editing site (A6G3), they replicate only two- to threefold less well than hexamer-length genomes in the absence of C (34). This was not because the rule of six did not apply; far from it. When minigenomes which are not of hexamer length are replicated in transfected cells under these conditions, genomes are generated in which nucleotides are inserted (or deleted) in the A6G3 purine run, which readjust their length to multiples of six (18), as schematized in Fig. 6. Pseudotemplated transcription was found to occur at the same purine run used for G insertions during mRNA synthesis, when the viral polymerase is copying the genome template during antigenome synthesis. This process (termed genome length correction) was found to differ from the G insertions during mRNA synthesis in three important respects. (i) It can delete as well as insert purines (and A residues as well as Gs). (ii) It can be detected at a significant frequency only when the genome is not of hexamer length. (iii) Only the A6G3 sequence itself is required, whereas the G insertions during mRNA synthesis require additional upstream sequences. Given that genomes which are precise multiples of six are preferentially replicated, this leads to a mechanism which corrects the genome length according to this rule.
FIG. 6.
Paramyxovirus genome length correction. The viral RNAs are shown as horizontal lines; the filled symbols at the extremities indicate 3′ ends. The direction of RNA synthesis is indicated by arrows. Unnatural nonhexamer-length genomes (e.g., 6n+5) can be introduced into cells via T7 RNA polymerase expression vectors. Although they replicate poorly, their constant production by the bacteriophage polymerase ensures that some antigenome synthesis occurs. Purine insertions take place during antigenome synthesis as well as during mRNA synthesis, and those which are now of hexamer length replicate more efficiently and accumulate, leading to correction of the genome length.
All negative-strand virus RNA polymerases which polyadenylate their mRNAs are thought to do so by stuttering on a short run of template U residues (4 to 7 nt long), and it was this that first suggested that the G insertions would similarly occur by pseudotemplated transcription (5, 36). However, while mRNA editing and polyadenylation are clearly related, they are also distinct. For example, DI H4 genomes which contain poly(A) (but not editing) sites are not corrected for hexamer length at this site, even in the absence of C protein expression in which significant replication occurs (34). Polymerase stuttering during antigenome synthesis appears to occur exclusively in the A6G3 purine run used for mRNA editing, as opposed to that (5′ ANTAAGA5) which is expanded in polyadenylation. The P gene editing site thus appears to have dual and complementary functions: to allow insertions during mRNA synthesis from hexamer-length templates and to allow insertions during antigenome synthesis from nonhexamer-length templates.
It is unclear, however, whether this latter function is an important feature of the virus replicative cycle or one whose importance is exaggerated by the use of reverse genetics. Whereas >20% of antigenomes made from nonhexamer-length genomes contain insertions, there is no evidence that insertions occur during antigenome synthesis from hexamer-length genomes (reference 18 and unpublished data), but frequencies of <5% are difficult to detect by limited primer extension. Should G insertions occur during normal antigenome synthesis, it is unclear whether full-length genomes with one or two G insertions would be automatically eliminated from the population simply on the basis of their altered P gene product expression. Recombinant SeV, which edit their P gene mRNA like PIV3 and therefore express roughly equal levels of P, V, and W proteins, are easy to prepare and they are remarkably robust (unpublished data). The hexamer rule might then be required to maintain the length of the P gene purine run against the instability which occurs during genome replication by preventing the inserted antigenomes from being copied back into genomes and thus selectively removing them from the population (Fig. 6). It is also possible that the additional requirement for certain viral polymerases to discriminate between the P gene editing site and other polypurine runs [like poly(A) sites], and to insert G residues by pseudotemplated transcription in a programmed virus-specific manner, was the driving force in enlisting the N subunit phase to help in this process. In either case, the proposed requirement for a given phase of the replication promoters relative to N subunits for efficient initiation would then have coevolved with the ability of these polymerases to edit their mRNAs.
ACKNOWLEDGMENTS
We thank Bert Rima (Belfast, Northern Ireland), Tom Barrett (Pirbright, United Kingdom), Yasuhiko Ito (Mie, Japan), Reay Paterson and Bob Lamb (Evanston, Ill.), and Martin Billeter (Zurich, Switzerland) for providing and discussing the nucleotide sequences. We are especially grateful to Cynthia Steinberger (Dept. of Molecular Biology, University of Geneva) for analyzing the entire sequences in depth for an overall hexamer phase and to Mike Schmid (Baylor University, Houston, Tex.) for interpretations of electron micrographs.
REFERENCES
- 1.Alberts B, et al. The molecular biology of the cell. 3rd ed. New York, N.Y: Garland Publishing; 1994. [Google Scholar]
- 1a.Baudin, F. (EMBL, Grenoble, France). Personal communication.
- 2.Blumberg B M, Giorgi C, Kolakofsky D. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell. 1983;32:559–567. doi: 10.1016/0092-8674(83)90475-0. [DOI] [PubMed] [Google Scholar]
- 3.Calain P, Curran J, Kolakofsky D, Roux L. Molecular cloning of natural paramyxovirus copy-back defective interfering RNAs and their expression from DNA. Virology. 1992;191:62–71. doi: 10.1016/0042-6822(92)90166-m. [DOI] [PubMed] [Google Scholar]
- 4.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattaneo R, Kaelin K, Baezko K, Billeter M A. Measles virus editing provides an additional cysteine-rich protein. Cell. 1989;56:759–764. doi: 10.1016/0092-8674(89)90679-x. [DOI] [PubMed] [Google Scholar]
- 6.Chuang J L, Jackson R L, Perrault J. Isolation and characterization of vesicular stomatitis virus PolR revertants: polymerase readthrough of the leader-N gene junction is linked to an ATP-dependent function. Virology. 1997;229:57–67. doi: 10.1006/viro.1996.8418. [DOI] [PubMed] [Google Scholar]
- 7.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curran J, Homann H, Buchholz C, Rochat S, Neubert W, Kolakofsky D. The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J Virol. 1993;67:4358–4364. doi: 10.1128/jvi.67.7.4358-4364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curran J, Boeck R, Lin-Marq N, Lupas A, Kolakofsky D. Paramyxovirus phosphoproteins form homo-trimers as determined by an epitope dilution method, via predicted coiled coils. Virology. 1995;214:139–149. doi: 10.1006/viro.1995.9946. [DOI] [PubMed] [Google Scholar]
- 10.Curran J. Reexamination of the Sendai virus P protein domains required for RNA synthesis: a possible supplemental role for the P protein. Virology. 1996;221:130–140. doi: 10.1006/viro.1996.0359. [DOI] [PubMed] [Google Scholar]
- 10a.Durbin A P, Siew J W, Murphy B R, Collins P C. Minimum protein requirements for transcription and RNA replication of a minigenome of human parainfluenza virus type 3 and evaluation of the rule of six. Virology. 1997;234:74–83. doi: 10.1006/viro.1997.8633. [DOI] [PubMed] [Google Scholar]
- 11.Egelman E H, Wu S S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enami M, Kohama T, Sugiura A. A measles virus subgenomic RNA: structure and generation mechanism. Virology. 1989;171:427–433. doi: 10.1016/0042-6822(89)90611-9. [DOI] [PubMed] [Google Scholar]
- 13.Engelhorn M, Stricker R, Roux L. Molecular cloning and characterization of a Sendai virus internal deletion defective RNA. J Gen Virol. 1993;74:137–141. doi: 10.1099/0022-1317-74-1-137. [DOI] [PubMed] [Google Scholar]
- 14.Finch J T, Gibbs A J. Observations on the structure of the nucleocapsids of some paramyxoviruses. J Gen Virol. 1970;6:141–150. doi: 10.1099/0022-1317-6-1-141. [DOI] [PubMed] [Google Scholar]
- 15.Fraenkel-Conrat H, Williams R. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc Natl Acad Sci USA. 1965;41:690–695. doi: 10.1073/pnas.41.10.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamaguci M, Yoshida T, Nishikawa K, Naruse H, Nagai Y. Transcriptive complex of Newcastle disease virus. I. Both L and P proteins are required to constitute an active complex. Virology. 1983;128:105–117. doi: 10.1016/0042-6822(83)90322-7. [DOI] [PubMed] [Google Scholar]
- 17.Harty R N, Palese P. Mutations within noncoding terminal sequences of model RNAs of Sendai virus: influence on reporter gene expression. J Virol. 1995;69:5128–5131. doi: 10.1128/jvi.69.8.5128-5131.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hausmann S, Jacques J-P, Kolakofsky D. Paramyxovirus RNA editing and the requirement for length. RNA. 1996;2:1033–1045. [PMC free article] [PubMed] [Google Scholar]
- 19.Heggeness M H, Scheid A, Choppin P W. Conformation of the helical nucleocapsids of paramyxoviruses and vesicular stomatitis virus reversible coiling and uncoiling induced by changes in salt concentration. Proc Natl Acad Sci USA. 1980;77:2631–2635. doi: 10.1073/pnas.77.5.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horikami S M, Curran J, Kolakofsky D, Moyer S A. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keene J D, Thornton B J, Emerson S U. Sequence-specific contacts between the RNA polymerase of vesicular stomatitis virus and the leader RNA gene. Proc Natl Acad Sci USA. 1981;78:6191–6195. doi: 10.1073/pnas.78.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolakofsky D, Curran J, Pelet T, Jacques J-P. Paramyxovirus P gene mRNA editing. In: Benne Rob., editor. RNA editing. Chichester, England: Ellis Horwood; 1993. [Google Scholar]
- 22a.Krystal, Mark. Personal communication.
- 23.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, et al., editors. Fields Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Press; 1996. [Google Scholar]
- 23a.Murphy S K, Parks G D. Genome nucleotide lengths that are divisible by six are not essential but enhance DI RNAs of the paramyxovirus SV5. Virology. 1997;232:145–157. doi: 10.1006/viro.1997.8530. [DOI] [PubMed] [Google Scholar]
- 24.Namba K, Pattanayek R, Strubbs G. Visualization of protein-nucleic acid interactions in a virus: refined structure of intact tobacco mosaic virus at 2.9 Å resolution by X-ray fiber diffraction. J Mol Biol. 1989;208:307–325. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- 25.Park K H, Huang T, Correia F F, Krystal M. Rescue of a foreign gene by Sendai virus. Proc Natl Acad Sci USA. 1991;88:1–5. doi: 10.1073/pnas.88.13.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pattnaik A K, Ball L A, LeGrone A, Wertz G W. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology. 1995;206:760–764. doi: 10.1016/S0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 28.Perrault J, Clinton G M, McClure M A. RNP template of vesicular stomatitis virus regulates transcription and replication functions. Cell. 1983;35:175–185. doi: 10.1016/0092-8674(83)90220-9. [DOI] [PubMed] [Google Scholar]
- 29.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Christiansen G, Billeter M A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samal S K, Collins P L. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider H, Kaelin K, Billeter M A. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in cultured cells. Virology. 1997;227:314–322. doi: 10.1006/viro.1996.8339. [DOI] [PubMed] [Google Scholar]
- 32.Sidhu M S, Crowley J, Lowenthal A, Karcher D, Mennona J, Udem S A, Dowling P. Defective measles virus in human subacute sclerosing panencephalitis brain. Virology. 1994;202:631–641. doi: 10.1006/viro.1994.1384. [DOI] [PubMed] [Google Scholar]
- 33.Sidhu M S, et al. Rescue of synthetic measles virus minireplicons: measles genomic termini direct efficient expression and propagation of a reporter gene. Virology. 1995;208:800–807. doi: 10.1006/viro.1995.1215. [DOI] [PubMed] [Google Scholar]
- 34.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Tapparel, C., D. Maurice, and L. Roux. The activity of the Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 35.Thomas D, Newcomb W W, Brown J C, Wall J S, Hainfeld J F, Trus B L, Steven A C. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J Virol. 1985;54:598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas S M, Lamb R A, Patterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner D R, Joyce L E, Butler P J G. The tobacco mosaic virus assembly origin RNA: functional characteristics defined by directed mutagenesis. J Mol Biol. 1988;203:531–547. doi: 10.1016/0022-2836(88)90190-8. [DOI] [PubMed] [Google Scholar]
- 38.Wilson T M A, Watkins P A C. Cotranslational disassembly of a cowpea strain (Cc) of TMV: evidence that viral RNA-protein interactions at the assembly origin block ribosome translocation in vitro. Virology. 1985;145:346–349. doi: 10.1016/0042-6822(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 39.Wilson T M A, McNicol J W. A conserved, precise RNA encapsidation pattern in tobamovirus particles. Arch Virol. 1995;140:1677–1685. doi: 10.1007/BF01322541. [DOI] [PubMed] [Google Scholar]