Abstract
Human papillomavirus type 16 (HPV-16) is strongly associated with the development of cervical cancer. Studies of model systems with animal papillomaviruses have demonstrated the importance of neutralizing antibodies in preventing papillomavirus-associated disease. The assessment of neutralizing antibody responses against HPV-16, previously hampered by the lack of a viral source, was enabled by the recent propagation of an HPV-16 stock in xenografted severe combined immunodeficiency (SCID) mice. HPV-16 infection of an immortalized human keratinocyte cell line was demonstrated by detection of an HPV-16-specific spliced mRNA amplified by reverse transcriptase PCR. Infection was blocked by preincubation of the virus with antiserum generated against HPV-16 virus-like particles (VLPs) composed of the major capsid protein, L1. To examine potential cross-neutralizing activity among the different genital HPV types, rabbit antisera to L1 VLPs corresponding to HPV-6, -11, -18, -31, -33, -35, -39, and -45 were assayed for the ability to block the HPV-16 infection of cultured cells. Antiserum raised against HPV-33 L1 VLPs was the only heterologous antiserum which inhibited HPV-16 infection. Thus, a neutralization assay for HPV-16 may help to characterize the components required to compose a broadly efficacious genital HPV vaccine.
Human papillomaviruses (HPVs) are the most common sexually transmitted viral pathogens in the United States (26). “Low-risk” HPVs such as HPV-6 and -11 are associated with the production of benign genital warts, while “high-risk” types such as HPV-16 and -18 are known to be a major causative factor in the development of cervical cancer. The association of cervical carcinogenesis and HPV infection is indicated by strong epidemiological evidence and the detection of HPV DNA in more than 93% of cervical cancers from all geographic areas (5). Of the high-risk types, HPV-16 is the most prevalent, being present in 50% of cervical tumor specimens worldwide. Other high-risk HPV types include HPV-18, -31, -33, and -45.
Due to the morbidity and mortality associated with the high-risk HPV types, there is keen interest in developing prophylactic HPV vaccines. Results obtained with several different animal models (canine oral papillomavirus, cottontail rabbit papillomavirus [CRPV], and bovine papillomavirus type 4 [BPV-4]) established the feasibility of developing vaccines to prevent papillomavirus disease (7, 19, 35). These animal studies demonstrated the protective efficacy of the major papillomavirus capsid component, the L1 protein. When expressed in eukaryotic cells, the L1 proteins of many different papillomavirus types self-assemble into virus-like particles (VLPs) that are antigenically and morphologically similar to authentic papillomavirions (16, 18, 31). Animals immunized with L1 VLPs were protected from subsequent papillomavirus challenge. Successful vaccination required that the VLPs be composed of the L1 protein of the challenge virus, and immunity was found to be generally type specific. In both the canine oral papillomavirus and CRPV animal models, passive transfer of immune serum from VLP-immunized animals to naive animals conferred protection from subsequent challenge with the homologous papillomavirus, suggesting that antibodies serve as a major protective component against papillomavirus infection (7, 35).
The results with animal models provide a strong rationale for the development of VLP-based vaccines to prevent HPV-induced genital warts and cervical cancer. However, HPV vaccine development has been hindered by the high degree of species specificity exhibited by these viruses, which has made direct evaluation of vaccine efficacy in animals impossible. Also, difficulties in the propagation of HPV stocks have hampered the examination of neutralizing antibody responses against authentic HPVs.
One notable exception is the low-risk HPV-11, which has been propagated with a xenograft system in a sufficient quantity to allow direct evaluation of neutralizing antibodies (12, 14, 20). Antisera generated against HPV-11 VLPs have been shown to contain high titers of HPV-11-neutralizing antibodies, as assessed by the abrogation of condyloma growth in the xenograft system. Recently, a method was developed to study antibody-mediated neutralization of HPV-11 in vitro (34). In this assay, HPV-11 infection of cultured human keratinocytes was determined by the appearance of an HPV-11-specific mRNA detected by reverse transcriptase PCR (RT-PCR). Preincubation of the virus with antibodies which had previously been shown to neutralize HPV-11 in the xenograft assay prevented HPV-11 infection of the keratinocytes, as demonstrated by the inability to detect HPV-11-specific transcripts.
The lack of a reliable source of virus has prevented the direct evaluation of neutralizing antibodies specific for the high-risk HPV-16. Researchers have relied on surrogate assays, such as inhibition of VLP-mediated hemagglutination, to study the functional activity of antisera generated against HPV-16 VLPs (28). Recently, HPV-16 has been propagated with a SCID mouse xenograft system (2). In the present study, we demonstrate that an HPV-16 stock prepared from the xenografted condylomas can infect an immortalized keratinocyte cell line in vitro, as measured by the appearance of an HPV-16-specific transcript. HPV type-specific antibodies inhibited HPV-16 infection in vitro, thus providing the first direct evidence of antibody-mediated neutralization of an authentic high-risk HPV. In addition, the potential for cross-protection among the high-risk and low-risk genital HPV types was assessed by examining the ability of antisera to VLPs of various heterologous HPV types to neutralize HPV-16 infection.
MATERIALS AND METHODS
Isolation and propagation of HPV-16.
Our HPV-16 strain was isolated and propagated with the xenograft SCID mouse model (3, 20). The propagation process and the typing analyses of viral stock passages are described in detail elsewhere (2). In brief, single biopsy samples were obtained from 11 patients with clinical condylomata acuminata. Subsequent histologic diagnosis showed that two of the patients had mild to moderate intraepithelial neoplasia. The biopsy samples were ground in phosphate-buffered saline (PBS) with sand and a mortar and pestle. The preparation was submitted to low-speed centrifugation, and the supernatant was pelleted at 100,000 × g for 1 h at 4°C and resuspended in PBS. This viral suspension was used to infect neonatal human foreskin fragments that were each implanted under the renal capsule of three SCID mice (3, 20). Twelve weeks later, the mice were sacrificed and the grafts were collected. One of the six grafts showed histologic evidence of intraepithelial neoplasia and was prepared as described above to make a lysate for a second passage. Twelve SCID mice were grafted under the renal capsule with neonatal foreskin fragments (one per kidney) that had been incubated in the lysate. The mice were sacrificed 19 weeks later. Five of 15 retrieved grafts had evidence of HPV infection by histology, and one of them contained HPV capsid protein by immunocytochemistry. The remaining tissue samples from the grafts were used to prepare a viral lysate as described above. HPV capsids were demonstrated after negative staining by electron microscopy. The viral DNA was extracted from the lysate and subjected to PCR analysis with the MY09/MY11 primer pair (21). The PCR fragment was cloned and sequenced. The DNA sequence was identical to a reference strain of HPV-16 (32), except at seven nucleotide positions. Five of the base changes resulted in no amino acid substitution, one caused a conservative change (threonine to serine [A6801T]), and one yielded a nonconservative substitution (threonine to proline [A6693C]). The viral lysate was used for a third passage of the virus. Single infected grafts were implanted under each renal capsule as well as under the skin of each flank of 24 SCID mice. The animals were sacrificed 27 weeks later, and the renal and subcutaneous grafts were collected to prepare a viral lysate as described above. The virus stock was typed by amplification of viral DNA with the MY09/MY11 primers and hybridization with oligonucleotide probes specific for the following HPV types: 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51 to 59, 66, 68, 73, MM4, LVX-82/MM7, and MM8 (1, 17, 22). Positive results were obtained only with the HPV-16-specific probes. The MY09/MY11 amplicon was also typed by digestion with the restriction enzymes EcoRI and PstI, which resulted in a banding pattern consistent with HPV-16 (23). This virus stock was designated HPV-16Rochester-1k/ur3 and was used in the present experiments.
Antisera to VLPs.
Polyclonal rabbit antisera specific for HPV-11, -16, and -18 L1 VLPs have been described previously (30). Antisera to HPV-6 L1 VLPs were kindly provided by S.-J. Ghim. To generate antibodies against HPV-31, -33, -35, -39, and -45 L1 VLPs, genomic clones of HPV-31 and HPV-35 were obtained from the American Type Culture Collection (Rockville, Md.); HPV-33 and HPV-39 DNAs were provided by M. Favre and G. Orth (Institut Pasteur, Paris, France); and cloned HPV-45 DNA was provided by A. Lorincz (Digene Diagnostics, Inc., Silver Spring, Md.). PCR amplification of the L1 sequences and generation of recombinant baculoviruses carrying genes encoding L1 were carried out as previously described (30). VLPs composed of HPV-31, -33, -35, -39, and -45 L1 were purified from recombinant baculovirus-infected Sf9 or High Five cells on CsCl density gradients. Total protein concentrations were determined with a commercial assay (bicinchoninic acid; Pierce Chemical Co., Rockford, Ill.). For each HPV VLP type, a New Zealand White rabbit was immunized intramuscularly at two sites with an emulsion of 50 μg of L1 protein in complete Freund’s adjuvant and given a booster injection 2 weeks later with an emulsion prepared with the same VLP type and incomplete Freund’s adjuvant. The reactivity of the antisera against the homotypic VLPs used for immunization was determined by enzyme-linked immunosorbent assay (ELISA), with the titer being defined as the greatest dilution which yielded an optical density value greater than 0.1.
HPV-16 VLP ELISA. (i) Coating antigen.
ELISAs were carried out with VLPs containing the HPV-16 L1 sequence variant corresponding to our virus stock. DNA was extracted from the second-passage HPV-16 virus stock as described above. The entire L1 gene was amplified by PCR and cloned into a baculovirus transfer vector by methods similar to those previously described (31). The DNA sequence of the full L1 clone was determined. The sequence was identical to that of the MY09/MY011 amplicon in the overlapping region and contained 11 additional nucleotide substitutions outside the region amplified by the MY09/MY011 primer pair. In total, this HPV-16 L1 variant contained nine amino acid differences from the prototype: histidine to tyrosine (C5862T), threonine to asparagine (C6163A), asparagine to threonine (A6178C), histidine to aspartic acid (C6240G), glycine to serine (G6252A), threonine to alanine (A6432G), threonine to proline (A6693C), threonine to serine (A6801T), and leucine to phenylalanine (G7058T). The transfer vector containing the HPV-16 L1 gene was used to generate a recombinant baculovirus as previously described (31). VLPs were purified from recombinant baculovirus-infected High Five cells as described above.
(ii) ELISA protocol.
HPV-16 L1 VLPs were diluted in PBS to 0.01 mg/ml and dispensed in 0.1-ml aliquots to 96-well microtiter plates. PBS without VLPs was dispensed to control wells. After being coated for 16 h at 4°C, the plates were blocked with blocking solution (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.) for 2 h at room temperature. Threefold serial dilutions of anti-HPV-VLP sera were made in PBS containing 1% bovine serum albumin and 10% (vol/vol) wild-type baculovirus-infected cell culture supernatant to reduce the background (30). After a 90-min room temperature incubation, plates were washed and anti-rabbit immunoglobulin G-alkaline phosphate conjugate (Kirkegaard & Perry Laboratories, Inc.) diluted 1:2,000 in blocking solution was added to the wells. Following incubation and washing, specific binding was detected with the alkaline phosphate substrate. Specific absorbance was calculated by subtracting the absorbance values obtained with PBS alone from those obtained with antigen. Averages of duplicate wells were calculated as the final absorbance values.
HPV-16 in vitro infection and neutralization.
HaCaT cells, an immortalized human keratinocyte cell line (6), were kindly supplied by N. Fusenig. Cells were grown to 90 to 100% confluency in 154/HKGS medium (Cascade Biologics, Inc., Portland, Oreg.) supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml) in 24-well plates. HPV-16 stock was sonicated for 25 s on ice, diluted into 154/HKGS medium in round-bottom polypropylene tubes, and incubated for 1 h at 37°C. Medium was aspirated from the HaCaT cells, and 0.5 ml of diluted virus was added per well. As a control, one well of cells on each plate received 0.5 ml of medium without virus. For antibody-mediated neutralization, antisera were diluted in 154/HKGS and incubated with a fixed quantity of the HPV-16 stock in a final volume of 0.5 ml in round-bottom polypropylene tubes for 1 h at 37°C prior to addition to the HaCaT cells. Fresh medium was added to each well of cells 4 days postinfection, and on day 7, total cellular RNA was extracted with Tri Reagent (Molecular Research Center, Inc., Cincinnati, Ohio) according to the manufacturer’s recommendations. Final RNA pellets were resuspended in 20 μl of diethylpyrocarbonate-treated water and quantified by spectrophotometry.
Detection of HPV-16 and cellular β-actin-spliced mRNA by RT-PCR.
Reverse transcription reactions were performed with a First Strand cDNA kit (Boehringer Mannheim, Indianapolis, Ind.) with 2 μg of total RNA as the template and oligo(dT) as the primer in a final volume of 20 μl. Nested PCR was needed to detect HPV-16 E1^E4 cDNA. The first round of amplification was carried out with 25% of the cDNA from each reverse transcription reaction and 5′-TGGAAGACCTGTTAATGGGCACAC-3′ (located at bases 797 to 818 in the HPV-16 genomic sequence) as the forward outside (FO) primer and 5′-TATAGACATAAATCCAGTAGACAC-3′ (located at bases 3826 to 3849 in the HPV-16 genomic sequence) as the reverse outside (RO) primer for 40 cycles of PCR. Ten percent of the first-round PCR mixture was used for nested reactions with 5′-GGAATTGTGTGCCCCATCTGTTC-3′ (located at bases 823 to 845 in the HPV 16 genomic sequence) as the forward nested primer (FN) and 5′-GTTCACGTTGACATTCACTATC-3′ (located at bases 3766 to 3787 in the genomic sequence) as the reverse nested primer (RN) for 35 PCR cycles. First-round and nested PCRs were set up with Hot Wax beads (1.5 mM) and pH 9.5 buffer (InVitrogen, San Diego, Calif.) with 200 μM deoxynucleoside triphosphates (dNTPs), 125 ng each of the forward and reverse primers, and 2.5 U of Taq polymerase (Perkin-Elmer, Foster City, Calif.) in a final volume of 50 μl. The temperature profile for both first-round and nested PCRs was 80°C for 5 min, 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min.
All cDNA samples were used in separate PCRs with primers specific for spliced cellular β-actin mRNA. The PCR primers were described by Smith et al. (34). Amplification of the β-actin spliced message was achieved with 125 ng of forward primer (5′-GATGACCCAGATCATGTTTG-3′) and 125 ng of reverse primer (5′-GGAGCAATGATCTTGATCTTC-3′) with 12.5% of the total cDNA as the template in 10 mM Tris-HCl (pH 8.3) buffer containing 50 mM KCl, 1.5 mM MgCl2, and 1% gelatin, with 200 μM dNTPs and 2.5 U of Taq polymerase in a final volume of 50 μl for 35 PCR cycles. The temperature profile for amplification was 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min.
All PCR products were separated by electrophoresis on a 2% agarose gel and visualized by ethidium bromide fluorescence.
DNA sequencing.
The E1^E4 nested PCR product was purified with Qiaquick-spin columns (Qiagen, Chatsworth, Calif.). The concentration of the column eluate was determined by spectrophotometry and was then diluted to achieve a final concentration of 125 ng/μl. Sequencing reactions were carried out with the Dye Terminator DNA sequencing reaction mix (Perkin-Elmer, Foster City, Calif.) with 125 ng of PCR product as template and 3 pmol of FN or RN primers. Samples were subjected to cycle sequencing according to the manufacturer’s recommendation. Extension products were purified with Centri Sep columns (Princeton Separations, Adelphia, N.J.) and dried under vacuum. Samples were resuspended in 4 μl of sequencing buffer (formamide containing 8.3 mM EDTA) and electrophoresed on a 4.2% acrylamide–8 M urea sequencing gel with the ABI 373 automated sequencer (ABI, Foster City, Calif.). Sequence data were analyzed with the Laser Gene program (DNA Star, Madison, Wis.).
RESULTS
HPV-16 infects human keratinocytes in vitro.
An HPV-16 stock (HPV-16Rochester-1K/ur3) that represented the third passage in xenografts of a viral lysate originally derived from patients with a history of condylomata acuminata was used to infect the human keratinocyte cell line HaCaT. Since HPV-16 was not expected to progress through an infectious cycle in cultured cells, an RT-PCR strategy was designed to detect an HPV-16-specific E1^E4 mRNA as a marker for infection (Fig. 1). E1^E4 mRNA species have been demonstrated to be very abundant in HPV-1-, HPV-6-, and HPV-11-induced condylomas (8, 9, 24, 25), as well as in an HPV-16-transformed rodent cell line (36, 37). In addition, an HPV-11-specific E1^E4 transcript was successfully used as a direct marker for HPV-11 infection in both the xenograft system and in cultured human cells (4, 33, 34).
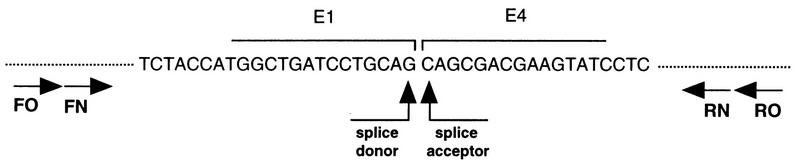
FIG. 1.
RT-PCR strategy for the detection of HPV-16 E1^E4 mRNA. Total cellular RNA obtained from HPV-16-exposed HaCaT cells was reverse transcribed, and the resultant cDNA was amplified by PCR with the FO and RO primers as described in Materials and Methods. Nested PCR with the FN and RN primers resulted in the amplification of a 487-bp product. Nucleotide sequence analysis of the PCR product revealed the splice donor (nucleotide 881)/acceptor (nucleotide 3356) site of HPV-16 E1^E4 mRNA.
HPV-16 stock was diluted and added to cultured HaCaT cells. After 7 days in culture, total RNA was extracted from the cells and was used for cDNA synthesis. Nested primers were then used to amplify an HPV-16 E1^E4 cDNA. A 487-bp PCR product consistent with the projected size of an HPV-16 E1^E4 transcript was amplified from the RNA of the cells incubated with virus (Fig. 2, lane 1). In contrast, no similar PCR product was detected with RNA isolated from control HaCaT cells which had not been exposed to virus (Fig. 2, lane 2). The inability to amplify the 487-bp product from the control cellular RNA was not due to poor RNA recovery or failed reverse transcription, since the cDNA sample was successfully used in a separate PCR to detect spliced β-actin mRNA (Fig. 2, lane 2).
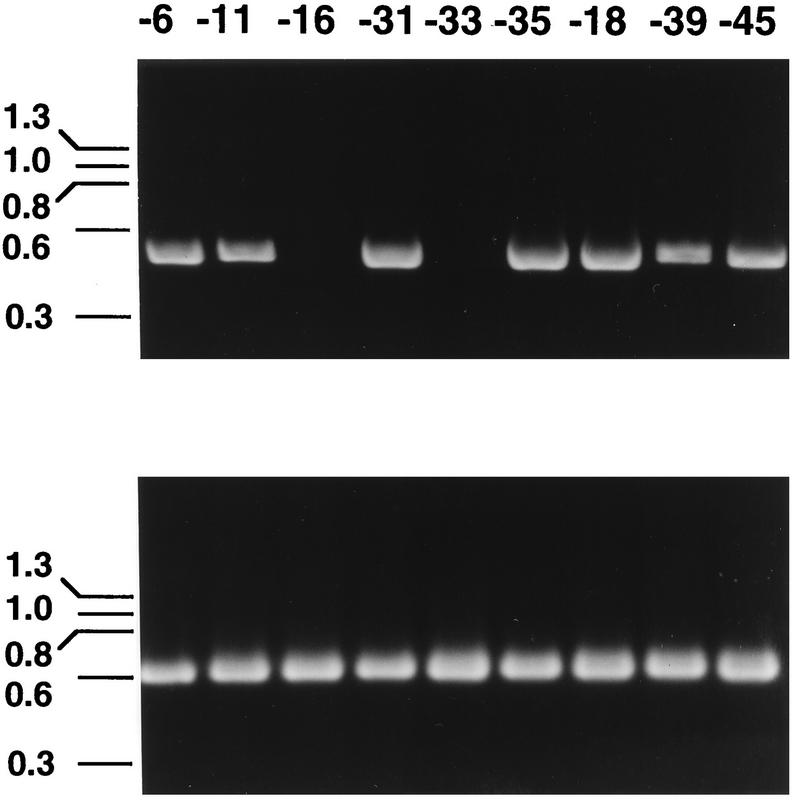
FIG. 2.
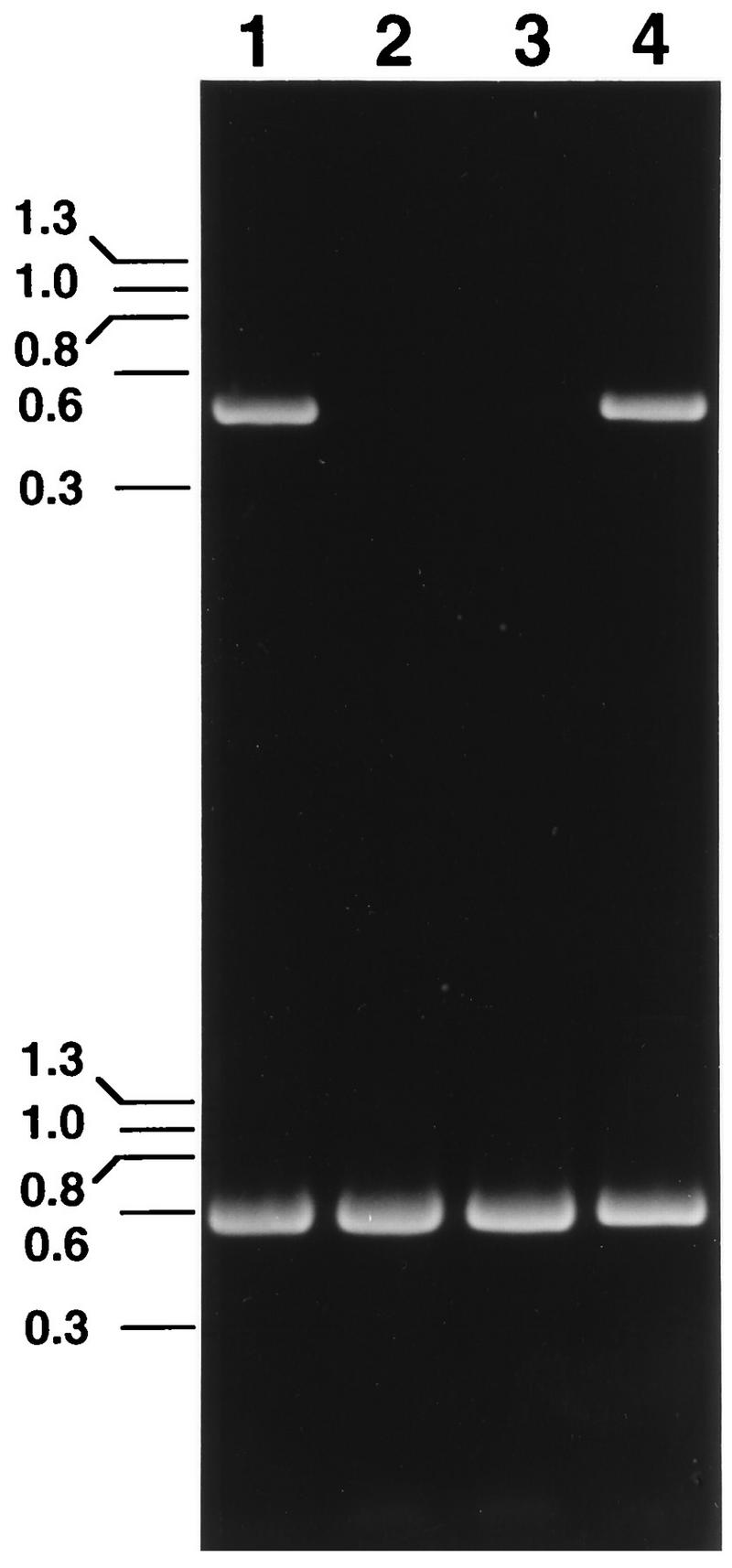
HPV-16 infection of HaCaT cells documented by the appearance of an HPV-16-specific E1^E4 mRNA. cDNA samples obtained by reverse transcription of total RNA isolated from cells infected with HPV-16 (lane 1), uninfected cells (lane 2), cells infected with HPV-16 preincubated with HPV-16 L1 VLP antiserum (lane 3), and cells infected with HPV-16 preincubated with normal control serum (lane 4) were amplified with primers specific for HPV-16 E1^E4 (top) or β-actin (bottom).
The identity of the 487-bp HPV-16-specific PCR product was confirmed by nucleotide sequence analysis. The DNA sequence of this PCR product represented HPV-16 nucleotides 823 to 3787, with a deletion spanning nucleotides 881 to 3356, consistent with an HPV-16 E1^E4 spliced mRNA species (Fig. 1) (32).
The results presented in Fig. 2 were obtained with the HPV-16 virus stock diluted 1:104. However, identical results were obtained with dilutions of virus stock from 1:102 to 1:106. The 487-bp HPV-16 product was not amplified from cells which had been exposed to the virus stock diluted to 1:107 (data not shown). Repetitive titration of the viral stock resulted in consistent detection of the HPV-16 spliced message with viral dilutions of ≤1:106.
Neutralization of HPV-16 in vitro infection.
No cytopathic changes were associated with HPV-16 infection of HaCaT cells. However, the ability to detect an HPV-16-specific mRNA following exposure of HaCaT cells to the viral stock indicated that the virus entered the cells and began its replication cycle at least to the point of expression of an E1^E4 transcript. To determine if specific antibodies could neutralize HPV-16 infection, the virus stock was preincubated with polyclonal anti-HPV-16 L1 VLP serum or normal control serum prior to addition to the HaCaT cells. Virus neutralization was demonstrated by the inability to detect the E1^E4 spliced message in virus-exposed cells following preincubation of the virus stock with a 1:100 dilution of the anti-HPV-16 L1 VLP serum (Fig. 2, lane 3). In contrast, the HPV-16 transcript was detected in cells incubated with virus mixed with control serum (Fig. 2, lane 4).
Polyclonal antisera were also generated against L1 VLPs corresponding to certain low-risk (types 6 and 11) and high-risk (types 18, 31, 33, 35, 39, 45) HPVs and were screened by ELISA against homotypic VLPs. Each of the antisera reacted strongly with homotypic VLPs with titers of ≥1:121,500. This panel of anti-VLP sera was tested for HPV-16-neutralizing activity by preincubation of a 1:100 dilution of each serum sample with the HPV-16 stock prior to exposure to the cells. As shown in Fig. 3, none of the heterotypic VLP antisera inhibited HPV-16 infection, except anti-HPV-33 L1 VLP. Additional experiments conducted with lower (1:20) dilutions of antisera confirmed that the HPV-6, -11, -18, -31, -35, -39, and -45-specific antibodies were unable to neutralize HPV-16.
FIG. 3.
HPV-16 neutralization by heterotypic genital HPV L1 VLP antisera. HPV-16 stock diluted 1:104 was preincubated with 1:100 dilutions of anti-HPV L1 VLP serum samples prior to addition to HaCaT cells. HPV-16 E1^E4 (top)- and cellular β-actin (bottom)-specific RT-PCR products obtained with RNA isolated from cells infected with HPV-16, which had been preincubated with anti-HPV-6, -11, -16, -31, -33, -35, -18, -39, and -45 L1 VLP sera, are shown.
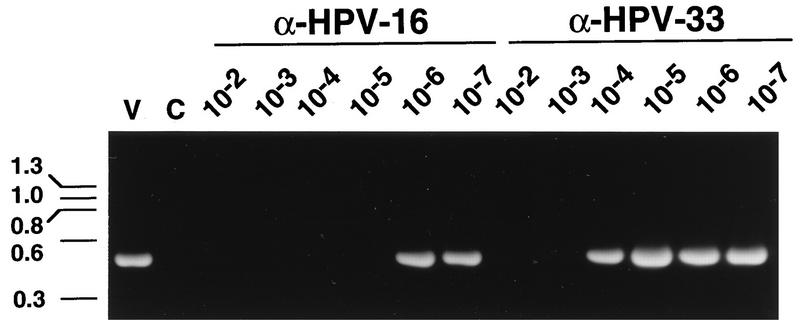
A quantitative assessment of the relative potency of HPV-16-neutralizing activity in the anti-HPV-16 and anti-HPV-33 VLP antisera was carried out by incubation of the HPV-16 stock with serial log10 dilutions of the serum samples. The anti-HPV-16 VLP serum inhibited the detection of the E1^E4 mRNA at dilutions of ≤1:105, while the anti-HPV-33 VLP serum only neutralized HPV-16 at dilutions of ≤1:103 (Fig. 4).
FIG. 4.
Titration of the HPV-16-neutralizing activity in the anti (α)-HPV-16 VLP and the anti-HPV-33 VLP sera. HPV-16 stock diluted 1:104 was preincubated with serial log10 dilutions of anti-HPV-16 L1 VLP or anti-HPV-33 L1 VLP antiserum prior to addition to HaCaT cells. RT-PCR products obtained with HPV-16-specific primers are shown. Lanes are labeled with the reciprocal dilution of antiserum used in the experiment. Lane V represents RT-PCR product obtained with RNA isolated from cells infected with HPV-16 preincubated with a 1:100 dilution of normal serum. Lane C represents the RT-PCR product obtained with RNA from uninfected cells.
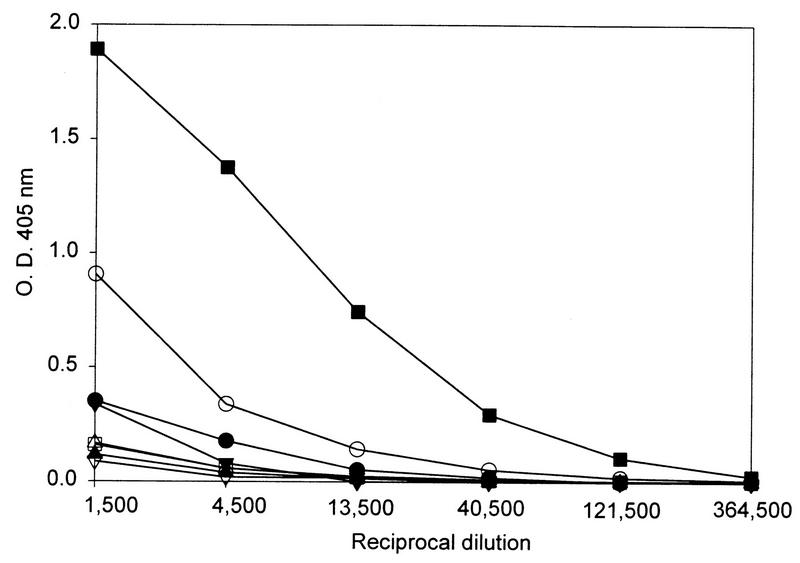
The differential neutralizing activity of the anti-HPV-16 VLP serum and the anti-HPV-33 VLP serum against HPV-16 was reflected in the relative ability of the two antisera to bind HPV-16 VLPs in an ELISA (Fig. 5). The anti-HPV-33 VLP serum exhibited binding to HPV-16 L1 VLPs, although to a much lesser extent than the homotypic anti-HPV-16 VLP serum. All of the remaining heterotypic antisera reacted very weakly with HPV-16 VLPs. Thus, with this panel of antisera, the relative ability to neutralize HPV-16 correlated with binding to HPV-16 L1 VLPs.
FIG. 5.
Reactivity of the anti-HPV VLP sera against HPV-16 L1 VLPs. ELISA plates were coated with HPV-16 VLPs and incubated with threefold serial dilutions of antisera to the following VLP types: HPV-11 (•), HPV-16 (▪), HPV-18 (▴), HPV-31 (▾), HPV-33 (○), HPV-35 (□), HPV-39 (▵), and HPV-45 (▿). Specific binding was detected with an anti-rabbit immunoglobulin G secondary antibody reagent as described in Materials and Methods. O.D., optical density.
DISCUSSION
We have developed an RT-PCR-based in vitro assay which can be used to study the early stages of HPV-16 infection and to examine antibody-mediated virus neutralization. The assay is relatively rapid and highly reproducible, and thus it is amenable to the evaluation of large numbers of serum samples for HPV-16-neutralizing antibodies. Since the assay is semiquantitative, it can be used to derive end point titers of HPV-16-neutralizing antibodies. The sensitivity of RT-PCR allows the HPV-16 stock to be used at relatively high dilutions (1:104 to 1:106), thereby conserving the virus, which is in limited supply. The amount of virus utilized in the assay is described as a viral stock dilution, since there is no methodology for determining the number of infectious HPV-16 particles. It is anticipated that a different viral stock would contain a different number of infectious particles and would require titration to maintain assay reproducibility.
The appearance of the spliced HPV-16-specific transcript in cells which had been cultured with the virus indicated that the initial stages of infection had been successfully accomplished: cell attachment and entry, uncoating, translocation, and transcription. This is an important consideration in studies of antibody-mediated virus neutralization, since it is not known which steps in the infection process are inhibited by antibodies. Roden et al. found that monoclonal antibodies to BPV-1 could neutralize BPV-1 either by inhibiting cell binding or by blocking a subsequent step in the infection pathway (29). Christensen et al. also reported antibody neutralization of BPV-1, CRPV, and HPV-11 at a step in the infectious cycle subsequent to virus attachment (10). Thus, the RT-PCR-based in vitro infectivity assay for HPV-16 represents a significant improvement over surrogate assays, such as VLP-mediated hemagglutination inhibition, which only detect antibodies that inhibit cell attachment and therefore may underestimate the virus-neutralizing potential of an antiserum sample (28).
Using the RT-PCR-based assay, we demonstrated that HPV-16 was neutralized by antibodies raised against HPV-16 L1 VLPs. This result was anticipated, since HPV-16 L1 VLPs had previously been shown to elicit production of antibodies which inhibited both HPV-16 L1 VLP-mediated hemagglutination and infection of cultured cells by HPV-16 pseudovirions (27, 28). However, neutralization of authentic HPV-16 virions lends further support to the potential application of VLPs as prophylactic HPV vaccines.
Whereas HPV-16 is the most prevalent high-risk HPV type, broad protection against cervical cancer would require that a vaccine target multiple HPV types (e.g., HPV-16, -18, -31, -33, and -45). Evaluation of the ability of different HPV VLPs to elicit production of cross-neutralizing antibodies is important in determining the ultimate composition of a broadly efficacious vaccine. The HPV-16 in vitro infectivity assay was used to test antisera against heterotypic HPV L1 VLPs for neutralizing activity against the virus. Antisera to the low-risk HPV-6 and -11 and the high-risk HPV-18, -31, -35, -39, and -45, each containing high titers of homotypic antibodies as measured by ELISA, all failed to neutralize HPV-16 infection. This result is in agreement with previous findings which demonstrated that antibody responses to the genital HPVs are largely type specific (27, 28, 30). In this regard, our observation that anti-HPV-33 L1 VLP antibodies neutralized HPV-16 is somewhat surprising. Previously, cross-neutralization had only been observed consistently with very closely related virus types, such as HPV-6 and -11, which possess L1 amino acid sequence identity of >90% (11). HPV-16 neutralization by antiserum raised against HPV-33 VLPs was not suggested by in vitro infectivity assays with HPV-16 pseudovirions (27). Weak cross-neutralization between HPV-16 and HPV-33 was seen in some hemagglutination assays with HPV-33 and -16 VLPs but was not consistently observed, thereby suggesting that detection of potential neutralizing activity was below the sensitivity of the assay (28). In contrast, by the RT-PCR-based assay, HPV-16 neutralization by anti-HPV-33 VLP antiserum was reproducible and titratable. Thus, the difference between our current result and previous results obtained by surrogate neutralization assays may indeed relate to differences in assay sensitivity. Alternatively, the discrepancy could be attributable to differences in the quantity and/or quality of the anti-HPV-33 VLP antibodies used in the different assays.
The amino acid sequence of HPV-16 L1 shares greater homology with HPV-31 L1 and HPV-35 L1 than with HPV-33 L1 (83.1, 82.8, and 79.7% amino acid sequence identity, respectively). However, amino acid sequence identity may not correlate strictly with structural similarity. It is well established that neutralizing antibodies raised against intact authentic HPV capsids and recombinant papillomavirus VLPs primarily recognize conformation-dependent epitopes on viral particles (12–15). Thus, the HPV-16-neutralizing capability of the anti-HPV-33 antibodies may reflect a greater degree of structural similarity between HPV-16 and HPV-33 than might be predicted by amino acid sequence comparisons.
Our current results suggest that HPV-33 and HPV-16 share a neutralizing epitope or epitopes. However, due to the lack of an HPV-33 stock, direct evaluation of the neutralizing activity of the HPV-33 and -16 L1 VLP antisera against HPV-33 was not possible. The anti-HPV-33 L1 VLP serum was not as potent as the anti-HPV-16 L1 VLP antiserum at neutralizing HPV-16, suggesting that HPV-33 and HPV-16 may possess both common and distinct neutralization sites similar to those previously reported for HPV-6 and HPV-11 (11). The ability of the anti-HPV-33 VLP serum to neutralize HPV-16 correlated with its ability to bind HPV-16 L1 VLPs in an ELISA. Interestingly, when the anti-HPV-16 L1 VLP serum was assayed for binding to HPV-33 L1 VLPs, no reactivity was detected (data not shown). Unckell et al. have reported neutralization of HPV-33 pseudovirions with an anti-HPV-33 VLP serum but not with an anti-HPV-16 VLP serum (38). Complete assessment of cross-neutralizing activities among the different genital HPV types will require the generation of additional infectious viral stocks and the development of the respective quantitative infectivity assays.
ACKNOWLEDGMENTS
This research was supported in part by a grant (NO1-AI-35159) from the National Institute for Allergy and Infectious Diseases to W.B. and a grant from the American Cancer Society (ACS IRG-18) to R.C.R.
REFERENCES
- 1.Bauer H M, Greer C E, Manos M M. Detection of genital human papillomavirus using PCR. In: Herrington C S, McGee J O, editors. Diagnostic molecular pathology: a practical approach. Oxford, United Kingdom: Oxford University Press; 1992. pp. 131–152. [Google Scholar]
- 2.Bonnez, W., C. Da Rin, C. Borkhuis, K. de Mesy Jensen, R. C. Reichman, and R. C. Rose. Isolation and propagation of human papillomavirus type 16 in human xenografts implanted in the severe combined immunodeficiency mouse. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 3.Bonnez W, Rose R C, Da Rin C, Borkhuis C, de Mesy Jensen K L, Reichman R C. Propagation of human papillomavirus type 11 in human xenografts using the severe combined immunodeficiency (SCID) mouse and comparison to the nude mouse model. Virology. 1993;197:455–458. doi: 10.1006/viro.1993.1611. [DOI] [PubMed] [Google Scholar]
- 4.Bonnez W, Rose R C, Reichman R C. Antibody-mediated neutralization of human papillomavirus type 11 (HPV-11) infection in the nude mouse: detection of HPV-11 mRNAs. J Infect Dis. 1992;165:376–380. doi: 10.1093/infdis/165.2.376. [DOI] [PubMed] [Google Scholar]
- 5.Bosch F X, Manos M M, Munoz N, Sherman M, Jansen A M, Petro J, Schifman M H, Moreno V, Kurman R, Shah K V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 6.Boukamp P, Petrussevska R T, Breikreutz D, Hornung J, Markham A, Fusenig N E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitburd F, Kirnbauer R, Hubbert N L, Nonnenmacher B, Trin-Dinh-Desmarquet C, Orth G, Schiller J T, Lowy D R. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J Virol. 1995;69:3959–3963. doi: 10.1128/jvi.69.6.3959-3963.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow L T, Nasseri M, Wolinsky S M, Broker T R. Human papillomavirus types 6 and 11 mRNAs from genital condylomata acuminata. J Virol. 1987;61:2581–2588. doi: 10.1128/jvi.61.8.2581-2588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow L T, Reilly S S, Broker T R, Taichman L B. Identification and mapping of human papillomavirus type 1 RNA transcripts recovered from plantar warts and infected epithelial cell cultures. J Virol. 1987;61:1913–1918. doi: 10.1128/jvi.61.6.1913-1918.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen N D, Cladel N M, Reed C A. Postattachment neutralization of papillomaviruses by monoclonal and polyclonal antibodies. Virology. 1995;207:136–142. doi: 10.1006/viro.1995.1059. [DOI] [PubMed] [Google Scholar]
- 11.Christensen N D, Reed C A, Cladel N M, Hall K, Leiserowitz G S. Monoclonal antibodies to HPV-6 L1 virus-like particles identify conformational and linear epitopes on HPV-11 in addition to type-specific epitopes on HPV-6. Virology. 1996;22:477–486. doi: 10.1006/viro.1996.0554. [DOI] [PubMed] [Google Scholar]
- 12.Christensen N D, Kreider J W. Antibody-mediated neutralization in vivo of infectious papillomavirus. J Virol. 1990;64:3151–3156. doi: 10.1128/jvi.64.7.3151-3156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen N D, Kreider J W. Neutralization of CRPV infectivity by monoclonal antibodies that identify conformational epitopes on intact virions. Virus Res. 1991;21:169–179. doi: 10.1016/0168-1702(91)90031-p. [DOI] [PubMed] [Google Scholar]
- 14.Christensen N D, Kreider J W, Cladel N M, Patrick S D, Welsh P A. Monoclonal antibody-mediated neutralization of infectious human papillomavirus type 11. J Virol. 1990;64:5678–5681. doi: 10.1128/jvi.64.11.5678-5681.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghim S, Christenen N D, Kreider J W, Jenson A B. Comparison of neutralization of BPV-1 infection of C127 cells and bovine fetal skin xenografts. Int J Cancer. 1991;49:285–289. doi: 10.1002/ijc.2910490224. [DOI] [PubMed] [Google Scholar]
- 16.Hagensee M E, Yaegashi N, Galloway D A. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J Virol. 1993;67:315–322. doi: 10.1128/jvi.67.1.315-322.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildesheim A, Schiffman M H, Gravitt P E, Glass A G, Greer C E, Zhang T, Scott D R, Rush B B, Lawler P, Sherman M E, Kurman R J, Manos M M. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- 18.Kirnbauer R, Booy F, Cheng N, Lowy D R, Schiller J T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirnbauer R, Chandrachud L M, O’Neil B W, Wagner E R, Grindlay G J, Armstrong A, McGarvie G M, Schiller J T, Lowy D R, Campo M S. Virus-like particles of bovine papillomavirus type 4 in prophylactic and therapeutic immunization. Virology. 1996;219:37–44. doi: 10.1006/viro.1996.0220. [DOI] [PubMed] [Google Scholar]
- 20.Kreider J W, Howett M K, Leure-Dupree A E, Zaino R J, Weber J A. Laboratory production in vivo of infectious human papillomavirus type 11. J Virol. 1987;61:590–593. doi: 10.1128/jvi.61.2.590-593.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manos M M, Ting Y, Lewis A J, Broker T R, Wolinsky S M. Use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells. 1989;7:209–214. [Google Scholar]
- 22.Manos M M, Waldman J, Zhang T Y, Greer C G, Eichinger M H, Schiffman M H, Wheeler C M. Epidemiology and partial nucleotide sequence of four novel genital human papillomaviruses. J Infect Dis. 1994;170:1096–1099. doi: 10.1093/infdis/170.5.1096. [DOI] [PubMed] [Google Scholar]
- 23.Meyer T, Arndt R, Stockfleth E, Flammann H T, Wolf H, Reischl U. Strategy for typing human papillomaviruses by RFLP analysis of PCR fragments and subsequent hybridization with a generic probe. BioTechniques. 1995;19:632–639. [PubMed] [Google Scholar]
- 24.Nasseri M, Hirochika R, Broker T R, Chow L T. A human papillomavirus type 11 transcript encoding an E1^E4 fusion protein. Virology. 1987;159:433–439. doi: 10.1016/0042-6822(87)90482-x. [DOI] [PubMed] [Google Scholar]
- 25.Palermo-Dilts D A, Broker T R, Chow L T. Human papillomavirus type 1 produces redundant as well as polycistronic mRNAs in plantar warts. J Virol. 1990;64:3144–3149. doi: 10.1128/jvi.64.6.3144-3149.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phelps W C, Alexander K A. Antiviral therapy for human papillomaviruses: rationale and prospects. Ann Intern Med. 1995;123:368–382. doi: 10.7326/0003-4819-123-5-199509010-00008. [DOI] [PubMed] [Google Scholar]
- 27.Roden R B S, Greenstone H L, Kirnbauer R, Booy F P, Jessie J, Lowy D R, Schiller J T. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J Virol. 1996;70:5875–5883. doi: 10.1128/jvi.70.9.5875-5883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roden R B S, Hubbert N L, Kirnbauer R, Christensen N D, Lowy D R, Schiller J T. Assessment of the serological relatedness of genital human papillomaviruses by hemagglutination inhibition. J Virol. 1996;70:3298–3301. doi: 10.1128/jvi.70.5.3298-3301.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roden R B S, Weissinger E M, Henderson D W, Booy F, Kirnbauer R, Mushinski J F, Lowy D R, Schiller J T. Neutralization of bovine papillomavirus by antibodies to L1 and L2 capsid proteins. J Virol. 1994;68:7570–7574. doi: 10.1128/jvi.68.11.7570-7574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose R C, Bonnez W, Da Rin C, McCance D J, Reichman R C. Serological differentiation of human papillomavirus types 11, 16 and 18 using recombinant virus-like particles. J Gen Virol. 1994;75:2445–2449. doi: 10.1099/0022-1317-75-9-2445. [DOI] [PubMed] [Google Scholar]
- 31.Rose R C, Bonnez W, Reichman R C, Garcea R L. Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J Virol. 1993;67:1936–1944. doi: 10.1128/jvi.67.4.1936-1944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seedorf K, Krämmer G, Dürst M, Suhai S, Röwekamp W G. Human papillomavirus type 16 DNA sequence. Virology. 1985;145:181–185. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- 33.Smith L H, Foster C, Hitchcock M E, Isseroff R. In vitro HPV-11 infection of human foreskin. J Invest Dermatol. 1993;101:292–295. doi: 10.1111/1523-1747.ep12365409. [DOI] [PubMed] [Google Scholar]
- 34.Smith L H, Foster C, Hitchcock M E, Leiserowitz G S, Hall K, Isseroff R, Christensen N D, Kreider J W. Titration of HPV-11 infectivity and antibody neutralization can be measured in vitro. J Invest Dermatol. 1995;105:1–7. doi: 10.1111/1523-1747.ep12321173. [DOI] [PubMed] [Google Scholar]
- 35.Suzich J S, Ghim S-J, Palmer-Hill F J, White W I, Tamura J K, Bell J A, Newsome J A, Bennett Jenson A, Schlegel R. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc Natl Acad Sci USA. 1995;92:11553–11557. doi: 10.1073/pnas.92.25.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka A, Noda T, Yajima H, Hatanaka M, Ito Y. Identification of a transforming gene of human papillomavirus type 16. J Virol. 1989;63:1465–1469. doi: 10.1128/jvi.63.3.1465-1469.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniguchi A, Yasumoto S. A major transcript of human papillomavirus type 16 in transformed NIH 3T3 cells contains polycistronic mRNA encoding E7, E5, and E1^E4 fusion gene. Virus Genes. 1990;3:221–233. doi: 10.1007/BF00393182. [DOI] [PubMed] [Google Scholar]
- 38.Unckell F, Streeck R E, Sapp M. Generation and neutralization of pseudovirions of human papillomavirus type 33. J Virol. 1997;71:2934–2939. doi: 10.1128/jvi.71.4.2934-2939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]