Abstract
A synthetic peptide, DP178, containing amino acids 127 to 162 of the human immunodeficiency virus type 1 (HIV-1) gp41 Env glycoprotein, is a potent inhibitor of virus infection and virus mediated cell-to-cell fusion (C. Wild, T. Greenwell, and T. Matthews, AIDS Res. Hum. Retroviruses 9:1051–1053, 1993). In an effort to understand the mechanism of action of this peptide, we derived resistant variants of HIV-1IIIB and NL4-3 by serial virus passage in the presence of increasing doses of the peptide. Sequence analysis of the resistant isolates suggested that a contiguous 3-amino-acid sequence within the amino-terminal heptad repeat motif of gp41 was associated with resistance. Site-directed mutagenesis studies confirmed this observation and indicated that changes in two of these three residues were necessary for development of the resistant phenotype. Direct binding of DP178 to recombinant protein and synthetic peptide analogs containing the wild-type and mutant heptad repeat sequences revealed a strong correlation between DP178 binding and the biological sensitivity of the corresponding virus isolates to DP178. The results are discussed from the standpoints of the mechanism of action of DP178 and recent crystallographic information for a core structure of the gp41 ectodomain.
The envelope glycoprotein of human immunodeficiency virus type 1 (HIV-1) is composed of two subunits, gp120 and gp41. These glycoproteins are proteolytic cleavage products of the gp160 envelope precursor and associate with each other through noncovalent interactions. During the course of viral infection, gp120 and gp41 are involved in the fusion process between membranes of HIV-1 and host cells expressing CD4 and certain coreceptors (2, 4, 13, 15–17). After receptor binding, structural rearrangements of the gp41 subunit are thought to play a role in membrane fusion (23, 31, 39, 40) in a poorly understood process which has often been compared to that used by the hemagglutinin HA2 protein of influenza (7, 9, 54).
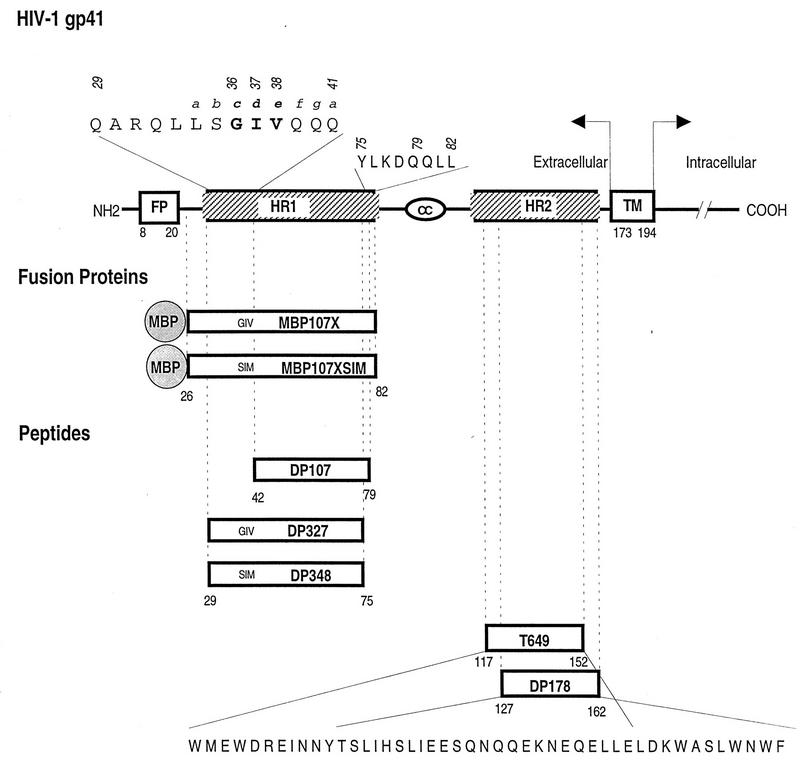
A model for the secondary structure of the HIV-1 gp41 glycoprotein has been described by Gallaher et al. (21). Several features of this model are shown in Fig. 1. The membrane-spanning or transmembrane domain (TM) has been mapped to residues 173 to 194 (the numbering is that used for the HXB2 viral clone [44]) (19) and divides gp41 into an ectodomain (extracellular) and an endodomain (intracellular). At the N terminus of gp41 is a hydrophobic fusion peptide sequence, FLGFLG, which has been proposed to act as an insertional sequence involved in the initial penetration of the target cell membrane (18, 20, 25). On the carboxyl side of the fusion peptide is a leucine zipper-like 4-3 repeat (14). This region is labeled heptad repeat 1 (HR1) in Fig. 1. A peptide, DP107, designed from this region (residues 42 to 79) was found to adopt a coiled-coil structure in solution and block HIV-1 infection (50). A second predicted helical region as described by the Gallaher model (21, 51) is adjacent to the membrane-spanning domain. This region is also predictive of a 4-3 repeat structure and is referred to as heptad repeat 2 (HR2) in Fig. 1. We have previously reported that a peptide, DP178, modeled from this region (residues 127 to 162) is a potent inhibitor of infection and virus-induced cell-to-cell fusion (48, 51). Antiviral activity has also been described for several overlapping peptides (26–28, 34, 35). In other experiments with several peptide and fusion protein models, evidence suggestive of interactions between HR1 and HR2 has been reported (11, 27, 49). Other recent studies have shown that two protein fragments overlapping the same regions associate with each other in an antiparallel fashion to form α-helical trimers of heterodimers (10, 28, 45–47).
FIG. 1.
Schematic representation of the HIV-1 gp41, recombinant proteins, and synthetic peptide reagents used in this study. The amino acids are represented by the one-letter code and numbered starting at the first amino acid of the HXB2 gp41. The positions of the amino acids according to a helical-wheel assignment are represented above the amino acid by an italicized lowercase letter. FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain.
In view of the apparent association of the HR1/HR2 peptide models, it has been speculated that the potent antifusion activity of DP178 might be mediated by binding to the HR1 sequence of gp41 (10, 11, 27, 28, 30, 49). In this report, we describe further experiments to test this possibility by using a genetic approach. Briefly, we derived HIV-1IIIB and NL4-3 progeny viruses which were resistant to the DP178 peptide. Mapping of the determinants involved was consistent with an HR1 site of action, and the results support the proposed association of the two heptad repeat regions in the ectodomain of gp41.
MATERIALS AND METHODS
Peptide synthesis.
Peptides were synthesized on an Applied Biosystems 431A peptide synthesizer by using Fast Moc chemistry and purified by reverse-phase high-pressure liquid chromatography (HPLC) (50). The peptides were acetylated at the N terminus and amidated at the C terminus, and their identities were confirmed by mass spectrometry.
Cells and parental viruses.
The human T-lymphoblastoid cell line CEM-4 was maintained in RPMI 1640 medium containing 10% fetal calf serum. HIV-1IIIB and the NL4-3 virus (derived by transfection of the infectious molecular clone pNL4-3 into CEM-4 cells) were used to generate the DP178-resistant viruses (178-14 and re4, respectively). HIV-1IIIB was obtained from R. Gallo (National Institutes of Health), and pNL4-3 was obtained through the AIDS Research and Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (1). HIV-1IIIB virus stock was prepared from acutely infected CEM-4 cells. NL4-3 virus stock was generated by transfection of CEM-4 with proviral DNA.
Transfection of proviral HIV-1 clones into CEM-4.
Proviral DNAs (3 μg) were incubated with 15 μl of Lipofectamine (GIBCO-BRL) in 200 μl of serum-free RPMI 1640 medium for 15 to 45 min at room temperature. About 2 × 106 CEM-4 cells were added to the DNA mixture to a final volume of 1 ml, and the mixture was incubated for 4 to 5 h at 37°C under 5% CO2. Complete RPMI 1640 containing 10% fetal calf serum was then added, and the cells were cultured at 37°C for 5 to 7 days. Supernatants of the transfected cells were monitored for virus production on the basis of reverse transcriptase (RT) activity.
Generation of DP178-resistant virus.
CEM-4 cells (106 cells) were infected with 20 50% tissue culture infective doses (TCID50) of cell-free HIV-1IIIB or NL4-3 virus stock derived from filtered culture supernatants. The virus was allowed to adsorb to 5 × 106 cells/ml for 1 h at 37°C. The culture was then diluted to 2 × 105 cells/ml in DP178-containing media at a final concentration of 100 ng/ml. Every other day, the cell concentration was adjusted to 2 × 105 cells/ml by the addition of peptide-containing medium. Samples of cell supernatant were collected every other day and evaluated for virus production on the basis of RT activity. Supernatants containing the highest level of RT activity were then used for subsequent rounds of infection by following the same protocol but with twofold-higher concentrations of DP178 at each passage of the virus (i.e., 200, 400, and 800 ng/ml). For the DP178-resistant virus pool, the infection was performed and maintained in the presence of DP178; the peptide was removed from the medium 48 h before the virus pool was harvested.
Virus infectivity assay.
Serial fourfold dilutions of each virus were incubated in duplicate with various concentrations of the DP178 peptide. The virus-peptide mixtures were then added to CEM-4 cells (final volume, 60 μl) in 96-well microtiter plates. Fresh peptide-free medium was added every other day. On day 8 postinfection, supernatants from each well were tested for the presence of RT activity as a criterion for successful infection. The infectious titer for each DP178-treated culture was calculated from the Reed and Muench formula (38) and compared to that for untreated cultures. The surviving virus fraction (Vn/V0) represents the TCID50 of peptide-treated virus (Vn), divided by the TCID50 of virus in the absence of peptide (V0). Multiplicity curves were generated by a plot of the surviving titer (Vn/V0) as a function of the peptide concentration.
Fusion assay.
About 104 CEM-4 cells chronically infected with HIV-1IIIB were mixed with 7 × 104 uninfected Molt4 cells in 96-well plates (half-area Costar cluster plates) in 100 μl of culture medium in the presence or absence of peptide. At 24 h later, multinucleated giant cells (syncytia) were estimated by microscopic examination at ×40 magnification. Uninhibited cultures yielded 90 to 120 syncytia per well for each virus tested.
RT assay.
The RT activity was determined as previously described (22, 52). Supernatants from cell culture were made 1% (vol/vol) Triton X-100. A 10-μl sample of supernatant was added to 50 μl of RT cocktail (75 mM KCl, 2 mM dithiothreitol, 5 mM MgCl2, 5 μg of poly(rA) per ml, 0.03 absorbance unit at 260 nm of oligo(dT)12–18 per ml, 0.05% Triton X-100, 50 mM Tris [pH 7.8], 10 μCi of [32P]dTTP) in a 96-well U-bottom microtiter plate and incubated at 37°C for 90 min. After the incubation, 40 μl of the reaction mixture was applied to a DE81 Whatman filter (96-well manifold [Schleicher & Schuell]) saturated with 2× SSC (0.3 M NaCl, 0.03 M sodium citrate). The filter was rinsed three times under vacuum and washed twice for 10 min in a tray containing 2× SSC. The filter was drained, and radioactivity was determined with a Matrix 9600 direct beta counter (Packard). In addition, the membrane was exposed to film overnight.
Cell lysates.
Cell pellets (107 cells) were incubated in lysis buffer (50 mM KCl, 10 mM Tris-HCl [pH 8], 2.5 mM MgCl2, 0.5% Tween 20, 0.5% Nonidet P-40, 150 mg of proteinase K per ml) at 37°C for 3 h and 100°C for 15 min. Debris was removed by centrifugation. The cell lysates were used in the PCRs.
DNA amplification, cloning, and sequencing.
Infected cell lysates (1/10 final dilution) were prepared as described above and mixed with 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 3.0 mM MgCl2, 0.1% Triton X-100, 200 μM each deoxynucleoside triphosphate, 40 ng each of HIV-1 env-specific primers 7198 (tggaggaggagatatgaggg) and 8302 (ctaccaagcctcctactatc), and 2.0 U of Taq DNA polymerase (Promega) in a final volume of 50 μl. The DNA was denatured for 6 min at 95°C and amplified by 35 cycles of PCR (95°C for 30 s, 52°C for 30 s, and 72°C for 4 min). The polymerization was concluded by a 10-min incubation at 72°C. The PCR product was visualized with ethidium bromide on a 2% Metaphor (FMC BioProducts) agarose gel. The DNA fragments were extracted from the agarose with phenol-chloroform, cut with BamHI and NheI, and substituted into pNL4-3 between the unique NheI and BamHI restriction sites. Four clones (pNL-178-3, pNL-178-4, pNL-178-6, and pNL-178-7) were obtained. The sequences of the inserted fragments were determined by using the dideoxy method.
Generation of the cloned mutant viruses.
The NL-GIV, NL-SIV, NL-GIM, NL-SIM, and NL-DIM cloned viruses were generated by site-directed mutagenesis of pNL4-3. Site-directed mutagenesis was performed with the Amersham Sculptor in vitro mutagenesis system kit. An HXB3 NheI-BamHI env fragment from HXB3 (nucleotides 6470 to 8118; a gift from B. Cullen) was subcloned into M13mp18. The single-stranded DNA was mutagenized in four separate reactions with four different oligonucleotides to generate the indicated amino acid changes. The resultant sequences of the insert DNAs were confirmed by dideoxy sequencing, and the fragments were substituted into pNL4-3.
Fusion protein construction, expression, and purification.
Fusion proteins containing a fragment of the HIV-1 gp41 region fused to the C terminus of the Escherichia coli maltose binding protein (MBP) were prepared as previously described (11, 42). To generate the MBP107X fusion protein, a fragment of the HXB3 env gene encoding amino acids 26 to 82 of gp41 was constructed by PCR amplification of plasmid pgTAT (29). The fragment was purified and cloned into the pMAL-p2 vector (New England BioLabs, Beverly, Mass.) at the XmnI and EcoRI sites immediately downstream of the malE gene, which encodes MBP. To generate the MBP107X-SIM protein, an analogous fragment was amplified from the plasmid pNL-SIM (described above) and similarly cloned into pMAL-p2. MBP107X-SIM differs from MBP107X at two residue positions: G36S and S38M. The MBP2* (MBP alone) was purchased from New England BioLabs. DNA sequences were confirmed by dideoxy sequencing. Recombinant proteins were expressed and purified as previously described (42).
Labeling of DP178.
DP178 peptide (30 μg) was labeled with 1 mCi of Na125I (Amersham) with Iodobeads (Pierce). The peptide was separated from the free Na125I by fractionation on a G10 column (Pharmacia). The fractions containing the peptide were collected, pooled, and diluted 100-fold in phosphate-buffered saline (PBS; GibcoBRL) containing 2% bovine serum albumin (BSA).
Binding assay.
A 100-μl volume of MBP fusion proteins or peptides at 2 μg/ml was incubated overnight in Immulon Removawell strips (Dynatech Laboratories, Inc.) at 4°C. The wells were blocked with 100 μl of 5% BSA in PBS for 1 h at 37°C. Serial dilutions of the 125I-DP178 were added to the wells in triplicate, and the wells were incubated for 2 h at 37°C. The wells were then washed three times with 2% BSA in PBS, and individual wells were counted in a gamma radiation counter.
RESULTS
Selection of viruses resistant to DP178.
HIV-1 isolates resistant to DP178 were derived by repeated passage of the uncloned HIV-1IIIB through the CEM-4 cell line in the presence of increasing concentrations of the peptide. We began the process with a DP178 concentration of 100 ng/ml, which is sufficient to reduce the wild-type HIV-1IIIB infectious titer by 95 to 98% (48, 51). About 20 infectious units of the virus was added to the host cells 1 h before the peptide was added. The peptide was maintained in the cultures by the addition of fresh peptide-containing medium every other day. Culture supernatants were routinely monitored for virus production on the basis of RT activity. Supernatants containing peak levels of RT activity were repassaged through the CEM-4 cells at a twofold-higher peptide concentration. After five such passages of virus over a period of 6 weeks, we obtained a virus population, referred to as 178-14, which was resistant to DP178 at a concentration of 1 μg/ml, about 10-fold greater than that required to completely inhibit the parental HIV-1IIIB.
DP178 resistance determinants contained within the envelope gene.
Direct sequence analysis of the envelope region of the resistant virus revealed several substitutions not previously reported for HIV-1IIIB or LAI clones. These changes were limited to the amino-terminal region of gp41. To test if the gp41 ectodomain of the envelope was involved in the resistance phenotype, a PCR fragment derived from the resistant 178-14 was substituted for the analogous region of pNL4-3. The fragment included Env sequences from the gp120 V3 through the gp41 intracellular domain (NheI to BamHI). Several proviral clones were obtained and transfected into CEM-4 cells. High-titer virus stocks were prepared, and one of the chimeric virus clones, NL178-3, was found to be resistant to DP178 at concentrations as high as 10 μg/ml (Table 1; Fig. 1). Sequence analysis of this clone showed an unusual sequence involving two substitutions at the amino terminus of the HR1 region of gp41. These substitutions were at positions 36 (glycine to serine, G36S) and 38 (valine to methionine, V38M) to yield an SIM sequence in place of the wild-type GIV sequence. As indicated in Table 1, several other clones derived from the 178-14 virus stock contained only one of these substitutions (G36S), and those clones exhibited intermediate sensitivity to the peptide, requiring 0.5 to 1 μg of DP178 per ml for a 10-fold reduction in virus titer. No other sequence differences were noted in the NheI-BamHI fragment contained within the resistant NL178-3 envelope in comparison to wild-type HIV-1IIIB clones.
TABLE 1.
Comparison of amino acid sequence and virus phenotype
| Viral clone | Amino acid sequencea | DP178 phenotype |
|---|---|---|
| HXB3 | Q L L S G I V Q Q Q | Sensitive |
| NL178-4/5/7 | S I V | Sensitive |
| NL178-3 | S I M | Resistant |
| NLGIV | G I V | Sensitive |
| NLSIV | S I V | Sensitive |
| NLGIM | G I M | Sensitive |
| NLSIM | S I M | Resistant |
| NLDIM | D I M | Resistant |
| re4 | D T V | Resistant |
| NL4-3 | Q L L S D I V Q Q Q | Sensitive |
The sequence from positions 32 to 41 is shown.
Resistance determinants mapped by site-directed mutagenesis.
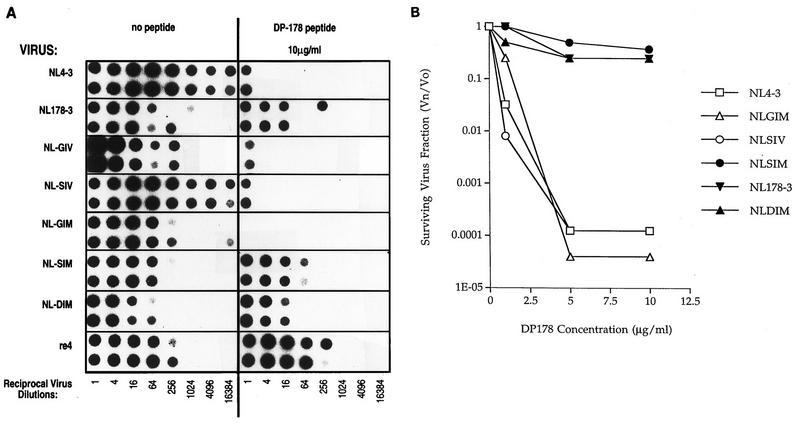
The above results suggested that two mutations in gp41 are involved in the DP178 resistance phenotype, and this possibility was tested by site-directed mutagenesis. A review of the envelope sequences in the Los Alamos database (33) showed that residues 36 to 38 are very highly conserved in that only three viruses with sequences other than GIV were noted in 196 sequences reported. These include BZ126A (EIV), CA1 (GIR), and the chimeric virus clone NL4-3 (DIV). It is somewhat surprising that NL4-3 contained a variant DIV (G36D) mutation, since this substitution has not been noted in other reported sequences for the HIV-1IIIB env donor of the pNL4-3 chimeric plasmid. The uncommon D36 residue does not render the NL4 virus resistant to the peptide, as indicated in Fig. 2. Nevertheless, for the purpose of the mutagenesis experiments, we preferred to base the site-directed changes on an envelope background that contained the more common GIV sequence. We therefore cloned into M13 for mutagenesis an NheI-BamHI fragment derived from HXB3, which is an infectious virus clone also derived from HIV-1IIIB and contains the normal GIV sequence. The mutagenized fragments were subcloned into pNL4-3. The following five NL4-3 mutant clones containing specific substitutions were constructed: wild-type pNL/HXB3 chimera (pNL-GIV), G36S (pNL-SIV), V38M (pNL-GIM), G36S and V38M (pNL-SIM), and G36D and V38M (pNL-DIM). In each case, virus was recovered from transfected CEM-4 cells and tested for susceptibility to the DP178 peptide. The data in Fig. 2A allows visualization by autoradiography of virus production from cells challenged with serial dilutions of virus on the basis of RT activity released to the supernatant of the cultures. In this experiment, each dilution of virus was added in duplicate to the host CEM-4 cells in the absence and presence of 10 μg of DP178 per ml. The titer of the NL4-3 and NL-SIV exceeded the greatest virus dilution tested (about 16,000) in the absence of peptide but was greatly reduced (to about 2) in the presence of this level of peptide. Thus, the relative reduction in infectious titer by the peptide for these isolates was about 10,000-fold. In contrast, the apparent virus titers shown in Fig. 2A for the NL178-3, SIM, and DIM virus clones were not substantially reduced by DP178 treatment. Figure 2B shows the results of a similar experiment but with the surviving titer for each virus plotted as a function of the peptide concentration. In both cases, the results are similar in that each of the G36 and V38 residues must be altered for complete resistance to DP178. At position 38, methionine is the only substitution that was noted. Position 36, however, was found to accommodate either the aspartic acid (as found in the wild-type pNL4-3 plasmid) or serine substitution (Table 1).
FIG. 2.
DP178 blockade of CEM-4 cell infection by NL4-3, NL178-3, NL-SIV, NL-GIM, NL-SIM, and NL-DIM viruses. Serial dilutions of the virus stocks were treated with no peptide (A), DP178 at 10 μg/ml (A), or DP178 at three concentrations (B) in duplicate. The cultures were incubated for 8 days, with fresh medium (without peptide) added every other day. On day 8, the medium was tested for the presence of RT activity as evidence of successful infection. The RT-associated radioactivity of each well was estimated by exposure of the filter to X-ray film (A). In another experiment (B), the radioactivity for each well was counted with a Matrix 9600. The counts were used to calculate the surviving virus fractions (Vn/V0 = TCID50 in the presence of peptide/TCID50 in absence of peptide) and are expressed as a function of the dose of DP178 (B).
Selection for a DP178-resistant NL4-3 virus.
The above sequences associated with resistance were derived from the uncloned HIV-1IIIB virus stock. To determine whether the same site might be involved in an independent selection beginning with an infectious molecular clone, we repeated the multiple-virus-passage strategy with NL4-3 and escalating doses of DP178. A peptide-resistant virus strain named re4 emerged after 5 weeks (Fig. 2A). As indicated above, the wild-type NL4-3 virus contains the unusual G36D substitution, so that the parental sequence for this selection contained a DIV sequence instead of the more common GIV residues. Sequence analysis of the resistant re4-derived virus following PCR revealed that the only change within the envelope region analyzed (V3 to the transmembrane domain of gp41) was an isoleucine-to-threonine change at the adjacent position 37 to yield a DTV sequence. It thus appeared that this particular site in the ectodomain of gp41 is particularly suited to mutations that yield both viable virus and resistance to the DP178 peptide.
Lack of DP178 binding to gp41 HR1 fragments containing substitutions that cause resistance.
We previously reported that the DP178 peptide binds to peptides and fusion proteins containing the HR1 region of gp41 and speculated that such an interaction might be related to its mechanism of action (11, 49). The resistant mutants described above, whose mutations also mapped to the HR1 domain, are consistent with this interpretation. To further test this hypothesis, we compared binding of the DP178 peptide to wild-type and escape-mutant forms of the HR1-containing peptide and protein models (Fig. 3 and 4).
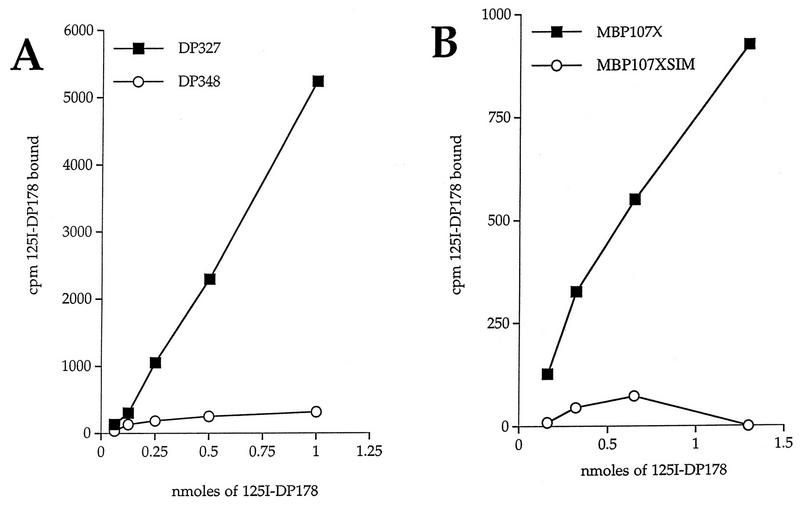
FIG. 3.
Direct binding of iodinated DP178 to HR1. Serial dilutions of 125I-DP178 at 106 cpm/ml were added to Removawells coated with either HR1-containing peptides (amino acids 29 to 75) or MBP fusion proteins (amino acids 26 to 82). (A) Binding of DP178 to DP327 (wild type) or to DP348 (SIM mutant). The background for each dilution was measured by performing the assay with a control peptide. (B) Binding of DP178 to MBP107X (wild type) or to MBP107XSIM (SIM mutant). The background for each dilution was measured by performing the assay with the control protein MBP2∗.
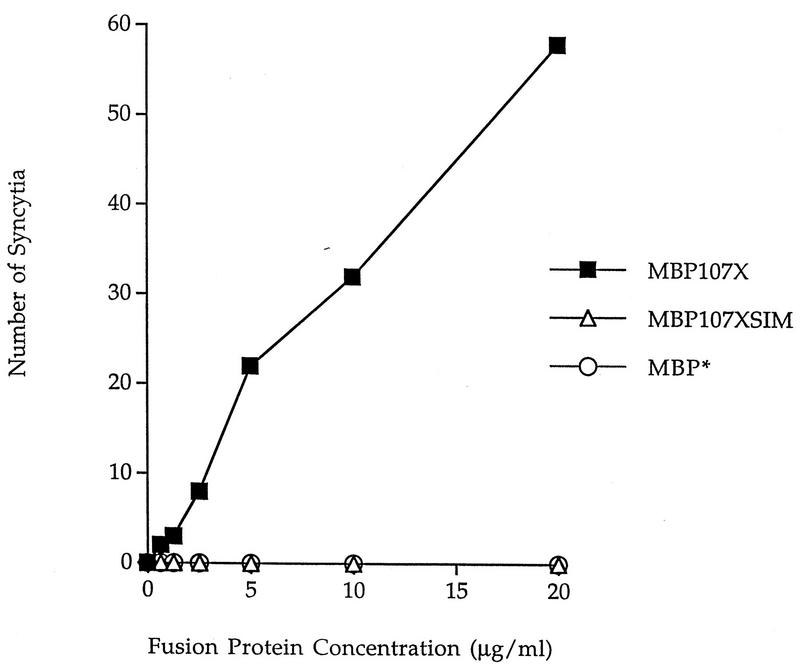
FIG. 4.
Abrogation of DP178 anti-cell fusion activity by coincubation with MBP107X but not MBP107XSIM. The assay for HIV-1-induced cell-to-cell fusion was carried out in the presence of 20 ng of DP178 per ml and the indicated concentrations of either MBP107X, MBP107XSIM, or MBP2∗. None of these MBPs affected syncytium formation when tested alone.
Two peptides were synthesized to include the HR1 envelope residues 29 to 75. One of these, labeled DP327, contained the wild-type GIV sequence, and the other, DP348, contained the SIM sequence associated with resistance (Fig. 1). Each of these peptides were coated on Immulon Removawells and tested for their ability to capture radioiodinated DP178. The results, shown in Fig. 3A, revealed a dose-dependent DP178 binding to the wild-type HR1 peptide, DP327, but only background binding to the DP348 peptide containing the SIM substitutions. The binding experiments were repeated with MBP fused to the HR1 domain (residues 26 to 82) (Fig. 1). As shown in Fig. 3B, binding of the iodinated DP178 again was apparent only with the wild-type GIV sequence, not with the SIM-containing fusion protein.
We tested the two MBP fusion proteins for their ability to abrogate inhibitory activity of DP178 in the cell-to-cell fusion assay (11). In this type of assay, both test components are in solution and differences cannot be ascribed to possible artifacts associated with the solid-phase binding experiments. The results comparing the wild-type and SIM variant fusion proteins are shown in Fig. 4. In this assay, about 80 syncytia were apparent in uninhibited cultures whereas no syncytia were detected in the presence of DP178 at 20 ng/ml. This concentration of DP178 is severalfold greater than the concentration required to completely block syncytium formation. In the experiment in Fig. 4, the DP178 concentration was held constant at 20 ng/ml while the HR1 fusion proteins were added to the same wells at increasing concentrations. Competition by the wild-type HR1 fusion protein, MBP107X, is evidenced by the increasing number of syncytia with increasing concentrations of the fusion protein. No competitive effect was noted with the mutant form of the fusion protein, MBP107XSIM. The results are in agreement with those of the direct-binding experiments (Fig. 3) and suggest that the fusion protein containing the parental but not the “escape mutant” HR1 sequence forms complexes with the DP178 peptide and prevents its antifusogenic activity.
Biological characteristics of wild-type and DP178-resistant viruses.
The above binding experiments suggest that the escape mutations noted in the HR1 domain impair binding of the DP178 peptide to the peptide-protein models of the HR1 domain. If this interaction also reflects an HR1-HR2 complex structure on the intact gp41, the resistant mutants might be expected to either contain compensatory changes in the DP178 HR2 domain or exhibit functional abnormalities indicative of a poor association between these two domains. As indicated above, we have not observed compensatory changes in the HR2 domain of any of the resistant mutants. Virus stability was evaluated on the basis of infectivity half-life estimates of virus preparations incubated at 37°C. Only minor differences were noted in that virus clones containing a substitution at the G36 position gave somewhat shorter half-life estimates (from 3.2 to 3.6 h) than did those with the wild-type glycine residue (about 6 h) at this position. On the basis of microscopic examination, the escape mutant isolates also did not exhibit obvious differences in their syncytium-forming capacities in the absence of peptide.
We also tested the resistant mutants for sensitivity to other inhibitory agents. These included soluble CD4, Leu3A antibody, several HIV-1-positive human sera, and the DP107 peptide (50). No significant differences between the parental and escape viruses were noted. Cell surface expression of the envelope glycoprotein was also probed by fluorescence-activated cell sorter analysis with HIV-1-positive sera and several monoclonal antibodies, including 2F5 (32) and Fab-d (11). Again, no differences were observed. We have previously reported that the Fab-d antibody binds a discontinuous epitope in gp41 whose formation is dependent on a complex between DP178 and a segment of the HR1 domain. Since the escape mutants bind this antibody, it is apparent that the HR2 region of the gp41 must still be able to associate with the mutant HR1 domain, even though the peptide models of these regions would not have predicted this result. Possible explanations for this inconsistency are discussed below.
DISCUSSION
The results of these experiments suggest that the site of action of the DP178 peptide is the HR1 region of the gp41 ectodomain. Each of the escape mutations described here, as well as several others in preliminary studies (38a) have mapped to the amino-terminal side of the HR1 domain as shown in Fig. 1 and Table 1. The substitutions noted directly affect DP178 binding, as evidenced by experiments with both fusion proteins and synthetic peptide models of the HR1 domain. These findings are consistent with a number of studies which demonstrate an association between the HR1 and HR2 domains to form a hetero-oligomeric complex (10, 28, 37, 46, 47). It has been argued that such an HR1-HR2 complex forms in the early stages of virus entry during structural rearrangements of the envelope to a fusion-competent form (10, 11, 28, 47). Presumably, binding of the inhibitory DP178 peptide interferes with the formation of the latter complex and traps the envelope in a fusion-inactive state (30).
Alternative targets and mechanisms of action for the DP178 peptide have been suggested. For example, Neurath et al. reported that DP178, as well as an overlapping peptide named SJ-2176, binds the fusion peptide sequence at the amino-terminal end of gp41 (34, 35). The fusion peptide used for those studies contained the N-terminal 24 amino acid residues of gp41. The GIV residues associated with escape from the antiviral effects of DP178 are in the predicted HR1 region at positions 36, 37, and 38, of gp41 and are not contained in the fusion peptide mimic studied in the latter report (34). None of the escape variants displayed substitutions in the fusion peptide sequence, and all of the DP178 escape mutants were also resistant to the SJ-2176 peptide (data not shown). These observations strongly argue in favor of the gp41 HR1 domain as the sensitive target for the antiviral effects of DP178. The same arguments, however, do not necessarily discount a possible binding of DP178 or SJ-2176 to the fusion peptide sequence as previously proposed (34). As indicated above, gp41 is thought to exist in at least two structural states (40), and binding of the HR2 peptide mimics to the fusion peptide sequence might reflect one of these structures while binding to the HR1 domain may represent the other. If that is the case, however, our results would suggest that it is the binding to the HR1 region that is associated with the antiviral effects of the peptide.
Other possible mechanisms of action of DP178 have included binding to and blockade of cell surface molecules, which may be used in the membrane fusion process. For example, several protein-peptide models of gp41 including a DP178-overlapping sequence have been reported to bind cell surface molecules (12, 36). Nevertheless, results presented here argue that the mechanism of action of DP178 is targeted directly at the virus and not at a putative cell surface component.
As discussed above, considerable experimental evidence suggests a high-affinity association between the two predictive heptad repeat regions of the gp41 ectodomain, HR1 and HR2 (Fig. 1). Most recently, this has been highlighted by the crystal structure for a complex formed between peptide-protein mimics of the HR1 and HR2 regions of gp41 (10, 46). In this structure, three HR1 peptides form a core superhelix and three HR2 peptides lie antiparallel within grooves created by the HR1 coiled-coil helix. Residues involved in formation of the central HR1 trimer were found in the expected a and d positions according to a classical helical-wheel assignment. Most contacts between the HR1 and HR2 heptad repeat peptides involved residues at the d and e or at the a and g positions of the inner HR1 trimer packed against residues a and d of the HR2 outer helix. The mutations found in the DP178-resistant viruses described in this report lie in the c, d, and e positions of the gp41 HR1 domain (Fig. 1). According to the crystal structure, these residues surround an asparagine (N145 according to the numbering in reference 46) in a d position of the HR2 (51a). It might be of interest that this particular asparagine has also been reported to be a critical residue for syncytium formation and virus infectivity (8).
The DP178-resistant mutants described in this report were derived from HIV-1IIIB and the related NL4-3 molecular clone. In each case, the resistance mutations mapped to the same GIV residues on the amino-terminal side of HR1. In preliminary experiments (data not shown), we have also derived a resistant variant of the primary isolate DH012 (41). Again, resistance was associated with an amino acid substitution at the same site to yield a GIG sequence in place of GIV. In general, the sequence of the HR1 region is highly conserved among HIV-1 isolates and mutations therein have commonly been found to disrupt envelope-mediated fusion events (8). It is now apparent that constraints on HR1 sequences include the requirements for formation of both the central triple helix and the attachment sites for the HR2 domain. It is possible that the GIV position is more variable and is one of the few HR1 sites that can be altered in a way that conserves both structure and function. However, the Los Alamos National Resource Laboratory Sequence Database (33) suggests that the GIV sequence is more constant than are other residues in this domain. We therefore conclude that the GIV-containing heptad is especially important for binding of the DP178 peptide. This idea is consistent with the results of binding studies of DP178 to various HR1-derived peptides and fusion proteins. For example, the DP107 peptide mimic of HR1 does not include the GIV-containing heptad and exhibits only low-affinity binding to DP178. These interactions can be visualized at the high peptide concentrations required for circular dichroism experiments (49) but not at the low peptide concentrations used in solid-phase binding experiments with radiolabeled peptides (data not shown). In contrast, HR1 peptides extended to the amino-terminal side of DP107 to include the GIV-containing heptad exhibit high-affinity binding in the solid-phase assays (Fig. 3). Thus, both the escape mutations and direct-binding experiments suggest that the GIV-containing heptad is a critical component of the high-affinity association of HR2-derived peptides with H1. This particular heptad also has an interesting influence on the structure of various HR1 peptide and protein models. Peptides and fusion proteins lacking this heptad exist as soluble helical oligomers that are best defined as tetrameric coiled coils (5, 27, 36, 42). Addition of HR2 peptide mimics such as DP178 tend to aggregate these otherwise well-ordered tetramers (49). In contrast, peptides and proteins extended to include the GIV-containing heptad, such as N51 (10), N36 (28), DP327, and MBP107X (Fig. 1), tend to form insoluble aggregates in the absence of HR2 peptides but form stable and well-ordered heteroduplex trimers in the presence of HR2 peptides (28). Thus, the inclusion of the GIV-containing heptad repeat in peptide models of the HR1 has a dramatic effect on both their solution structure and their ability to bind the HR2 peptide mimics. That the escape determinants map to the same heptad suggests that the site also plays a critical role in DP178 binding on the full-length envelope, not just the peptide and protein models of this region of the envelope glycoprotein.
One of the interesting features of the results reported here is the lack of compensatory changes in the envelope HR2 domain which encodes the DP178 sequence. There are several possible explanations for this observation. One is that mutations in the HR1 site which affect the binding of free HR2 peptides have a much reduced impact on binding the covalently attached HR2 site within gp41 because of the higher local concentration of the latter HR2 sequence. A second possible explanation is that other sites within HR1 play a more dominant role in the folding of the intact envelope glycoprotein. A candidate sequence, for example, has been highlighted recently (10, 46) as an especially deep cavity within the HR1 triplex. This site is at the opposite end (carboxy side) of the HR1 from the GIV site. Importantly, the region of the HR2 motif that “fits” into this deep cavity includes the WMEW sequence N-terminal to, and not contained within, the DP178 peptide. Consistent with this idea, we have found that the DP178-resistant viruses remain sensitive to an overlapping peptide, T649 (Fig. 1) (3), which contains the WMEW sequence (3a). This latter peptide exhibits potency similar to DP178 on prototypic virus isolates but is more active against primary isolates. Interestingly, it has been more difficult to derive virus resistant to the T649 peptide than to the DP178 peptide, although several such variants have been identified. Sequence analysis of the resistant viruses has revealed mutations within the HR1 sequence in the glutamine triad directly adjacent to and on the carboxyl side of the GIV site. No changes were noted at the C terminus of HR1, which contains the WMEW-accepting cavity. Although these observations remain to be confirmed by site-directed mutagenesis, the result highlights the importance of the GIV region of the HR1 domain in the binding of HR2 peptide mimics.
A mutation in the HR1 region of gp41 has also been associated with escape from neutralizing antibody (53). In this case, resistance was associated with distal conformational effects on gp120 rather than a direct effect on the antibody binding epitope. Again, the mutation had no obvious detrimental effect on virus growth; rather, the resultant virus was more cytopathic than the parental virus. In a subsequent study, these investigators reported that long-term culture of the antibody escape mutant yielded revertant virus which displayed a compensatory substitution in the HR2 domain of gp41 (43). We are also in the process of long-term culture of several of the DP178 variants described here but have so far not observed such compensatory changes.
In view of the antiviral potency of HR2-derived peptides such as DP178 and T649, these reagents are being considered as possible candidates in the treatment of HIV-1 infection (11, 34, 35, 48, 51). Experiments with the HuPBMC-SCID mouse model have demonstrated a DP178 antiviral effect in vivo (6), and the peptide is currently in phase I clinical trials (trial PHA043 TRI001) in humans (24). The results of the experiments described in this paper, i.e., the correlation between escape mutations in the gp41 HR1 and lack of peptide binding to HR1 analogs carrying these mutations, suggest that the mechanism of action of the inhibitory peptides is binding to the HR1 region and inhibition of HR1-HR2 complex formation within the gp41 protein. Small molecules with similar modes of action might also represent attractive drug candidates. The crystal structures which are now available for this hetero-oligomeric complex (10, 46), as well as the various HR1-HR2 binding assays described here and in earlier reports (11), might prove useful in the identification of novel drugs to treat HIV infection.
ACKNOWLEDGMENTS
This work was supported by NIH grants 2-RO1-AI30411-06A2 and 5-P30-AI28662.
pNL4-3 was obtained from Malcolm Martin through the AIDS Research and Reference Program, Division of AIDS, NIAID, NIH. We thank Dani Bolognesi, Celia C. LaBranche, and Don Wiley for helpful discussion and critical reading of the manuscript, and we thank Donna Davison, Susie Farmer, Teresa Greenwell, and Larry Stoltenberg for technical assistance. We thank Shawn Barney, Kelly Guthrie, and Dennis Lambert (Trimeris Inc.) for helpful discussions and for the gift of the T649 peptide.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5-A Rantes, Mip-1-Alpha, Mip-1-beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Barney S, Guthrie K, Davis-Rodhes D E, Medinas R, Bucy T, Hauser T, Erickson J, DiMassimo B, Venetta T, Lawless M K, Merutka G, Lambert D M. Synthetic peptides from conserved regions of HIV-1, HIV-2 and SIV fusion proteins are potent antivirals (inhibitors of viral fusion) 1996. p. A57. , abstr. 134. In 9th International Conference for Antiviral Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Barney, S., and L. T. Rimsky. Unpublished data.
- 4.Bates P. Chemokine receptors in HIV-1: an attractive pair. Cell. 1996;86:1–3. doi: 10.1016/s0092-8674(00)80070-7. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein H B, Tucker S P, Kar S R, Pherson S A M, Pherson D T M, Dubay J W, Lebowitch J, Compans R W, Hunter E. Oligomerization of the hydrophobic heptad repeat on gp41. J Virol. 1995;69:2745–2750. doi: 10.1128/jvi.69.5.2745-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black P L, Wood O, Broud D, Bacho M, Kunder S, Papermaster S, Lambert D, Barney S, Ussery M. XI International Conference on AIDS. 1996. T-20, a novel inhibitor of HIV-1 fusion, blocks recovery of infectious HIV-1 and inhibits viral load in vivo in the HuPBMC-SKID mouse model, abstr. TU.A.263; p. 221. [Google Scholar]
- 7.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of the influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 8.Cao J, Bergeron L, Helseth E, Thalis M, Repke H, Sodroski J. Effect of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr C M, Kim P S. A spring-loaded mechanism of the conformational change of influenza haemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 10.Chan D, Fass D, Berger J, Kim P. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen C, Matthews T J, McDanal C B, Bolognesi D P, Greenberg M L. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J Virol. 1995;69:3771–3777. doi: 10.1128/jvi.69.6.3771-3777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Dierich M P. Identification of a second site in the HIV-1 gp41 mediating binding to cells. Immunol Lett. 1996;52:153–156. doi: 10.1016/0165-2478(96)02603-x. [DOI] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, Rosa G L, Newman W, Girard N, Girard C, Sodroski J. The chemokine receptors CCR3 and CCR5 facilitate infection by primary isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Delwart E L, Mosialos G, Gilmore T. Retroviral envelope glycoproteins contain a leucine zipper-like repeat. AIDS Res Hum Retroviruses. 1990;6:703–706. doi: 10.1089/aid.1990.6.703. [DOI] [PubMed] [Google Scholar]
- 15.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Dimarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 16.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y X, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4(+) cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 Entry cofactor—functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 18.Freed E, Meyers D, Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc Natl Acad Sci USA. 1990;87:4650–4654. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabuzda D, Olshevsky U, Bertani P, Haseltine W A, Sodroski J. Identification of membrane anchorage domains of the HIV-1 gp160 envelope glycoprotein precursor. J Acquired Immune Defic Syndr. 1991;4:34–40. [PubMed] [Google Scholar]
- 20.Gallaher W R. Detection of a fusion peptide sequence in the transmembrane protein of the human immunodeficiency virus. Cell. 1987;80:327–328. doi: 10.1016/0092-8674(87)90485-5. [DOI] [PubMed] [Google Scholar]
- 21.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 22.Goff S, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hart T K, Kirsh R, Hellens H, Sweet R W, Lambert D M, Petteway S R, Leary J, Bugelski P. CD4 HIV-1 interactions: binding of soluble CD4 (sT4) to HIV-1 and HIV-1 infected cells induces shedding of envelope gp120. Proc Natl Acad Sci USA. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopkins, S., and M. Saag. Personal communication.
- 25.Kowalski M, Bergeron L, Dorfman T, Haseltine W, Sodroski J. Attenuation of human immunodeficiency virus type 1 cytopathic effect by mutations affecting the transmembrane glycoprotein. J Virol. 1991;65:281–291. doi: 10.1128/jvi.65.1.281-291.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert D M, Barney S, Lambert A L, Guthrie K, Medinas R, Davis D E, Bucy T, Erickson J, Merutka G, Petteway S R. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawless M K, Barney S, Guthrie K, Bucy T B, Petteway Jr S R, Merutka G. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry. 1996;35:13697–13708. doi: 10.1021/bi9606962. [DOI] [PubMed] [Google Scholar]
- 28.Lu M, Blacklow S C, Kim P S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 29.Malim M H, Hauber J, Fenrick R, Cullen B R. Immunodeficiency virus rev transactivator modulates the expression of the viral regulatory genes. Nature. 1988;331:181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- 30.Matthews T J, Wild C, Chen C, Bolognesi D P, Greenberg M L. Structural rearrangement in the transmembrane glycoprotein after receptor binding. Immunol Rev. 1994;140:93–104. doi: 10.1111/j.1600-065x.1994.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 31.Moore J P, Keating R A M, Weiss R A, Sattenteau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 32.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Rüker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers G, Korber B, Hahn B H, Jeang K, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N. Human retroviruses and AIDS. Los Alamos, N.M: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1995. [Google Scholar]
- 34.Neurath A R, Lin K, Strick N, Jiang S. Two partially overlapping antiviral peptides from the external portion of HIV-1 gp41, adjoining the transmembrane region, affect the glycoprotein 41 fusion domain. AIDS Res Hum Retroviruses. 1994;11:189–190. doi: 10.1089/aid.1995.11.189. [DOI] [PubMed] [Google Scholar]
- 35.Neurath A R, Strick N, Jiang S. Synthetic peptides and antipeptide antibodies as probes to study interdomain interactions involved in virus assembly: the envelope of the human immunodeficiency virus (HIV-1) Virology. 1992;188:1–13. doi: 10.1016/0042-6822(92)90729-9. [DOI] [PubMed] [Google Scholar]
- 36.Rabenstein M, Shin Y K. A peptide from the heptad repeat of human immunodeficiency virus gp41 shows both membrane binding and coiled-coil formation. Biochemistry. 1995;34:13390–13397. doi: 10.1021/bi00041a016. [DOI] [PubMed] [Google Scholar]
- 37.Rabenstein M D, Shin Y. HIV-1 gp41 tertiary structure studied by EPR Spectroscopy. Biochemistry. 1996;35:13922–13928. doi: 10.1021/bi961743t. [DOI] [PubMed] [Google Scholar]
- 38.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 38a.Rimsky, L. T., and S. Barney. Unpublished data.
- 39.Sattentau Q J. CD4 activation of HIV-1 fusion. Int J Cell Cloning. 1992;10:323–332. doi: 10.1002/stem.5530100603. [DOI] [PubMed] [Google Scholar]
- 40.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing macrophage/T cell line tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shugars D C, Wild C T, Greenwell T K, Matthews T J. Biophysical characterization of recombinant proteins expressing the leucine zipper-like domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1996;70:2982–2991. doi: 10.1128/jvi.70.5.2982-2991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern T L, Reitz M S, Robert-Guroff M. Spontaneous reversion of human immunodeficiency virus type 1 neutralization-resistant variant HXB2thr582: in vitro selection against cytopathicity highlights gp120-gp41 interactive regions. J Virol. 1995;69:1860–1867. doi: 10.1128/jvi.69.3.1860-1867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- 45.Weissenhorn W, Calder L J, Dessen A, Laue T, Skehel J, Wiley D C. Assembly of a rod-shaped chimera of a trimeric GCN4 zipper and the HIV-1 gp41 ectodomain expressed in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6065–6069. doi: 10.1073/pnas.94.12.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weissenhorn W, Dessen A, Harrisson S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 47.Weissenhorn W, Wharton S A, Calder L J, Earl P L, Moss B, Aliprandis E, Skehel J J, Wiley D C. The ectodomain of HIV-1 env subunit gp41 forms a soluble, alpha-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 1996;15:1507–1514. [PMC free article] [PubMed] [Google Scholar]
- 48.Wild C, Greenwell T, Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus mediated cell-cell fusion. AIDS Res Hum Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 49.Wild C, Greenwell T, Shugars D, Rimsky-Clarke L, Matthews T. The inhibitory activity of an HIV type 1 peptide correlates with its ability to interact with a leucine zipper structure. AIDS Res Hum Retroviruses. 1995;11:323–325. doi: 10.1089/aid.1995.11.323. [DOI] [PubMed] [Google Scholar]
- 50.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wild C, Shugars D, Greenwell T, McDanal C, Matthews T. Peptides corresponding to a predictive alpha helical domain of HIV-1gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a.Wiley, D. Personal communication.
- 52.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson C, Reitz M S, Aldrich K, Klasse P J, Bomberg J, Gallo R C, Robert-Guroff M. The site of an immune-selected point mutation in the transmembrane protein of the human immunodeficiency virus type 1 does not constitute the neutralization epitope. J Virol. 1990;64:3240–3248. doi: 10.1128/jvi.64.7.3240-3248.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Y G, King D S, Shin Y. Insertion of a coiled-coil peptide from influenza virus haemagglutinin into membranes. Science. 1994;266:274–276. doi: 10.1126/science.7939662. [DOI] [PubMed] [Google Scholar]