Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV) gene transcription in the BC-1 cell line (KSHV and Epstein-Barr virus coinfected) was examined by using Northern analysis with DNA probes extending across the viral genome except for a 3-kb unclonable rightmost region. Three broad classes of viral gene transcription have been identified. Class I genes, such as those encoding the v-cyclin, latency-associated nuclear antigen, and v-FLIP, are constitutively transcribed under standard growth conditions, are unaffected by tetradecanoylphorbol acetate (TPA) induction, and presumably represent latent viral transcripts. Class II genes are primarily clustered in nonconserved regions of the genome and include small polyadenylated RNAs (T0.7 and T1.1) as well as most of the virus-encoded cytokines and signal transduction genes. Class II genes are transcribed without TPA treatment but are induced to higher transcription levels by TPA treatment. Class III genes are primarily structural and replication genes that are transcribed only following TPA treatment and are presumably responsible for lytic virion production. These results indicate that BC-1 cells have detectable transcription of a number of KSHV genes, particularly nonconserved genes involved in cellular signal transduction and regulation, during noninduced (latent) virus culture.
Kaposi’s sarcoma-associated herpesvirus (KSHV; also designated human herpesvirus 8) is the most recently identified human herpesvirus (6, 7) and is associated with Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL; also known as body cavity-based lymphoma [BCBL]) (3), and a subset of multicentric Castleman’s disease (34). Epidemiologic studies of KSHV relying on PCR detection of specific viral DNA in lesions and tissues as well as subsequent serological assays suggest that this virus largely fulfills Hill’s criteria for causation in KS (26).
KSHV can be directly cultured to high copy number in PEL-derived cell lines, yet the efficiency of transmission to other cell lines and serial propagation of the virus remain low (9, 21). The first reported and characterized KSHV cell line, designated BC-1 (HBL-6), is coinfected with Epstein-Barr virus (EBV) (4, 10). This cell line exhibits a clonal immunoglobulin heavy-chain rearrangement characteristic of B-cell origin and contains an average of 40 to 60 KSHV genome copies per cell (4). Similar to some Burkitt’s lymphoma-derived cell lines, the EBV LMP1 and EBNA-2 genes responsible for lymphoblastoid immortalization are not expressed in BC-1 (21). However, unlike EBV-infected Burkitt’s lymphoma cell lines (16), c-myc rearrangements are not present in either the cell line or the parental tumor (10). Other cell lines derived from PEL have been subsequently established; some of these are EBV negative, indicating that EBV coinfection is not necessary for in vitro cultivation of KSHV (1, 2, 29, 31).
The entire nucleotide sequence, except for a 3-kb unclonable region, of the KSHV genome in the BC-1 cell line was recently determined (30). It consists of a double-stranded long unique DNA of approximately 140.5 kb flanked by high-G+C terminal repeat (TR) units. Pulsed-field gel electrophoresis analysis of encapsidated DNA from KSHV particles derived from an EBV-negative BCBL-1 cell line demonstrated that the viral genome is approximately 165 kb long (28, 29); however, the genome of the KSHV strain infecting BC-1 cells is approximately 270 kb in size. The larger size estimate for the KSHV genome in the BC-1 cell line is due to a duplication of a coding segment which is inserted into the TR region (30). While KSHV genome transmission from BC-1 cells has been detected by PCR analysis (17, 21), a formal demonstration that this strain is infection competent has not been achieved. In line with this observation, TR analysis of KSHV in BC-1 is consistent with the virus being under tight latent replication control under standard BC-1 culture conditions (30). Both BC-1 and HBL-6 (4, 10) cell lines were independently established from the same parental tumor and have identical TR polymorphic patterns, suggesting monoclonal expansion of the virus in these cell lines. Treatment of BC-1 cells with either sodium butyrate or 12-O-tetradecanoylphorbol-13-acetate (TPA) induces viral gene expression characteristic for late lytic infection phase (18). No indication for significant viral late gene expression is present in BC-1 in the absence of chemical treatment, providing additional evidence for tight latency control of KSHV in this cell line (18). Other cell lines, such as BCBL-1 and BCP-1, may contain a minority cell population undergoing spontaneous lytic replication, thus complicating the search for genes expressed during virus latency.
Sequence analyses for KSHV from the BC-1 cell line (30) as well as from a KS lesion (22) demonstrate that a large portion of the KSHV genome, represented by blocks of genes encoding viral replication and structural proteins, is conserved among herpesviruses (5, 21, 24, 30). These areas demonstrate colinear homology with two gammaherpesviruses with transforming potential, EBV and herpesvirus saimiri (HVS). In between the conserved gene blocks, divergent regions contain a number of unique viral proteins, some of which can mimic cell cycle regulation and signal transduction proteins (30). No KSHV genes with sequence homology to the EBV genes implicated in cell immortalization and oncogenesis (such as EBNA-1, -2, and -3, LMP1 and LMP2, or gp350/220) (15) or the two genes implicated in HVS transformation (STP and TIP) (13, 14) were detected by sequence analysis (30). This does not exclude the possibility that unique KSHV oncoproteins with distinct amino acid composition and structure exist.
Transcription of herpesvirus genes is generally tightly regulated and takes place sequentially. Herpesvirus gene transcription typically dichotomizes into a latent phase which occurs while the viral genome is maintained in an episomal form and a lytic phase which takes place in a cascade fashion during productive (lytic) infection. Treatment of BC-1 cells with TPA induces lytic EBV replication which is reflected by an eightfold increase in EBV DNA, whereas only a 1.3- to 1.4-fold increase in KSHV DNA takes place (21). Butyrate treatment preferentially increases KSHV DNA replication over that of EBV in BC-1 (18). In contrast, other PEL/BCBL cell lines demonstrate a 15-fold increase in KSHV DNA following TPA treatment and have detectable virion production (29), suggesting that the KSHV strain in BC-1 may be replication defective. Nevertheless, comparative Northern analysis of mRNA extracted from BC-1 and TPA-induced BC-1 indicates that treatment with TPA induces extensive viral transcription which likely corresponds to the active lytic phase of the viral life cycle.
Zhong et al. (38) previously performed a KSHV gene transcription survey using hybridization of viral genomic fragments with radiolabeled cDNA probes derived from KS tissue as well as from a cell line infected with KSHV alone (BCBL-1). This method provides an important initial screen for viral gene transcription; however, it has limited sensitivity allowing only the detection of highly transcribed mRNAs and does not provide information on mRNA size or alternative transcription patterns. Two highly abundant transcripts, T0.7 and T1.1, were identified by this method and subsequently characterized by Northern analysis.
To provide a map of KSHV actively transcribed regions in the BC-1 cell line, we surveyed gene transcription by using Northern hybridization. This map may prove useful to investigators as an initial transcriptional characterization of the KSHV genome. The primary aim of this study was to identify potential coding sequences expressed during latency which will allow subsequent finer mapping and characterization studies. The BC-1 cell line is ideal for transcription studies in BCBL since KSHV is under tight latent control in standard growth conditions (18). While BC-1 is EBV coinfected, which may affect KSHV gene transcription, this type of coinfection occurs naturally in most BCBL tumors and hence reflects the biologic status of the virus in vivo. Transcripts which are found in the BC-1 cell line under standard growth conditions are likely to encode latency-associated proteins which may be important in maintaining the latent viral genome and/or transforming virus-infected cells.
MATERIALS AND METHODS
Cell cultures.
BC-1 (4) and P3HR1 (obtained from the American Type Culture Collection) cells were grown at 37°C in RPMI 1640 (GibcoBRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (GibcoBRL) in the presence of 5% CO2. To induce lytic gene transcription, cells were exposed to TPA (20 ng/ml; Sigma Chemical Co., St. Louis, Mo.) and harvested after 48 h.
Northern analysis.
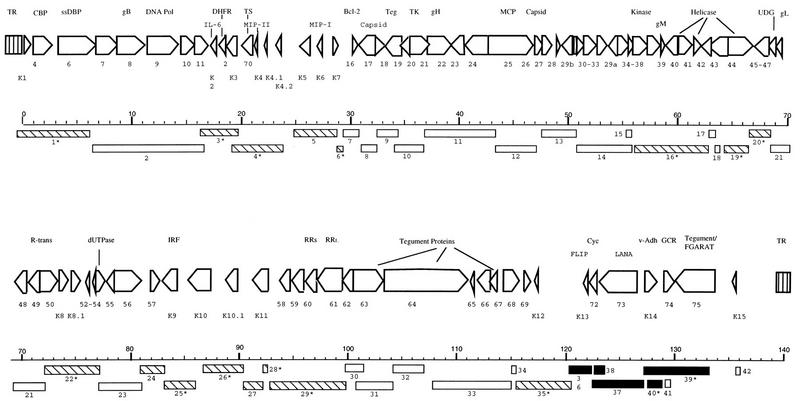
Total RNA was extracted by the RNAzol method (Tel-Test, Friendswood, Tex.) followed by mRNA selection using a PolyATract mRNA isolation kit (Promega, Madison, Wis.). Five hundred nanograms of the poly(A)-selected mRNA was loaded per lane on formaldehyde 1% agarose gel and transferred onto nylon membranes (GeneScreen; NEN Research Products, Boston, Mass.). Probes extending over the cloned genome (derived from plasmid subclones and PCR products) were labeled by random priming using synthetic hexanucleotide primers (RediPrime DNA labeling system; Amersham International, Amersham, England) and [32P]dCTP. The positions of the probes are shown in Fig. 1, and their precise locations are listed in Table 1. Hybridization was performed in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–50% formamide–5× Denhardt’s solution–2% sodium dodecyl sulfate–10% dextran sulfate–100 μg of denatured sheared salmon sperm DNA per ml at 42°C. β-Actin probe was used as a standard for the amount of the RNA loaded.
FIG. 1.
Locations of KSHV Northern hybridization probes. Arrow boxes above the kilobase marker indicate identified ORFs along with their designated numbers and putative functions. The boxes below the kilobase marker indicate DNA probes (1 to 42) that were used for Northern analysis. Dark boxes represent probes detecting class I transcription, hatched boxes represent probes detecting class II transcription, and empty boxes represent probes detecting class III transcription. Asterisks indicate probes which hybridize with more than one transcription category. Abbreviations: TR, terminal repeat; CBP, complement binding protein; ssDBP, single-stranded binding protein; gB, glycoprotein B; Pol, polymerase; IL-6, interleukin-6; DHFR, dihydrofolate reductase; TS, thymidylate synthase; MIP, macrophage inflammatory protein; Teg, tegument protein; TK, thymidine kinase; MCP, major capsid protein; UDG, uracil DNA glucosidase; IRF, interferon regulatory factor; RRS, ribonucleotide reductase, small; RRL, ribonucleotide reductase, large; FLIP, FLICE-inhibitory protein; Cyc, cyclin; LANA, latency-associated nuclear antigen; Adh, adhesion molecule; GCR, G-protein-coupled receptor; FGARAT, N-formylglycinamide ribotide amidotransferase.
TABLE 1.
Transcription mapping of KSHV in BC-1 cells with and without TPA
| Probe no. | ORFs encoded on probe | Position (bp) | Probe size (kb) | mRNAs (kb) identified in BC-1a
|
Transcription class | |
|---|---|---|---|---|---|---|
| Without TPA induction | With TPA induction | |||||
| 1 | K1,4 (v-CBP), 6 | TR (366 bp) | 6.715 | 6.0+ | 6.0 | II |
| 1–6349 | 4.0 | III | ||||
| 3.0 | III | |||||
| 1.8 | III | |||||
| 1.0 | III | |||||
| 2 | 7, 8, 9, 10, 11 | 6900–16600 | 9.7 | − | 7.0 | III |
| 6.0 | III | |||||
| 2.5 | III | |||||
| 1.0 | III | |||||
| 3 | 11, K2 (v-IL-6), 2 (DHFR), K3 (IEI) | 16200–19700 | 3.5 | 8.0 | III | |
| 6.0 | III | |||||
| 3.1 | III | |||||
| 2.0 | III | |||||
| 1.0 ++ (K2[v-IL-6]) | 1.0 | II | ||||
| 4 | K3 (IE1), 70 (TS), | 19130–23906 | 4.776 | 4.0 | III | |
| K4 (v-MIP-II), K4.1 | 2.5 | III | ||||
| 2.3+ | 2.3 | II | ||||
| 2.0 | III | |||||
| 1.5 | III | |||||
| 0.8 ++ (K4 [vMIP-II]) | 0.8 | II | ||||
| 5 | K5 (IE1), K6 (v-MIP-I), | 24839–28888 | 4.049 | 4.0 + | 4.0 | II |
| K7 (nut-1) | 1.3 + | 1.3 | II | |||
| 1.1 +++ (T1.1/nut1) | 1.1 | II | ||||
| 6 | K7 (nut-1) | 28889–29676 | 0.787 | 9.0 | III | |
| 1.1 ++++ (T1.1/nut-1) | 1.1 | II | ||||
| 7 | 16 (v-Bc12), 17 | 29677–30840 | 1.163 | − | 9.0 | III |
| 7.0 | III | |||||
| 3.5 | III | |||||
| 2.0 | III | |||||
| 1.0 | III | |||||
| 8 | 17 | 30841–32361 | 1.52 | − | 7.0 | III |
| 6.0 | III | |||||
| 3.5 | III | |||||
| 2.0 | III | |||||
| 1.0 | III | |||||
| 9 | 17, 18, 19 | 32362–34401 | 2.039 | − | 7.0 | III |
| 6.0 | III | |||||
| 1.0 | III | |||||
| 10 | 19, 20, 21 | 34021–36882 | 2.861 | − | 7.0 | III |
| 6.0 | III | |||||
| 11 | 21, 22, 23, 24, 25 | 36883–43171 | 6.288 | − | 7.0 | III |
| 6.0 | III | |||||
| 3.0 | III | |||||
| 12 | 25, 26 | 43172–47193 | 4.021 | − | 7.0 | III |
| 2.0 | III | |||||
| 13 | 26, 27, 28, 29b, 30 | 47517–50637 | 3.12 | − | 7.0 | III |
| 2.0 | III | |||||
| 14 | 30, 31, 32, 33, 29a, 34, 35 | 50638–55722 | 5.084 | − | 8.0 | III |
| 6.0 | III | |||||
| 3.0 | III | |||||
| 15 | 34, 35 | 55269–55819 | 0.55 | − | 4.5 | III |
| 3.0 | III | |||||
| 1.1 | III | |||||
| 16 | 36, 37, 38, 39, 40, 41, 42 | 55820–62901 | 7.081 | 4.5 | III | |
| 3.0 | III | |||||
| 2.5 + | 2.5 | II | ||||
| 1.4 | III | |||||
| 0.5 | III | |||||
| 17 | 42, 43 | 62902–63403 | 0.501 | − | 9.0 | III |
| 6.0 | III | |||||
| 4.5 | III | |||||
| 3.0 | III | |||||
| 2.5 | III | |||||
| 1.4 | III | |||||
| 18 | 43 | 63404–63793 | 0.389 | − | 9.0 | III |
| 4.5 | III | |||||
| 3.0 | III | |||||
| 19 | 43, 44 | 64037–66502 | 2.465 | 6.0 + | 6.0 | II |
| 4.5 | III | |||||
| 3.0 | III | |||||
| 2.7 | III | |||||
| 20 | 44, 45 | 66503–68324 | 1.821 | 9.0 | III | |
| 4.0 | III | |||||
| 2.5+ | 2.5 | II | ||||
| 1.5 +++ | 1.5 | II | ||||
| 21 | 45, 46, 47, 48, 49 | 68325–72191 | 3.866 | − | 8.0 | III |
| 2.5 | III | |||||
| 1.5 | III | |||||
| 0.8 | III | |||||
| 22 | 49, 50, K8, K8.1, 52 | 72192–77087 | 4.895 | 7.0 | III | |
| 4.0 | III | |||||
| 1.8 + | 1.8 | II | ||||
| 1.5 + | 1.5 | II | ||||
| 23 | 52, 53, 54, 55, 56 | 77088–81046 | 3.958 | − | 3.0 | III |
| 2.0 | III | |||||
| 1.3 | III | |||||
| 24 | 56, 57 | 81047–83278 | 2.231 | 2.0 + | 2.0 | II |
| 25 | 57, K9 (v-IRF1), K10 | 83279–86279 | 3.0 | 2.0 | III | |
| 1.5 ++ (v-IRF) | 1.5 | II | ||||
| 26 | K10 | 86879–90158 | 3.279 | 3.0 | III | |
| 2.0 + | 2.0 | III | ||||
| 1.5 | III | |||||
| 27 | K10.1, K11 | 90159–92110 | 1.951 | 2.0 + | 2.0 | II |
| 28 | K11 | 92111–92468 | 0.357 | 4.5 | III | |
| 3.0 | III | |||||
| 2.0 + | 2.0 | II | ||||
| 29 | K11, 58, 59 60, 61 | 92665–99864 | 7.199 | 6.0 | III | |
| 3.0 | III | |||||
| 2.0 + | 2.0 | II | ||||
| 30 | 61, 62, 63 | 99865–101232 | 1.367 | − | 2.0 | III |
| 1.7 | III | |||||
| 31 | 62, 63, 64 | 100784–104075 | 3.291 | − | 2.0 | III |
| 1.7 | III | |||||
| 32 | 64 | 104076–107008 | 2.932 | − | 5.0 | III |
| 3.0 | III | |||||
| 2.0 | III | |||||
| 1.7 | III | |||||
| 1.2 | III | |||||
| 33 | 64, 65, 66, 67, 67.5, 68 | 107940–115051 | 7.111 | − | 7.0 | III |
| 5.0 | III | |||||
| 4.0 | III | |||||
| 2.5 | III | |||||
| 1.2 | III | |||||
| 34 | 68 | 115052–115707 | 0.655 | − | 9.0 | III |
| 7.0 | III | |||||
| 6.0 | III | |||||
| 4.0 | III | |||||
| 35 | 68, 69, K12 (kaposin) | 115708–120725 | 5.017 | 4.0 | III | |
| 0.7 ++++ (T0.7/kaposin) | 0.7 | II | ||||
| 36 | K13 (v-FLIP) | 120601–122590 | 1.989 | 6.0 + (v-FLIP, v-Cyc, LANA) | 6.0 | I |
| 2.0 + (v-FLIP, v-Cyc) | 2.0 | I | ||||
| 37 | K13 (v-FLIP), | 122591–127392 | 4.801 | 6.0 ++ (v-FLIP, v-Cyc, LANA) | 6.0 | I |
| 72 (v-Cyc), 73 (LANA) | 2.0 ++ (v-FLIP, v-Cyc) | 2.0 | I | |||
| 38 | 72 (v-Cyc) | 122792–123565 | 0.773 | 6.0 ++ (v-FLIP, v-Cyc, LANA) | 6.0 | I |
| 2.0 ++ (v-FLIP, v-Cyc) | 2.0 | I | ||||
| 39 | K14, 74 (V-GCR), 75 | 127393–133381 | 5.988 | 6.0 + (v-FLIP, v,Cyc, LANA) | 6.0 | I |
| 4.5 +++ | 4.5 | I | ||||
| 3.0 | III | |||||
| 2.5 | III | |||||
| 2.0 + (v-FLIP, v-Cyc) | 2.0 | I | ||||
| 40 | K14 (v-Adh) | 127882–128928 | 1.046 | 4.5 + | 4.5 | I |
| 2.5 | III | |||||
| 1.5 | III | |||||
| 1.2 | III | |||||
| 41 | 74 (v-GCR) | 129513–129985 | 0.472 | − | 9.0 | III |
| 2.5 | III | |||||
| 42 | K15 | 135976–136278 | 0.302 | − | 9.0 | III |
| 6.0 | III | |||||
Specific genes identified with specific sequence probes are indicated in paretheses. Relative transcription in BC-1 without TPA: −, none; +, low; ++, moderate; +++, high; ++++, very high. For abbreviations, see the legend to Fig. 1.
RESULTS
Because signal intensity may vary between probes, depending on factors such as probe length, transcript length, exposure time, and specific activity, we have provided a semiquantitative estimate of relative probe hybridization only for genes expressed without TPA induction. Each gel assayed Northern hybridization for BC-1 under TPA-induced and noninduced conditions as well as P3HR1, to control for these factors for each probe. Whereas probe conditions may account in part for the variation in hybridization intensities between probes, this technique allowed accurate estimation of relative TPA-induced versus noninduced intensity of transcript hybridization for genes encompassed by any given probe. Because of the large numbers of bands present after BC-1 TPA induction, no attempt was made to semiquantitate relative intensities of TPA-induced transcripts. No hybridization signals were detected for P3HR1 mRNA, with or without TPA treatment, using any of the KSHV probes, indicating no apparent cross-hybridization to P3HR1-encoded EBV transcripts. Since the EBV strain in P3HR1 is deleted for EBNA-2 and partially deleted for EBNA-LP (15), cross-hybridization to this region of EBV cannot be excluded although KSHV does not encode sequence homologs to these EBV genes.
KSHV gene transcription with and without TPA.
In agreement with the findings of Renne et al. and Zhong et al. for the BCBL-1 cell line (29, 38) and viral protein analysis using BC-1 (18), Northern analysis indicates that under standard growth conditions, BC-1 cells exhibit a relatively restricted pattern of gene transcription. As indicated in Table 1, genes which are transcribed in BC-1 without TPA induction include the 1.1- and 0.7-kb polyadenylated mRNA transcripts (T1.1 and T0.7) (29, 36, 38, 39) as well as 6.0- and 2.0-kb transcripts which represent alternative transcripts encoding open reading frame (ORF) K13 (v-FLIP [see the legend to Fig. 1 for abbreviations]) and ORF 72 (v-cyclin) (27, 33). The 6.0-kb transcript additionally encodes ORF 73 (LANA), which is spliced out in the 2.0-kb transcript (33). We designate the 6.0- and the 2.0-kb transcripts as latent transcript 1 (LT1) and LT2, respectively. These results confirm immunoblot studies of LANA expression which demonstrate that this protein is not TPA inducible and not phosphonoacetic acid inhibitable and is thus likely to be expressed during viral latency (12). A 4.5-kb transcript (LT3) encoded by a region corresponding to the ORF K14 and ORF 75 genes is also transcribed without TPA treatment.
In contrast to the studies by Renne et al. and Zhong et al. (29, 38), additional transcripts are detectable without TPA induction by the Northern hybridization. These include the 1.0-kb ORF K2 (v-IL-6), 0.8-kb ORF K4 (v-MIP-II), and 1.5-kb ORF K9 (v-IRF) transcripts (19, 25) encoding genes which are homologous to cellular cytokine and intracellular signal transduction proteins. Additional transcripts expressed without TPA induction were detected with probes encoding ORFs K1, 4, and 6 (probe 1), ORFs K3, 70, K4, and K4.1 (probe 4), K5, K6, and K7 (probe 5), ORFs 36 to 42 (probe 16), ORFs 43 and 44 (probe 19), ORFs 44 and 45 (probe 20), ORFs 49, 50, K8, K8.1, and 52 (probe 22), ORFs 56 and 57 (probe 24), ORF K10 (probe 26), ORFs K10.1 and 11 (probe 27), ORF K11 (probe 28), ORFs K11, 58, 59, 60, and 61 (probe 29), ORFs K14, 74, and 75 (probe 39), and ORF K14 (probe 40). All of these transcripts demonstrated increased hybridization intensities after TPA treatment (Table 1). The majority of the transcripts which are expressed during presumed viral latency, but are inducible during lytic phase gene transcription, lie in nonconserved regions of the genome (Fig. 1). Several bands which may span multiple probe sequences and/or are polycistronic were identified. These potentially include a 2.0-kb transcript detected by probes 26 to 29 and a 4.5-kb transcript detected by probes 39 and 40. The present study cannot resolve this question, which will require high-resolution transcript mapping of start and stop sites for these mRNAs.
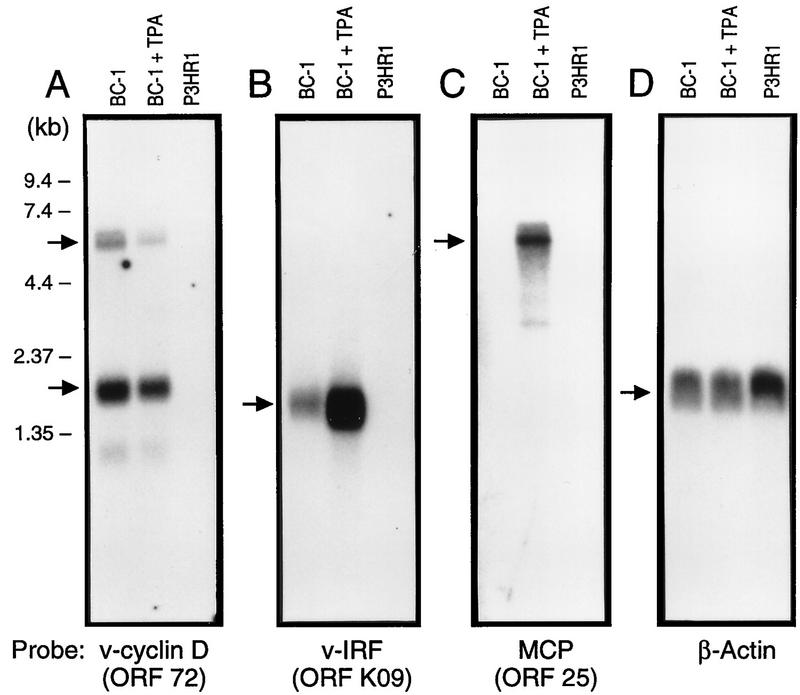
Most transcripts which are only detectable after TPA induction lie in regions which are conserved among herpesviruses and encode structural or lytic replication-related genes. Probes 11 and 12, for example, encompass the 7.0-kb ORF 25 (major capsid protein [MCP]) transcript which is only detectable after TPA induction (Fig. 2C).
FIG. 2.
Northern hybridization of mRNA from BC-1, TPA-induced BC-1, and P3HR1 cell lines demonstrating the three classes of viral gene transcription. mRNA (500 ng per lane) was separated on 1% agarose-formaldehyde gel and transferred to nylon filters. The filters were hybridized with four DNA probes: (A) v-cyclin D, which represents class I transcription; (B) v-IRF, which represents class II transcription; (C) MCP, which represents class III transcription; and (D) β-actin to control for RNA integrity and equivalent loading.
DISCUSSION
The pattern of gene transcription found in BC-1 cells can be divided into three classes of gene transcription: I, II, and III. Class I (Fig. 2A) includes mRNAs that are detected in the BC-1 cell line under standard growth conditions and are not induced by TPA. This does not exclude the possibility that TPA decreases class I gene transcription, since cellular gene transcription may be inhibited by TPA treatment in KSHV-infected cells (16a, 18) and our blots were standardized for equivalent amounts of mRNA. The relative abundance of KSHV class I transcripts compared to cellular transcripts, however, does not appear to be affected by TPA treatment.
Only three class I transcripts were identified, all of which are encoded on the right side of the genome. Two overlapping transcripts, LT1 and LT2, encode 6.0- and 2.0-kb mRNAs which are detected by probes 36 to 39. Both transcripts originate at the same initiation site within the probe 39 region at nucleotide 127870 (33). ORF 72- and ORF K13-specific probes (36 and 37) hybridize with both LT1 and LT2, suggesting polycistronic (i.e., multigenic transcripts) transcription of both genes. An ORF 73 probe hybridizes only to the 6.0-kb (LT1) transcript, and subsequent cDNA mapping studies show that LT2 originates as a spliced product which excises the ORF 73 gene (33). All three ORFs have corresponding sequence homologs in HVS (23, 37). ORF K13 (122710 to 122144) encodes a homolog of viral inhibitor of Fas-mediated apoptosis (37). ORF 72 (123566 to 122793) encodes a functional cyclin D homolog that can substitute for human cyclin D by phosphorylating the retinoblastoma tumor suppressor protein (8). ORF 73 (127296 to 123808) encodes LANA (27), a highly immunogenic protein that is expressed in PEL-derived cell lines and is being used in immunofluorescence and Western assay-based serological tests (12). The third class I transcript (LT3) is an mRNA of 4.5 kb detected with probes 39 and 40. Unlike LT1 and LT2, LT3 has not been characterized. The ORF 74-specific probe (41) hybridizes only with TPA-induced transcripts of 9.0 and 2.5 kb, indicating that ORF 74 is not encoded by LT3; however, it remains to be seen whether ORF K14 (v-Adh) or ORF 75 (FGARAT) is encoded by LT3. The fact that the abundance of the above transcripts is not affected by TPA treatment supports the notion that they represent true latent-phase mRNAs. These mRNAs were not detected previously in BCBL-1 (29, 38), probably due to a limited sensitivity of the method used.
The second class of gene transcription, class II (Fig. 2B), includes mRNAs which are detected in variable abundance (high, moderate, and low) in the BC-1 cell line under standard growth conditions and are induced to higher levels of transcription by TPA. The most highly abundant mRNAs in this class were previously found in the BCBL-1 cell line and are designated T1.1 (nuclear transcript 1 [nut-1]) and T0.7 (ORF K12 [kaposin]) (38) (probe 6, [1.1-kb mRNA] and probe 35 [0.7-kb mRNA], respectively). T1.1 (28622 to 29002) localizes to high-molecular-weight ribonucleoprotein complexes and may function in modulating RNA splicing events (39). This transcript was detected in 0.5 to 1% of KSHV-infected cells in KS lesions by in situ hybridization and colocalizes with ORF 25 (MCP) (35). T0.7 (118101 to 117919) appears to encode a 60-amino-acid membrane protein (38) and was found by in situ hybridization to be expressed in the majority of the KS spindle cells (35). mRNAs which are transcribed at moderate levels without TPA treatment in class II include the cytokines v-IL-6 (ORF K2; probe 3), v-MIP-II (ORF K4; probe 4), and v-IRF (ORF K9; probe 25) (19, 25). The sizes of these transcripts are 1.0, 0.8, and 1.5 kb, respectively. Similarly, probe 20 hybridizes with 1.5- and 2.0-kb mRNAs from BC-1 cells which are still induced by TPA. It should be noted that class II transcripts, which appear to be expressed during both latent and lytic virus replication cycles, tend to be clustered among the nonconserved regions of the genome which encode signal transduction and regulatory protein homologs. Transcripts which demonstrate low levels of expression in this class are listed in Table 1.
The third class of transcripts, class III (Fig. 2C), includes mRNAs that are detected in BC-1 only following TPA induction. These transcripts most likely encode lytic genes which are transcribed during active infection and are necessary for efficient viral replication and virion particle production. Some examples include transcripts of the ORF 25 (MCP; probe 12), ORF 6 (DNA polymerase; probe 2), and ORF 21 (gH; probe 11). TPA may also induce EBV-encoded genes, such as lytic transactivator proteins, which may affect KSHV gene transcription. KSHV gene transcription in the BC-1 cell line may be affected by EBV gene expression, and thus these results may not be the same for PEL/BCBL cell lines infected with KSHV alone. Our results are consistent with those of Lagunoff and Ganem, who showed that the ORF K1 gene behaves as a class III gene in that 1.35- and 3-kb transcripts which hybridize with the ORF K1 probe are induced only after TPA treatment (16a). While we did not localize expression in this region, these transcripts are probably identical to the 1.8- and 3-kb class III transcripts found with probe 1 (Table 1). The high abundance of conserved structural and metabolic gene transcripts after TPA induction and the inability to detect them in the absence of TPA treatment provide support for the notion that KSHV is under tight latent control in BC-1 under standard growth conditions.
The present study was designed to provide a preliminary survey of potential coding sequences for latently associated mRNAs in PEL that can help direct more detailed expression studies. Our results suggest that true latent transcription is restricted in PEL as in KS lesions. However, a number of genes are expressed, generally at low transcription levels, in PEL without TPA treatment and are inducible with TPA (class II). Interestingly, this category of transcription includes many unique KSHV genes (e.g., the viral cytokines and v-IRF) which may play a role in abrogating cellular pathways to control viral infection and in cell transformation (11, 20).
Several important caveats to this study should be mentioned. Very low levels of expression may not have been detected under our conditions; we have previously described low level transcription of ORF16 (v-Bcl2) in BC-1 (32), and thus this gene may fall into our class II transcription category on more detailed examination. Further, the fact that transcription of class II mRNAs is induced by TPA raises the possibility that these are not authentic latent transcripts; rather, they may represent highly abundant lytic-phase mRNAs and/or relatively stable mRNAs which accumulate in a minor subpopulation of the infected cells undergoing spontaneous lytic reactivation in culture. However, phosphonoformic acid treatment to inhibit DNA polymerase activity did not affect abundance of ORF K2 (v-IL-6) or ORF K9 (v-IRF) class II transcripts in either TPA-treated or untreated cells (data not shown), implying that detection of these class II transcripts in TPA-uninduced cells is not caused by late lytic mRNA expression in a minority population of cells. The inability to detect transcription of conserved late genes such as the major capsid protein, a marker for late lytic infection (probes 11 and 12), and the fact that class II transcripts are largely expressed from nonconserved regions which do not encode structural genes argue against this interpretation. It is more likely that class II transcripts represent constitutively expressed mRNAs which are induced to higher levels during the active phase of the infection. It is not likely that KSHV subgenomic duplication in BC-1 results in a double gene dosage or loss of regulatory control allowing expression of class II transcripts during latency since most of the genes in class II (e.g., ORF K2 [v-IL-6]) are not part of the BC-1 genomic duplication. Finally, another caveat is that alternative splicing events may occur in which transcription of the TPA-induced class II transcripts results from more than one promoter: one which is constitutive and allows gene expression in standard growth condition and another which is TPA inducible. Finer mapping of those transcripts may validate this possibility.
This study provides a working map of KSHV gene transcription in BC-1 during presumed latency and lytic replication which may prove useful for future investigations on the regulation of individual genes. This mapping study provides a more sensitive assessment of KSHV transcription than previously determined by using cDNA hybridization, allowing us to identify additional genes transcribed in the absence of TPA induction. These genes may play a role in maintaining the latent KSHV episome and may contribute to KSHV-related pathogenesis.
ACKNOWLEDGMENTS
We thank G. Evans and M. Davis for help in preparation of the manuscript.
This work was supported by Public Health Service grant CA-68939 from the National Cancer Institute and a James S. McDonnell Foundation award.
REFERENCES
- 1.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 2.Boshoff, C., S.-J. Gao, L. Healy, S. Matthews, A. Thomas, R. Warnke, J. Strauchen, E. Matutes, R. A. Weiss, P. S. Moore, O. W. Kamel, and Y. Chang. In vivo characterization of KSHV positive primary effusion lymphoma (PEL) cells. Submitted for publication.
- 3.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences are present in AIDS-related body cavity based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 4.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two AIDS-related lymphoma cell lines containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 5.Cesarman E, Nador R G, Bai F, Bohenzky R A, Russo J J, Moore P S, Chang Y, Knowles D M. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;265:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y, Moore P S. Kaposi’s sarcoma (KS)-associated herpesvirus and its role in KS. Infect Agents Dis. 1996;5:215–222. [PubMed] [Google Scholar]
- 8.Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden-Kent D, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 9.Foreman K E, Friborg J J, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi’s sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 10.Gaidano G, Cechova K, Chang Y, Moore P S, Knowles D M, Dalla-Favera R. Establishment of AIDS-related lymphoma cell lines from lymphomatous effusions. Leukemia. 1996;10:1237–1240. [PubMed] [Google Scholar]
- 11.Gao S-J, Boshoff C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (v-IRF) is an oncogene that inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1986. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 12.Gao S-J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion of antibodies to Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens prior to onset of Kaposi’s sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 13.Jung J U, Desrosiers R C. Identification and characterization of the herpesvirus saimiri oncoprotein STP-C488. J Virol. 1991;65:6953–6960. doi: 10.1128/jvi.65.12.6953-6960.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung J U, Trimble J J, King N W, Biesinger B, Fleckenstein B W, Desrosiers R C. Identification of transforming genes of subgroup A and C strains of Herpesvirus saimiri. Proc Natl Acad Sci USA. 1991;88:7051–7055. doi: 10.1073/pnas.88.16.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieff E, Liebowitz D. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, editors. Virology. Vol. 2. New York, N.Y: Raven Press; 1990. pp. 1889–1920. [Google Scholar]
- 16.Klein G. Epstein-Barr virus strategy in normal and neoplastic B cells. Cell. 1994;77:791–793. doi: 10.1016/0092-8674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 16a.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 17.Mesri E A, Cesarman E, Arvanitakis L, Rafii S, Moore M A S, Posnett D N, Knowles D M, Asch A S. Human herpesvirus-8/Kaposi’s sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J Exp Med. 1996;183:2385–2390. doi: 10.1084/jem.183.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 20.Moore, P. S., and Y. Chang. KSHV-encoded oncogenes and oncogenesis. J. Natl. Cancer Inst., in press. [DOI] [PubMed]
- 21.Moore P S, Gao S-J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, McGeoch D J, Pellett P, Chang Y. Primary characterization of a herpesvirus-like agent associated with Kaposi’s sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholas J, Cameron K R, Honess R W. Herpesvirus saimiri encodes homologues of G protein-coupled receptors and cyclins. Nature. 1992;355:362–365. doi: 10.1038/355362a0. [DOI] [PubMed] [Google Scholar]
- 24.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo H G, Reitz M S, Hayward G S. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H G, Hayward G S, Reitz M S. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 26.Olsen, S. J., and P. S. Moore. Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV8) and the etiology of KS. In H. Friedman, P. Medveczky, and M. Bendinelli (ed.), Molecular immunology of herpesviruses. Molecular immunology of herpesviruses, in press. Plenum Publishing, New York, N.Y.
- 27.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington S, Moore P S, Schulz T F. The 226- to 234-kilodalton latent nuclear antigen (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 30.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Said W, Chien K, Takeuchi S, Tasaka T, Asou H, Cho S K, Cesarman V S de, E, Knowles D M, Koeffler H P. Kaposi’s sarcoma-associated herpesvirus (KSHV or HHV8) in primary effusion lymphoma: ultrastructural demonstration of herpesvirus in lymphoma cells. Blood. 1996;87:4937–4943. [PubMed] [Google Scholar]
- 32.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi’s sarcoma-associated herpesvirus encodes a functional Bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 33.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1997. Unpublished data.
- 34.Soulier J, Grollet L, Oskenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M-F, Clauvel J-P, Raphael M, Degos L, Sigaux F. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 35.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun R, Lin S-F, Gradoville L, Miller G. Polyadenylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thome M, Schneider P, Hofman K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J-L, Schröter M, Scaffidl C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 38.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong W D, Ganem D. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:1207–1212. doi: 10.1128/jvi.71.2.1207-1212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]