Abstract
Sequence analysis of the right variable genomic region of the pathogenic African swine fever virus (ASFV) isolate E70 revealed a novel gene, UK, that is immediately upstream from the previously described ASFV virulence-associated gene NL-S (L. Zsak, Z. Lu, G. F. Kutish, J. G. Neilan, and D. L. Rock, J. Virol. 70:8865–8871, 1996). UK, transcriptionally oriented toward the right end of the genome, predicts a protein of 96 amino acids with a molecular mass of 10.7 kDa. Searches of genetic databases did not find significant similarity between UK and other known genes. Sequence analysis of the UK genes from several pathogenic ASFVs from Europe, the Caribbean, and Africa demonstrated that this gene was highly conserved among diverse pathogenic isolates, including those from both tick and pig sources. Polyclonal antibodies raised against the UK protein specifically precipitated a 15-kDa protein from ASFV-infected macrophage cell cultures as early as 2 h postinfection. A recombinant UK gene deletion mutant, ΔUK, and its revertant, UK-R, were constructed from the E70 isolate to study gene function. Although deletion of UK did not affect the growth characteristics of the virus in macrophage cell cultures, ΔUK exhibited reduced virulence in infected pigs. While mortality among parental E70- or UK-R-infected animals was 100%, all ΔUK-infected pigs survived infection. Fever responses were comparable in E70-, UK-R-, and ΔUK-infected groups; however, ΔUK-infected animals exhibited significant, 100- to 1,000-fold, reductions in viremia titers. These data indicate that the highly conserved UK gene of ASFV, while being nonessential for growth in macrophages in vitro, is an important viral virulence determinant for domestic pigs.
African swine fever (ASF) is a highly lethal and economically significant disease of domestic pigs for which there is no vaccine or disease control strategy other than animal quarantine and slaughter. The causative agent of ASF, a large enveloped double-stranded DNA virus (ASFV), is the sole member of an unnamed family of animal viruses (7, 10, 16). Although the icosahedral morphology of the ASFV virion resembles those of iridoviruses, both the ASFV genomic organization, which includes terminal cross-links and inverted terminal repeats, and the cytoplasmic replication strategy indicate a close relationship to the Poxviridae (20, 35, 41).
ASFV is the only known DNA arbovirus (7, 10, 16). In nature, the perpetuation and transmission of this virus involve the cycling of virus between two highly adapted hosts, Ornithodoros ticks and wild pig populations (warthogs and bushpigs) in sub-Saharan Africa (37, 38, 50, 54). In the warthog host, ASFV infection is subclinical, characterized by low viremia titers (39, 49).
In domestic pigs the severity of ASF ranges from a highly lethal hemorrhagic disease to subclinical infection, depending on contributing viral and host factors (9, 31, 39). ASFV infects cells of the mononuclear-phagocytic system, including highly differentiated fixed-tissue macrophages and specific lineages of reticular cells; affected tissues show extensive damage after infection with highly virulent viral strains (9, 26, 27, 31, 32). This ability to replicate and induce marked cytopathology in these cell types in vivo appears to be critical for ASFV virulence. The natures of viral and host factors responsible for the differing outcomes of infection with strains of high virulence and of lesser virulence are largely unknown.
High degrees of variation in genomic size and restriction pattern are observed among different ASFV isolates. Like poxviruses, variations within the ASFV genome are primarily localized to the terminal regions (4, 5, 53). Poxvirus genes located in the terminal variable regions are often nonessential for viral replication in cell culture, performing instead functions related to viral host range (30). ASFV terminal variable regions comprise the left 35-kb and the right 15-kb ends of the genome and contain at least five multigene families (MGF): MGF100, MGF110, MGF300, MGF360, and MGF530 (2, 12, 21, 51, 56). Variation within these regions, including gene deletion events, is observed during ASFV adaptation to monkey cell lines (4, 44) and appears to be associated with reduction of viral virulence (44). Given the similarities with poxviruses, it is likely that ASFV variable region genes are associated with important host range functions in either the pig or tick host.
Previously, we described an ASFV right variable region gene, NL-S, with similarity to the neurovirulence-associated gene (ICP34.5) of herpes simplex virus and demonstrated, using a viral gene deletion mutant, that NL-S, while being nonessential for replication in swine macrophages in vitro, is a significant viral virulence factor. Deletion of this gene from the European pathogenic isolate E70 resulted in almost complete attenuation of the virus in the domestic swine host (57). Consistent with a host range function, NL was found to be highly conserved among diverse pathogenic ASFV isolates, existing in either a long (184-amino-acid) or a short (70- to 72-amino acid) form (57).
Here, we describe a second ASFV right variable region gene, UK, associated with pig virulence. Our data indicate that (i) the UK gene is highly conserved among African and European ASFV isolates; (ii) UK is a novel gene, showing no similarity to other known genes in the current sequence databases; (iii) UK encodes a 15-kDa protein that is expressed in virus-infected macrophages at early times postinfection; and (iv) although it is nonessential for growth in porcine macrophage cell cultures, UK is a significant viral virulence determinant in domestic swine. Thus, the right variable region of the ASFV genome contains at least two genes, NL-S and UK, with functions involving pig virulence and swine host range.
MATERIALS AND METHODS
Cell culture and viruses.
Primary porcine macrophage cell cultures were prepared from defibrinated swine blood as previously described (19). Briefly, heparin-treated swine blood was incubated at 37°C for 1 h to allow sedimentation of the erythrocyte fraction. Mononuclear leukocytes were separated by flotation over a Ficoll-Paque (Pharmacia, Piscataway, N.J.) density gradient (specific gravity, 1.079). The monocyte/macrophage cell fraction was cultured in plastic Primaria (Falcon; Becton Dickinson Labware, Franklin Lakes, N.J.) tissue culture flasks containing RPMI 1640 medium with 30% L929 supernatant and 20% fetal bovine serum for 48 h (37°C in 5% CO2). Adherent cells were detached from the plastic with 10 mM EDTA in phosphate-buffered saline and then reseeded into Primaria T25 6- or 96-well dishes at a density of 5 × 106 cells per ml for use in assays 24 h later.
Pathogenic ASFVs used in this study were as follows. The tick isolates were Malawi Lil-20/1 (1983), Chiredzi/83/1 (1983), Crocodile/96/1 (1996), Crocodile/96/3 (1996), Pretoriuskop/96/5 (1996), Fairfield/96/1 (1996), and Wildebeeslaagte/96/1 (1996), and the pig isolates were E70 (1970), Brazil (1979), Cameroon (1982), Kerita (1967), Spencer (1951), Uganda 61 (1961), Victoria Falls (1967), Zimbabwe (1967), Tengani (1961), and Haiti 811 (1980).
DNA manipulation, cloning, and sequencing.
Viral DNAs were isolated from purified virions with proteinase K and by sodium dodecyl sulfate lysis followed by phenol extraction and ethanol precipitation (53). Southern blot, radiolabeling, and hybridization analyses were performed by standard methods (40). Plasmid DNA was prepared and manipulated essentially as described by Sambrook et al. (40).
The recombinant lambda clone LMw23 (14) from the pathogenic ASFV isolate Malawi Lil-20/1 genome was sequenced as previously reported (43).
An E70 genomic cosmid library was constructed as previously described (57). A cosmid clone, H7, representing the right terminus of the E70 genome was further subcloned, and a 5-kbp EcoRI-SalI fragment contained within it, H7E, was sequenced in its entirety with an Applied Biosystems Inc. model 370A automated DNA sequencer. DNA sequences were assembled by using Staden’s Sequence Assembly program (42) and analyzed by the FASTA method (36) as well as other phylogenetic programs (46, 47). Predicted protein sequences were analyzed by using the Genetics Computer Group (University of Wisconsin) computer programs (13) and SAPS software (6). Protein sequences were compared to those in the EMBL (release 51), GenBank (release 101), SwissProt (release 32), and PIR (release 52) databases by using the FASTA (36), BLAST (3), and MOST (48) computer programs. Proteins were aligned by using the Bestfit and Pileup computer programs from the Genetics Computer Group package with the Dayhoff Pam-250 symbol comparison table and a 0.5 cutoff value for peptide comparison.
PCR cloning and reverse transcription (RT)-PCR analysis.
PCR amplification of the UK gene region from various ASFVs was performed with low-molecular-weight DNA extracts from virus-infected cells as targets (23). The UK gene region, bracketed by open reading frame (ORF) MR on the left and a member of MGF363, ORF OR, on the right (Fig. 1A), was amplified with a primer pair derived from sequences flanking this region: forward primer 5′-CTTTCACCCCACGACTTCTTA-3′ and reverse primer 5′-CACTTGTAGAGTGGATGGCAT-3′ (nucleotides 729 and 1983 in the H7E clone, respectively). The UK region from ASFV isolate Haiti 811 was amplified with the following primer set: forward primer 5′-CTCCCGCCCCATGAATTCCTA-3′ and reverse primer 5′-TCATGCCACCATAAACCACAA-3′. PCR was performed for 40 cycles of thermal denaturation (96°C for 15 s), reannealing (50°C for 30 s), and extension (60°C for 30 s). Amplified products were cloned into the TA cloning vector pCR II (Invitrogen, San Diego, Calif.). Three or four PCR clones derived from independent amplifications of each isolate were sequenced completely with the T7 and Sp6 forward and reverse primers (40). The chromatogram traces were base called with Phred (version 0.961028), which also produced a quality file containing a probability of error at each base position. The sequence was assembled with Phrap (version 0.0.96731) with the quality files and default settings to produce a consensus sequence for each isolate.
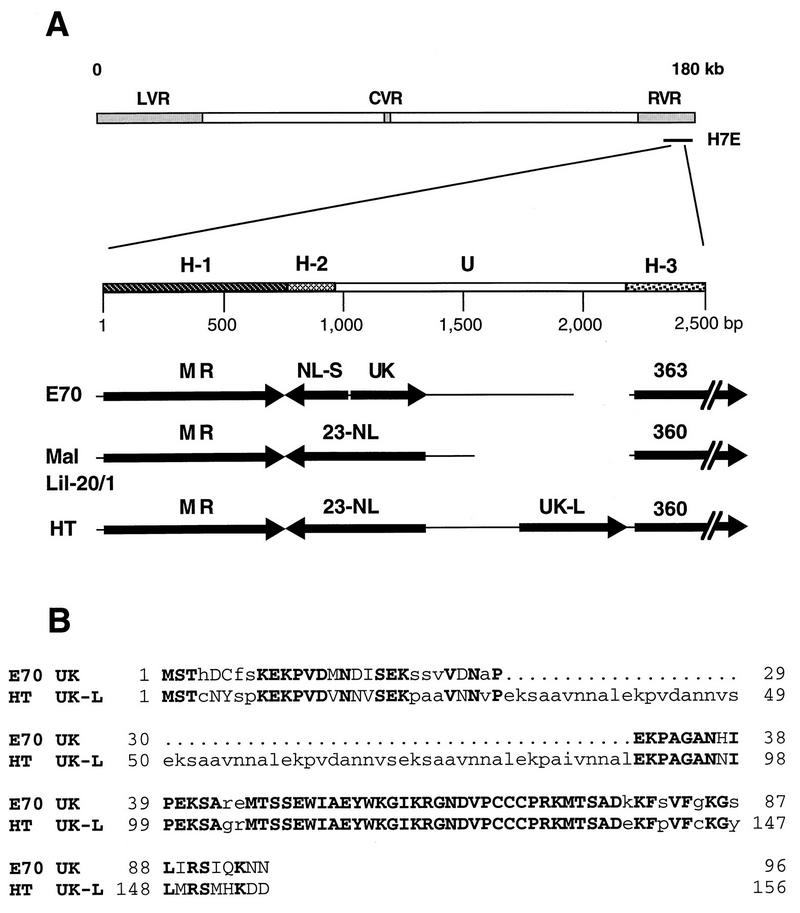
FIG. 1.
(A) Structural arrangement of the ASFV UK gene region in pathogenic virus isolates E70, Malawi Lil-20/1 (Mal Lil-20/1), and Haiti 811 (HT). H-1, ORF MR region; H-2, ORF NL carboxyl terminus region; U, unique region; H-3, MGF363/360 ORF OR region; LVR, left variable region; CVR, central variable region; RVR, right variable region. (B) Alignment of the predicted amino acid sequences encoded by the UK genes in the E70 and HT viruses. Identical residues are shown as uppercase boldface letters, while conservative amino acid substitutions are indicated by uppercase lightface letters. Periods denote missing amino acids.
RT-PCR was performed according to the protocols for RT of RNA and PCR amplification of cDNA provided with the GeneAmp thermostable rTth reverse transcriptase RNA PCR kit (Perkin-Elmer). Briefly, RNAs were extracted from ASFV-infected primary porcine macrophage cell cultures at 16 h postinoculation (multiplicity of infection [MOI], 10) with a Micro-Scale total RNA separator kit (Clontech, Palo Alto, Calif.) and treated with 100 U of DNase I (Boehringer Mannheim, Indianapolis, Ind.) per μg of RNA at 37°C for 90 min. One microgram of total infected-cell RNA was reverse transcribed for 15 min at 70°C by using 5 U of rTth DNA polymerase and a gene-specific downstream primer. Resulting cDNAs were then amplified by PCR with gene-specific primer pairs. For the NL-S gene, we used forward primer 5′-GTATGGGAAGCCGACGACATC-3′ and reverse primer 5′-TTACTGCTGCTCCAGTAGCTT-3′, and for the p72 gene, we used forward primer 5′-ATTTTAAGCCTTATGTTCCAG-3′ and reverse primer 5′-CTCTAAAAGGTGTTTGGTTGTC-3′. Non-reverse-transcribed RNA samples were used in the PCR as a control to ensure the absence of viral DNA contamination. PCR products were analyzed by gel electrophoresis and Southern blot hybridization by using PCR-generated DNA probes whose sequences were contained within the primary amplification products.
UK protein expression and immunoprecipitation.
The ORF UK was amplified by PCR and cloned into the expression vector pET 21a (Novagen, Madison, Wis.) with E70 genomic DNA as the template. PCR amplification was performed with a set of degenerate primers that created BglII sites at the 5′ and 3′ ends of the ORF: forward primer 5′-GTATAGTAGATCTTAGCATGT-3′ and reverse primer 5′-AAATATTAGATCTAACACGTT-3′ (nucleotides 2286 and 2689 in the H7E clone, respectively). Clones containing the ORF UK were identified by colony hybridization, and proper framing was confirmed by DNA sequencing. Escherichia coli BL21 (DE3) cells, transformed with the recombinant plasmid pET-UK, were grown in Luria-Bertani medium containing 100 mg of ampicillin per ml. Synthesis of UK protein and production of rabbit immune serum were performed as previously described (24, 33).
Primary porcine macrophage cell cultures were infected with ASFV Malawi Lil-20/1 and E70 isolates (MOI, 20), pulse-labeled for 2-h periods at various times postinfection with l-[35S]methionine in methionine-deficient RPMI 1640 medium, and immunoprecipitated with UK-monospecific antibodies as previously described (1).
Construction of ASF recombinant virus ΔUK and its revertant UK-R.
ASFV recombinant viruses were generated by homologous recombination between parental ASFV genomes and engineered recombination transfer vectors in swine macrophage cell cultures as previously described (57).
The recombination transfer vector for introducing the UK gene deletion in E70 was constructed by deleting a 257-bp HindIII-BamHI fragment from the cosmid subclone H7E (see Fig. 3A) and replacing it with the β-glucuronidase (GUS) reporter gene under the control of an ASFV late structural gene promoter, p72 (33). The BamHI site is a unique restriction site in the H7E clone, while the HindIII site was created by PCR site-directed mutagenesis at the UK start site (position 2303). This deletion removes all but 30 carboxyl-terminal nucleotides of the UK ORF. Transfection-infection assays were done as previously described (57). Recombinant viruses were plaque purified on macrophage cell cultures and analyzed and characterized by PCR and Southern blotting (57).
FIG. 3.
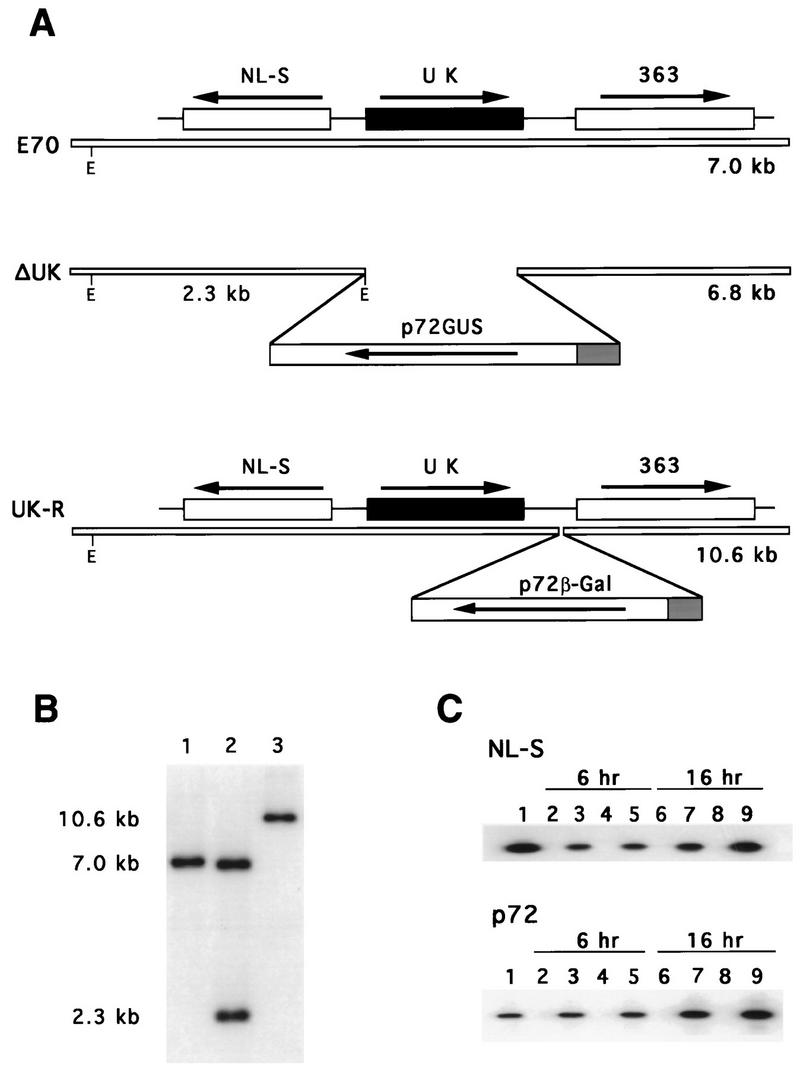
Characterizations of an ASFV UK gene deletion mutant, ΔUK, and its revertant, UK-R. (A) Diagram of the UK gene regions in the parental E70 isolate, the deletion mutant, ΔUK, and its revertant, UK-R. E, EcoRI site. (B) Southern blot analysis of E70 (lane 1), ΔUK (lane 2), and UK-R (lane 3). Purified viral DNAs were digested with EcoRI, electrophoresed, blotted, and hybridized with a DNA probe including UK gene sequences and flanking regions. Positions of molecular size markers are shown in kilobase pairs at the left. (C) RT-PCR amplification at 6 and 16 h postinfection of RNAs from macrophages infected with E70 (lanes 2, 3, 6, and 7) and ΔUK (lanes 4, 5, 8, and 9). One hundred nanograms of total RNA was used in the assay with either NL-S gene-specific or p72 gene-specific primers (lanes 3, 5, 7, and 9). PCR amplification from genomic DNAs (lane 1) and non-reverse-transcribed, DNase-treated RNA samples from macrophages infected with E70 (lanes 2 and 6) and ΔUK (lanes 4 and 8) were included as controls.
A revertant virus was constructed from the E70 UK gene deletion mutant ΔUK. A novel BglII site was created in the H7E clone by PCR site-directed mutagenesis at nucleotide 3090. A reporter cassette, p72β-Gal, containing the β-galactosidase gene was inserted into BglII-digested H7E to yield p72β-GalH7E. This construct was used in the transfection-infection experiment to restore the ORF UK in the E70 ΔUK genome. Putative revertants, GUS-negative and β-galactosidase-positive viruses, were purified by plaque assay on macrophage cell cultures and analyzed and characterized as described above.
Animal infections.
Yorkshire pigs (30 to 35 kg in experiments 1 and 2 and 60 to 70 kg in experiment 3) were inoculated intramuscularly with either 102 50% tissue culture infective doses (TCID50) of parental E70, recombinant ΔUK, or revertant UK-R viruses. A dose of 102 TCID50 of E70 represents a challenge of between 10 and 100 100% lethal doses (57). Clinical signs of ASF (fever [a rectal temperature greater than or equal to 40°C], anorexia, lethargy, shivering, cyanosis, and recumbency) were monitored daily. Blood samples were collected every other day for 30 days postinfection (DPI). Virus isolation and titration of ASFV in blood samples were performed as previously described (34). Virus titers were calculated by the method of Spearman-Karber and expressed as TCID50 (18).
Nucleotide sequence accession numbers.
The UK gene sequences were assigned GenBank accession no. AF015671 (E70), AF015666 (Brazil), AF015667 (Cameroon), AF015674 (Kerita), AF015677 (Spencer), AF015679 (Uganda 61), AF015680 (Victoria Falls), AF015681 (Zimbabwe), AF015678 (Tengani), AF015668 (Chiredzi/83/1), AF015669 (Crocodile/96/1), AF015670 (Crocodile/96/3), AF015676 (Pretoriuskop/96/5), AF015673 (K1/Fairfield/96/1), AF015675 (M1/Wildebeeslaagte/96/1), and AF015672 (Haiti 811).
RESULTS
A unique ASFV right variable region gene, UK, is conserved among most pathogenic virus isolates.
We described previously a highly conserved virulence-associated gene, NL-S, in the right variable region of several pathogenic ASFV isolates. A 5-kb fragment of clone H7E (Fig. 1A) from the right variable region of the European pathogenic isolate E70 was sequenced in its entirety, and the genetic content was analyzed and compared with the sequence from the same region of the African pathogenic isolate Malawi Lil-20/1. In contrast to the Malawi Lil-20/1 isolate, where the 23-NL gene was bracketed by the 23-MR ORF on the left and a member of MGF360 on the right, the E70 genome contained a novel ORF, UK, immediately upstream from NL-S and transcriptionally oriented toward the right end of the genome. Searches of the complete nucleotide sequence of the Malawi Lil-20/1 genome (15, 28) failed to identify UK gene sequences.
The sequence for the E70 UK gene begins 2,264 bases from the EcoRI restriction site and 1,057 bases from the start site of ORF 23-MR and extends for 288 bases on the positive strand of the H7E clone (Fig. 1A). ORF UK encodes a 96-amino-acid polypeptide (predicted molecular mass, 10.7 kDa; pI, 8.5) with no predicted signal sequence or membrane-spanning regions. A search of the Prosite database (release 13) identified two consensus protein kinase C phosphorylation motifs at amino acid residues 19 and 42; five casein kinase phosphorylation sites at residues 2, 23, 42, 47, and 74; one N-myristoylation site at residue 58; and no Asn glycosylation sites. The hydrophilic amino portion of UK contains four tandem repeats, each of which is 10 residues long (EKXXXXXXXX) and has a conserved charge distribution (−+00000000), which predict four flexibly linked alpha helices with high potential antigenicities. The carboxyl terminus is slightly hydrophobic. Searches of genetic databases found statistically significant similarity (P ≤ 0.001) between these repeats and repeats contained within the Trypanosoma cruzi chronic-phase antigenic protein (8), the Babesia bovis 80-kDa protein (11), and the Plasmodium falciparum ring-infected erythrocyte surface antigen (17). These proteins also contain 10-residue tandem repeats which have similar charge distributions and the same predicted secondary structure. Other regions of those proteins show no similarity to UK.
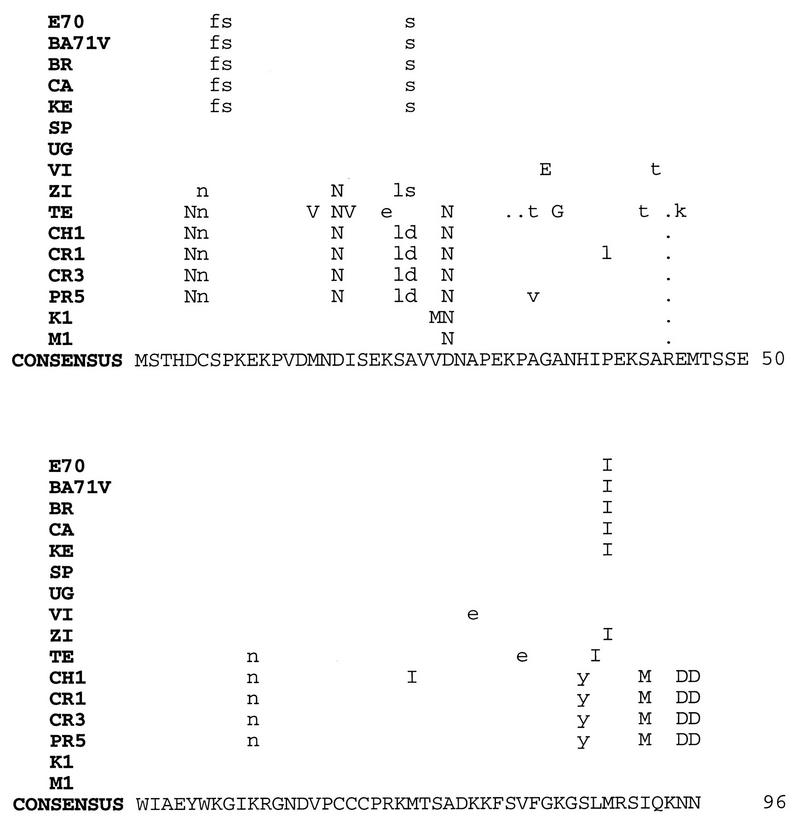
To assess the degree of UK gene conservation, 15 additional pathogenic viruses, representing African, European, and Caribbean isolates from both pig and tick sources, were examined by sequence analysis. The UK ORF from each virus was amplified by PCR, cloned, and sequenced completely. With the exception of the HT isolate, sequence analysis revealed an E70-type UK ORF for all isolates, encoding either 92, 95, or 96 amino acids (Fig. 2). Pathogenic isolates E70, BR, CA, and KE contained identical UK ORFs encoding 96 amino acids. The UK gene of the highly cell culture-adapted virus BA71V (55) was identical at the nucleotide level to UK of E70. Identical UK ORFs were found in the African isolates SP and UG. It is interesting that the African tick isolates formed a distinct, although not significantly different, group with UK ORFs encoding 95 amino acids with highly conserved sequence homology (99 to 100% identity over 95 residues).
FIG. 2.
Alignment of the predicted amino acid sequences encoded by UK ORFs from pathogenic ASFV isolates E70 (Spain, 1970), BR (Brazil, 1979), CA (Cameroon, 1982), KE (Kerita, 1967), SP (Spencer, 1951), UG (Uganda 61), VI (Victoria Falls, 1967), ZI (Zimbabwe, 1967), TE (Tengani, 1961), CH1 (Chiredzi/83/1, 1983), CR1 (Crocodile/96/1, 1996), CR3 (Crocodile/96/3, 1996), PR5 (Pretoriuskop/96/5, 1996), K1 (Fairfield/96/1, 1996), and M1 (Wildebeeslaagte/96/1, 1996) and from a highly cell culture-adapted European virus, BA71V (GenBank accession no. U18466). Differences in residues are shown above the consensus sequence. Conservative amino acid substitutions are indicated by uppercase letters. Periods denote missing amino acids.
The HT isolate contained a larger UK gene of 156 amino acids, UK-L (Fig. 1B). This longer form of UK is due entirely to the presence of six additional tandem repeats (EKXAXXNNXX), making a total of 10 tandem repeats in Haiti UK-L compared to only four repeats in E70 UK. The larger UK-L (predicted molecular mass, 16.9 kDa) is slightly more acidic (pI, 6.2), due to the six additional repeats, and 74% identical (94% similar) to the shorter E70 UK ORF but has the same secondary structure prediction and database matches as the E70 UK ORF. Interestingly, and unlike other described UK gene-containing virus genomes, the Haiti isolate encoded a long form of the NL gene (185 amino acids) with striking similarity to the Malawi 23-NL gene (92% identity and 96% similarity over 185 amino acids). Thus, the Haiti isolate has the only known ASFV genome containing both a long form of NL and the UK gene.
Among the UK genes sequenced, there were 59 polymorphic nucleotide sites with 134 changes and a 0.03 average change per nucleotide position with a 3.4 average corrected transition-transversion ratio. There was no significant difference among the isolates in overall relationship at the amino acid level (amino acid Poisson correction distance was estimated by a neighbor-joining branch-length test and a cluster test with 1,000 bootstrap samples [chi-square test result, 9.84; 10 degrees of freedom; P = 0.45]), although Haiti pig isolate HT was the most different from the others, encoding an extra six copies of the tandem repeat, while African domestic pig isolate TE encoded only 92 amino acids. At the nucleic acid level there was an indication that the UK locus was divided into a tick cluster and a pig cluster (Kimura 2 parameter distance estimate, Z = 2.817, P = 0.005). Given the small number of isolates examined, the significance of these apparent clusters remains to be determined. These data indicate that, with the exception of the Malawi Lil-20/1 isolate, the UK gene is highly conserved among diverse pathogenic ASFVs isolated from either domestic pigs or Ornithodoros ticks.
Construction and analysis of the recombinant ASFV UK gene deletion mutant, ΔUK, and its revertant, UK-R.
An ASFV UK gene deletion mutant and its revertant were constructed from the pathogenic European isolate E70 by homologous recombination between parental viral genomes and recombination transfer vectors in primary porcine macrophage cell cultures as described in Materials and Methods. The introduced deletion removed a 257-bp HindIII-BamHI fragment (Fig. 3A) which contained all but the carboxyl-terminal 30 nucleotides of UK and inserted in its place a 2.4-kb p72GUS reporter gene cassette. A revertant virus, UK-R, was constructed by restoring UK into the genome of the E70 UK gene deletion mutant, ΔUK, as described in Materials and Methods (Fig. 3A). Genomic DNAs from the parental virus E70, the null mutant ΔUK, and its revertant, UK-R, were analyzed by Southern blot hybridization (Fig. 3B). Viral DNAs were digested with EcoRI, gel electrophoresed, Southern blotted, and hybridized with the 32P-labeled 5-kb EcoRI-SalI fragment contained in clone H7E. The terminal EcoRI fragment in the right variable region of E70 was 7 kb long (Fig. 3B, lane 1). Novel EcoRI fragments of the predicted sizes 6.8 and 2.3 kb were observed for ΔUK as a result of a new EcoRI site, introduced with the p72GUS cassette (Fig. 3B, lane 2). As expected, a terminal EcoRI fragment of 10.6 kb was observed for UK-R; the net 3.6-kb size increase resulted from the insertion of the p72β-Gal reporter gene cassette (Fig. 3B, lane 3).
RT-PCR analysis of the adjacent NL-S gene indicated that the UK deletion introduced into ΔUK did not affect NL-S gene transcription in infected macrophage cell cultures (Fig. 3C). At both 6 and 16 h postinfection, comparable levels of NL-S transcription were observed in cells infected with parental E70 (Fig. 3C, lanes 3 and 7) and cells infected with ΔUK (Fig. 3, lanes 5 and 9) by RT-PCR analysis with NL-S-gene-specific and p72-gene-specific primers. Insertion of a p72β-Gal reporter cassette upstream from the 363 ORF did not affect the growth of the revertant UK-R in porcine macrophage cell cultures or virulence in pigs, indicating that neither insertions nor deletions upstream from the 363 gene resulted in any deleterious effect upon 363 gene function.
Expression of UK protein p15 in ASFV-infected swine macrophage cell cultures.
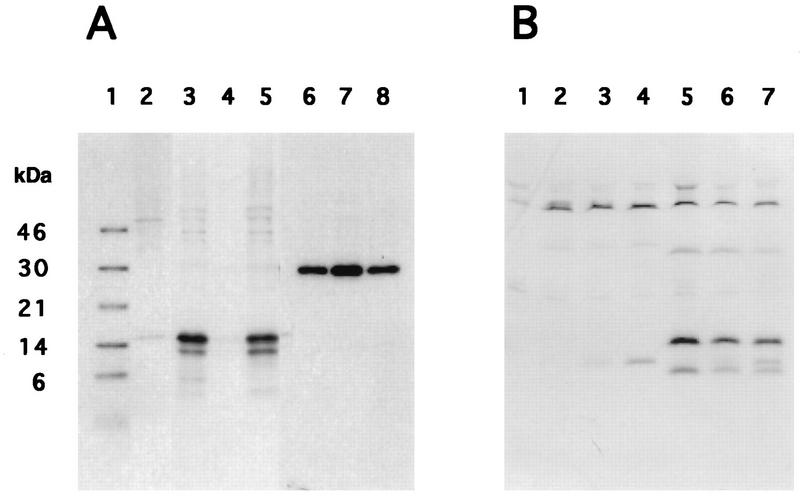
Monospecific rabbit antiserum specifically immunoprecipitated a major protein of approximately 15 kDa (p15) from both ASFV E70- and UK-R-infected swine macrophage cell cultures (Fig. 4A, lanes 3 and 5, respectively). A less intense band of approximately 13 kDa was also observed. The smaller polypeptide might represent a product of p15 proteolysis or a translation product initiated from an internal initiation codon present within the UK ORF (Fig. 2). No specific protein was immunoprecipitated from mock-infected or from ΔUK-infected cell extracts (Fig. 4A, lanes 2 and 4). As a control, monoclonal antibody recognizing a highly antigenic ASFV phosphoprotein, p30 (1), immunoprecipitated proteins of 30 kDa from E70-, ΔUK-, and UK-R-infected cell extracts (Fig. 4A, lanes 6 to 8, respectively) at comparable levels. Although the UK ORF contains seven potential serine or threonine phosphorylation sites, no detectable phosphorylation of p15 was observed when infected macrophage cell cultures were labeled with inorganic 32P (data not shown).
FIG. 4.
Expression of UK protein p15 in ASFV-infected porcine macrophage cell cultures. (A) Immunoprecipitation of cell extracts from mock-infected macrophages (lane 2) and macrophages infected with E70 (lanes 3 and 6), ΔUK (lanes 4 and 7), and UK-R (lanes 5 and 8) and labeled from 2 to 4 h postinfection was performed with either an anti-UK (lanes 2 to 5) or anti-p30 (lanes 6 to 8) monospecific rabbit antiserum. Lane 1 contains Rainbow 14C-methylated protein molecular mass markers (Amersham Life Science). (B) Time course of p15 expression. Mock-infected (lane 1), Malawi Lil-20/1-infected (lanes 2 to 4), and E70-infected (lanes 5 to 7) macrophage cell cultures were pulse labeled from 2 to 4 h (lanes 2 and 5), 4 to 6 h (lanes 3 and 5), and 6 to 8 h (lanes 4 and 7) postinfection, and then cell extracts were immunoprecipitated with anti-p15 antiserum.
Time course protein-labeling experiments demonstrated that p15 expression was most abundant between 2 to 4 h postinfection, with slightly decreased levels at later times (Fig. 4B, lanes 5 to 7). As expected, no specific protein was immunoprecipitated with the anti-UK antiserum from Malawi Lil-20/1-infected macrophage cell cultures (Fig. 4B, lanes 2 to 4).
UK is nonessential for growth of ASFV in swine macrophages in vitro.
Growth characteristics of ΔUK were compared to those of parental virus E70 and the revertant UK-R by infecting primary swine macrophage cell cultures (MOI, 1) and then titrating both extracellular and intracellular virus at various times postinfection. Growth kinetics and viral yields of ΔUK were statistically indistinguishable from those of E70 and UK-R (Fig. 5).
FIG. 5.
Growth characteristics of ASFV isolates E70, ΔUK, and UK-R in swine macrophage cell cultures. Porcine macrophage cell cultures were infected (MOI, 1) with E70, ΔUK, and UK-R viruses. At indicated times postinfection duplicate samples were collected and titrated for extracellular (A) and intracellular (B) virus yields. Data are the means and standard errors of results from two independent experiments.
UK is a significant virulence determinant in domestic swine.
To study the role of UK in viral virulence for domestic swine, Yorkshire pigs were infected intramuscularly with 102 TCID50 of the parental virus E70, the UK null mutant ΔUK, and its revertant, UK-R. A 102 TCID50 of ASFV E70 represents a challenge of between 10 and 100 100% lethal doses for both younger (30- to 35-kg) and older (60- to 70-kg) pigs (57). Data from three independent experiments are shown in Table 1. In contrast to E70- and UK-R-infected groups, where mortality was 100%, all ΔUK-infected animals survived infection. Times of onset of clinical disease were similar for animals infected with the three viruses. Animals infected with ΔUK were febrile for a 4- to 6-day period and exhibited transient lethargy for 2 to 3 days; however, other clinical signs and further disease progression were not observed. Animals infected with E70 or UK-R presented with clinical signs of ASF 3 to 4 DPI, and these symptoms progressed until death in all cases. Viremia titers in ΔUK-infected animals were significantly lower than those of E70- or UK-R-infected animals (Fig. 6). At 4 DPI, a 300- to 100,000-fold reduction of virus titer was observed for ΔUK-infected animals. Significant decreases of approximately 100-fold were evident at both 6 and 8 DPI. Viremias persisted in ΔUK-infected animals for periods of 30 to 42 DPI.
TABLE 1.
Swine survival and fever response following infection with E70, ΔUK, and UK-R ASFVs
| Expt | Group (n) | No. of pigs surviving/ total no. of pigs | Days to death | Fever
|
||
|---|---|---|---|---|---|---|
| Days to onset | No. of days of fever | Max temp (°C) | ||||
| 1 | E70 (4) | 0/4 | 8.0 ± 1.0 | 4.0 ± 0.5 | 4.0 ± 1.5 | 41.6 ± 0.2 |
| ΔUK (3) | 3/3 | 5.7 ± 0.7 | 4.5 ± 2.6 | 40.8 ± 0.4 | ||
| 2 | UK-R (5) | 0/5 | 7.8 ± 0.3 | 3.4 ± 0.4 | 4.4 ± 0.4 | 41.3 ± 0.2 |
| ΔUK (5) | 5/5 | 4.0 ± 0.0 | 5.8 ± 2.4 | 41.3 ± 0.3 | ||
| 3 | E70 (4) | 0/4 | 13.0 ± 3.8 | 3.5 ± 0.5 | 8.3 ± 2.3 | 41.9 ± 0.2 |
| ΔUK (4) | 4/4 | 4.0 ± 0.0 | 5.3 ± 0.8 | 41.4 ± 0.2 | ||
FIG. 6.
Viremia of E70-, ΔUK-, and UK-R-infected pigs during acute disease. Animal infections, blood sample collection, and virus titration were performed as described in Materials and Methods. Data are group mean titers with standard errors. Values significantly different from those of the E70 or UK-R group are indicated as follows: ∗, P = 0.0002; #, P = 0.05; @, P = 0.0001; and ¶, P = 0.002.
At 42 DPI, convalescent ΔUK-infected animals from experiment 1 were challenged with 104 TCID50 of parental E70 to assess the level of immunity conferred by ΔUK infection. Following challenge, all three animals remained clinically normal, with no detectable viremia, indicating that a solid level of homologous protective immunity had been induced following ΔUK infection.
DISCUSSION
Previously, we described a virulence-associated gene, NL-S, present in the right variable region of the ASFV E70 genome (57). Here, we describe a second virulence-associated gene, UK, present in the right variable region of the E70 genome.
UK is a novel gene with no similarity to other genes or known protein motifs in the current genetic databases. Except for the Malawi Lil-20/1 isolate, which lacked the gene, UK was highly conserved among the European, Caribbean, and African ASFV isolates examined here. In all but a single case, the genes encoded proteins of 92 to 96 amino acids with four tandem repeats, each of which had 10 residues with a conserved charge distribution, and the genes were present in a right-variable-region genomic arrangement (Fig. 1) that included the previously identified virulence-associated gene NL-S (57). The sole exception was the UK gene of the Haiti 811 isolate, which contained 10 tandem repeats and encoded a predicted protein of 156 amino acids. Interestingly, Haiti 811 is the only viral genome we have observed that contains both UK and the long form of the NL gene, 23-NL, in the right variable region (43). The significance of the number of repeats and their physicochemical relevance to UK protein function are unknown, and p15’s lack of similarity to other known proteins makes it difficult to speculate on possible function.
The UK-encoded protein, p15, is abundantly expressed at early times in virus-infected macrophage cell cultures. Its apparent molecular mass of 15 kDa was higher than the 10.7 kDa predicted by the primary sequence. This discrepancy may be due to posttranslational modifications or to aberrant gel mobility resulting from the presence of the tandem repeats (45). The protein does contain seven putative phosphorylation sites; however, no detectable phosphorylation of p15 was observed in macrophage cell cultures (58).
While it is highly conserved in most pathogenic ASFV isolates, UK was nonessential for viral replication in porcine macrophages in vitro. The E70 gene deletion mutant ΔUK exhibited wild-type replication kinetics and virus yields in macrophage cell cultures (Fig. 5). Although UK is clearly nonessential for replication in macrophages in vitro, it does appear that the gene affects ASFV replication in infected pigs (Fig. 6). Since differentiated macrophages and reticular cells are the major viral targets in vivo (9, 26, 27, 31, 32), p15 may perform a host range function in these cell types in vivo. This function might be a conditional one, required in cells only at specific stages of cell differentiation and/or activation but not required under in vitro cell culture conditions. It has been suggested that the stage of monocyte differentiation may influence cell susceptibility to ASFV infection (22, 29, 52). The fact that p15 is expressed early in virus-infected cells suggests a function involving early events in the virus-cell interaction.
The E70 UK gene deletion mutant ΔUK was significantly attenuated for domestic pigs when it was compared with either parental E70 or the revertant virus UK-R. Infection with ΔUK was characterized by less severe clinical symptoms, reduced viremia titers (100- to 1,000-fold), and no mortality. Although the exact mechanism responsible for viral attenuation is unknown, lower viremia titers for ΔUK-infected animals suggest a growth defect that might reduce tissue damage and allow time for an appropriate host immune response to be mounted. Alternatively, it is possible that ΔUK fails to infect or replicate in a yet-to-be-identified critical target cell during viral infection, thus preventing a lethal outcome.
Although remarkable, the degree of attenuation observed for ΔUK was not as marked as that observed when the NL-S gene was deleted from the E70 genome. Apart from a transient fever response, no evidence of clinical disease was observed in pigs following infection with the E70 NL-S gene deletion mutant ΔNL-S (57). We have constructed a double-gene-deletion mutant of E70, ΔNL-S/UK, which lacks both virulence-associated genes, NL-S and UK. Deletion of both genes did not result in further attenuation of E70; ΔNL-S/UK exhibited a disease and virulence phenotype indistinguishable from that of ΔNL-S (58).
While UK and NL-S genes may be necessary for E70 viral virulence in pigs, they are not alone sufficient. Highly passaged avirulent European ASFV isolates BA71V and MS44 contain both genes, and they show 100% amino acid identity with the E70 genes (55, 58). Thus, other viral determinants must play significant roles in ASFV virulence. Interestingly, the highly pathogenic African isolate Malawi Lil-20/1 does not contain a UK gene (15, 28). Either a UK-like function is provided by a second yet-to-be-identified Malawi Lil-20/1 gene or, alternatively, the need for this gene may depend on the complement of other virulence and swine host range genes contained by a given virus isolate.
To date, at least four conserved but nonessential genes have been identified in the right variable region of the ASFV genome, and two of these, NL-S and UK, have been associated with viral virulence and swine host range (25, 28, 57). It is likely that additional genes performing host range functions in either the swine or tick host will be identified within the terminal variable regions of the ASFV genome. Characterization of these and other host range genes may allow for rational design of engineered live-attenuated, host range-restricted ASFV vaccines.
ACKNOWLEDGMENTS
We thank Aniko Zsak, Rochelle Mireles, J. R. Emmanuelli, and the PIADC animal care staff for excellent technical assistance and Steven Kleiboeker, Glen Scoles, Thomas Burrage, and Stefan Swanepoel for providing African tick isolates of ASFV.
REFERENCES
- 1.Afonso C L, Alcaraz C, Brun A, Sussman M D, Onisk D V, Escribano J M, Rock D L. Characterization of p30, a highly antigenic membrane and secreted protein of African swine fever virus. Virology. 1992;189:368–373. doi: 10.1016/0042-6822(92)90718-5. [DOI] [PubMed] [Google Scholar]
- 2.Almendral J M, Almazán F, Blasco R, Viñuela E. Multigene families in African swine fever virus: family 110. J Virol. 1990;64:2064–2072. doi: 10.1128/jvi.64.5.2064-2072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Blasco R, Agüero M, Almendral J M, Viñuela E. Variable and constant regions in African swine fever virus DNA. Virology. 1989;168:330–338. doi: 10.1016/0042-6822(89)90273-0. [DOI] [PubMed] [Google Scholar]
- 5.Blasco R, de la Vega I, Almazán F, Agüero A, Viñuela E. Genetic variation of African swine fever virus: variable regions near the ends of the viral DNA. Virology. 1989;173:251–257. doi: 10.1016/0042-6822(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 6.Brendel V, Bucher P, Nourbakhsh I, Blaisdell B E, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown F. The classification and nomenclature of viruses: summary of results of meetings of the International Committee on Taxonomy of Viruses in Sendai, September 1984. Intervirology. 1986;25:141–143. doi: 10.1159/000150091. [DOI] [PubMed] [Google Scholar]
- 8.Buschiazzo A, Campetella D E. Sequence of the gene for a Trypanosoma cruzi protein antigenic during the chronic phase of human Chagas disease. Mol Biochem Parasitol. 1992;54:125–128. doi: 10.1016/0166-6851(92)90105-s. [DOI] [PubMed] [Google Scholar]
- 9.Colgrove G S, Haelterman E O, Coggins L. Pathogenesis of African swine fever in young pigs. Am J Vet Res. 1969;30:1343–1359. [PubMed] [Google Scholar]
- 10.Costa J V. African swine fever virus. In: Darai G, editor. Molecular biology of iridoviruses. Norwell, Mass: Kluwer Academic Publishers; 1990. pp. 247–270. [Google Scholar]
- 11.Dalrymple B P, Peters J M, Goodger B V, Bushell G R, Waltisbuhl D J, Wright I G. Cloning and characterisation of cDNA clones encoding two Babesia bovis proteins with homologous amino- and carboxy-terminal domains. Mol Biochem Parasitol. 1993;59:181–189. doi: 10.1016/0166-6851(93)90216-k. [DOI] [PubMed] [Google Scholar]
- 12.De la Vega I, Viñuela E, Blasco R. Genetic variation and multigene families in African swine fever virus. Virology. 1990;179:234–246. doi: 10.1016/0042-6822(90)90293-z. [DOI] [PubMed] [Google Scholar]
- 13.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon L. Molecular cloning and restriction enzyme mapping of an African swine fever virus isolate from Malawi. J Gen Virol. 1988;69:1683–1694. doi: 10.1099/0022-1317-69-7-1683. [DOI] [PubMed] [Google Scholar]
- 15.Dixon L K, Twigg S R F, Baylis S A, Vydelingum S, Bristow C, Hammond J M, Smith G L. Nucleotide sequence of a 55 kbp region from the right end of the genome of a pathogenic African swine fever virus isolate (Malawi LIL20/1) J Gen Virol. 1994;75:1655–1684. doi: 10.1099/0022-1317-75-7-1655. [DOI] [PubMed] [Google Scholar]
- 16.Dixon, L. K., D. L. Rock, and E. Viñuela. 1995. African swine fever-like viruses. Arch. Virol. 10(Suppl.):92–94.
- 17.Favaloro J M, Coppel R L, Corcoran L M, Foote S J, Brown G V, Anders R F, Kemp D J. Structure of the RESA gene of Plasmodium falciparum. Nucleic Acids Res. 1986;14:8265–8277. doi: 10.1093/nar/14.21.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finney D J. Statistical methods in biological assays. 3rd ed. New York, N.Y: Macmillan Publishing Co., Inc.; 1978. Assays based on quantal responses; pp. 394–398. [Google Scholar]
- 19.Genovesi E V, Villinger F, Gerstner D J, Whyard T C, Knudsen R C. Effect of macrophage-specific colony-stimulating factor (CSF-1) on swine monocyte/macrophage susceptibility to in vitro infection by African swine fever virus. Vet Microbiol. 1990;25:153–176. doi: 10.1016/0378-1135(90)90074-6. [DOI] [PubMed] [Google Scholar]
- 20.González A, Talavera A, Almendral J M, Viñuela E. Hairpin loop structure of African swine fever virus DNA. Nucleic Acids Res. 1986;14:6835–6844. doi: 10.1093/nar/14.17.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez A, Calvo V, Almazan F, Almendral J M, Ramirez J C, De la Vega I, Blasco R, Viñuela E. Multigene families in African swine fever virus: family 360. J Virol. 1990;64:2073–2081. doi: 10.1128/jvi.64.5.2073-2081.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess W R, DeTray D E. The use of leukocyte cultures for diagnosing African swine fever. Bull Epizoot Dis Afr. 1960;8:317–320. [Google Scholar]
- 23.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 24.Irusta P M, Borca M V, Kutish G F, Lu Z, Caler E, Carrillo C, Rock D L. Amino acid tandem repeats within a late viral gene define the central variable region of African swine fever virus. Virology. 1996;220:20–27. doi: 10.1006/viro.1996.0281. [DOI] [PubMed] [Google Scholar]
- 25.Kleiboeker, S. B., et al. Unpublished data.
- 26.Konno S, Taylor W D, Dardiri A H. Acute African swine fever. Proliferative phase in lymphoreticular tissue and the reticuloendothelial system. Cornell Vet. 1971;61:71–84. [PubMed] [Google Scholar]
- 27.Konno S, Taylor W D, Hess W R, Heuschele W P. Liver pathology in African swine fever. Cornell Vet. 1971;61:125–150. [PubMed] [Google Scholar]
- 28.Lu, Z., et al. Unpublished data.
- 29.Malmquist W A, Hay D. Hemadsorption and cytopathic effect produced by African swine fever virus in swine bone marrow and buffy coat cultures. Am J Vet Res. 1960;21:104–108. [PubMed] [Google Scholar]
- 30.Massung R F, Esposito J J, Liu L, Qi J, Utterback T R, Knight J C, Aubin L, Yuran T E, Parsons J M, Loparev V N, Selivanov N A, Cavallaro K F, Kerlavage A R, Mahy B W J, Venter J C. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature (London) 1993;366:748–751. doi: 10.1038/366748a0. [DOI] [PubMed] [Google Scholar]
- 31.Mebus C A. African swine fever. Adv Virus Res. 1988;35:251–269. doi: 10.1016/s0065-3527(08)60714-9. [DOI] [PubMed] [Google Scholar]
- 32.Moulton J, Coggins L. Comparison of lesions in acute and chronic African swine fever. Cornell Vet. 1968;58:364–388. [PubMed] [Google Scholar]
- 33.Neilan J G, Lu Z, Kutish G F, Zsak L, Burrage T G, Borca M V, Carrillo C, Rock D L. A BIR motif containing gene of African swine fever virus, 4CL, is nonessential for growth in vitro and viral virulence. Virology. 1997;230:252–264. doi: 10.1006/viro.1997.8481. [DOI] [PubMed] [Google Scholar]
- 34.Onisk D V, Borca M V, Kutish G, Kramer E, Irusta P, Rock D L. Passively transferred African swine fever virus antibodies protect swine against lethal infection. Virology. 1994;198:350–354. doi: 10.1006/viro.1994.1040. [DOI] [PubMed] [Google Scholar]
- 35.Ortin J, Enjuanes L, Viñuela E. Cross-links in African swine fever virus DNA. J Virol. 1979;31:579–583. doi: 10.1128/jvi.31.3.579-583.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 37.Plowright W, Parker J, Pierce M A. The epizootiology of African swine fever in Africa. Vet Rec. 1969;85:668–674. [PubMed] [Google Scholar]
- 38.Plowright W, Parker J, Pierce M A. African swine fever virus in ticks (Ornithodoros moubata, Murray) collected from animal burrows in Tanzania. Nature (London) 1969;221:1071–1073. doi: 10.1038/2211071a0. [DOI] [PubMed] [Google Scholar]
- 39.Plowright W, Thomson G R, Neser J A. African swine fever. In: Coetzer J A W, Thomson G R, Tustin R C, editors. Infectious diseases in livestock with special reference to South Africa. Vol. 1. Cape Town, South Africa: Oxford University Press; 1994. pp. 568–599. [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Sogo J M, Almendral J M, Talavera A, Viñuela E. Terminal and internal inverted repetitions in African swine fever virus DNA. Virology. 1984;133:271–275. doi: 10.1016/0042-6822(84)90394-5. [DOI] [PubMed] [Google Scholar]
- 42.Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982;10:2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sussman M D, Lu Z, Kutish G, Afonso C L, Roberts P, Rock D L. Identification of an African swine fever virus gene with similarity to a myeloid differentiation primary response gene and a neurovirulence-associated gene of herpes simplex virus. J Virol. 1992;66:5586–5589. doi: 10.1128/jvi.66.9.5586-5589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabarés E, Olivares I, Santurde G, Garcia M J, Martin E, Carnero M E. African swine fever virus DNA: deletions and additions during adaptation to growth in monkey kidney cells. Arch Virol. 1987;97:333–346. doi: 10.1007/BF01314431. [DOI] [PubMed] [Google Scholar]
- 45.Takano E, Maki M, Mori H, Hatanaka M, Marti T, Titani K, Kannagi R, Ooi T, Murachi T. Pig heart calpastatin: identification of repetitive domain structures and anomalous behavior in polyacrylamide gel electrophoresis. Biochemistry. 1988;27:1964–1972. doi: 10.1021/bi00406a024. [DOI] [PubMed] [Google Scholar]
- 46.Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 48.Tatusov R L, Altschul S F, Koonin E V. Detection of conserved segments in proteins: interactive scanning of sequence databases with alignment blocks. Proc Natl Acad Sci USA. 1994;91:12091–12094. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomson G R, Gainaru M D, Van Dellen A F. Experimental infection of warthog (Phacochoerus aethiopicus) with African swine fever virus. Onderstepoort J Vet Res. 1980;47:19–22. [PubMed] [Google Scholar]
- 50.Thomson G R, Gainaru M, Lewis A, Biggs H, Nevill E, Van Der Pypekamp M, Gerbes L, Esterhuysen J, Bengis R, Bezuidenhout D, Condy J. The relationship between ASFV, the warthog and Ornithodoros species in southern Africa. In: Wilkinson P J, editor. ASF, EUR 8466 EN. Proceedings of CEC/FAO Research Seminar. Luxembourg, Belgium: Commission of the European Communities; 1983. pp. 85–100. [Google Scholar]
- 51.Vydelingum S, Baylis S A, Bristow C, Smith G L, Dixon L K. Duplicated genes within the variable right end of the genome of a pathogenic isolate of African swine fever virus. J Gen Virol. 1993;74:2125–2130. doi: 10.1099/0022-1317-74-10-2125. [DOI] [PubMed] [Google Scholar]
- 52.Wardley R C, Wilkinson P J. The growth of virulent African swine fever virus in pig monocytes and macrophages. J Gen Virol. 1978;38:183–186. doi: 10.1099/0022-1317-38-1-183. [DOI] [PubMed] [Google Scholar]
- 53.Wesley R D, Tuthill A E. Genome relatedness among African swine fever virus field isolates by restriction endonuclease analysis. Prev Vet Med. 1984;2:53–62. [Google Scholar]
- 54.Wilkinson P J. African swine fever virus. In: Pensaert M B, editor. Virus infections of porcines. Amsterdam, The Netherlands: Elsevier Science Publishers; 1989. pp. 17–35. [Google Scholar]
- 55.Yáñez R J, Rodríguez J M, Nogal M L, Yuste L, Enríquez C, Rodriguez J F, Viñuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 56.Yozawa T, Kutish G F, Afonso C L, Lu Z, Rock D L. Two novel multigene families, 530 and 300, in the terminal variable regions of African swine fever virus genome. Virology. 1994;202:997–1002. doi: 10.1006/viro.1994.1426. [DOI] [PubMed] [Google Scholar]
- 57.Zsak L, Lu Z, Kutish G F, Neilan J G, Rock D L. An African swine fever virus virulence-associated gene NL-S with similarity to the herpes simplex virus ICP34.5 gene. J Virol. 1996;70:8865–8871. doi: 10.1128/jvi.70.12.8865-8871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zsak, L., et al. Unpublished data.