Abstract
Programmed ribosomal frameshifting is a molecular mechanism that is used by many RNA viruses to produce Gag-Pol fusion proteins. The efficiency of these frameshift events determines the ratio of viral Gag to Gag-Pol proteins available for viral particle morphogenesis, and changes in ribosomal frameshift efficiencies can severely inhibit virus propagation. Since ribosomal frameshifting occurs during the elongation phase of protein translation, it is reasonable to hypothesize that agents that affect the different steps in this process may also have an impact on programmed ribosomal frameshifting. We examined the molecular mechanisms governing programmed ribosomal frameshifting by using two viruses of the yeast Saccharomyces cerevisiae. Here, we present evidence that pokeweed antiviral protein (PAP), a single-chain ribosomal inhibitory protein that depurinates an adenine residue in the α-sarcin loop of 25S rRNA and inhibits translocation, specifically inhibits Ty1-directed +1 ribosomal frameshifting in intact yeast cells and in an in vitro assay system. Using an in vivo assay for Ty1 retrotransposition, we show that PAP specifically inhibits Ty1 retrotransposition, suggesting that Ty1 viral particle morphogenesis is inhibited in infected cells. PAP does not affect programmed −1 ribosomal frameshift efficiencies, nor does it have a noticeable impact on the ability of cells to maintain the M1-dependent killer virus phenotype, suggesting that −1 ribosomal frameshifting does not occur after the peptidyltransferase reaction. These results provide the first evidence that PAP has viral RNA-specific effects in vivo which may be responsible for the mechanism of its antiviral activity.
The maintenance of a correct reading frame is fundamental to the integrity of the translation process and, ultimately, to cell growth and viability. Although ribosomes translate mRNAs with great accuracy, a number of cases in which ribosomes are directed to shift a reading frame have been identified and characterized. These most often have occurred in double-stranded RNA (dsRNA) and plus-strand RNA viruses, as well as in a few bacterial cellular genes and the ornithine decarboxylase antizyme gene in mammals (for reviews, see references 4, 11, and 24). The study of these ribosomal frameshifts is important both because of their critical role in animal and plant pathogens and because of the information that they provide about the mechanisms by which a reading frame is normally maintained.
We have used two different viral systems (the L-A–M1 killer system and the Ty1 retrotransposable element) of Saccharomyces cerevisiae as models to study programmed ribosomal frameshifting. The 4.6-kb dsRNA genome of the yeast L-A virus contains two open reading frames. The 5′ gag gene encodes the major viral coat protein (Gag), and the 3′ pol gene encodes a multifunctional protein domain which includes the RNA-dependent RNA polymerase and a domain required for viral RNA packaging (12, 29). A −1 ribosomal frameshift event is responsible for the production of the Gag-Pol fusion protein (18, 21, 29). The M1 virus is a satellite of L-A, and its 1.6- to 1.8-kb dsRNA genome encodes a secreted killer toxin (reviewed in reference 8). The M1 plus strand is encapsidated and replicated in L-A-encoded viral particles. Ty1 is the yeast equivalent of a retrovirus which uses a ribosomal frameshift in the +1 direction for the production of its Gag and Gag-Pol proteins (reviewed in references 11 and 20). With their different frameshift mechanisms, these two viral systems constitute a powerful set of tools with which ribosomal frameshifting can be dissected. The efficiency of ribosomal frameshifting determines the ratio between viral Gag (structural) and Gag-Pol fusion (enzymatic) proteins, and the proper ratio is required for proper viral particle assembly. Changing the efficiency of ribosomal frameshifting upsets this stoichiometry, inhibiting virus maintenance (2, 15, 16, 32, 46). The efficiency, not the direction of the frameshift, is important (15).
Programmed ribosomal frameshifting in the −1 direction in viruses that infect eukaryotes requires a special sequence, X XXY YYZ (the 0 frame is indicated by spaces), called the slippery site (31). The simultaneous slippage of ribosome-bound A- and P-site tRNAs by 1 base in the 5′ direction still leaves their nonwobble bases correctly paired in the new reading frame. A second frameshift-promoting element (30), usually an RNA pseudoknot, is located immediately 3′ to the slippery site (5, 12, 40). The mRNA pseudoknot structure makes the ribosome pause over the slippery site and is thought to increase the probability of frameshifting (37, 42). The efficiency of −1 ribosomal frameshifting can be affected by the ability of the ribosome-bound tRNAs to unpair from the 0 frame, the ability of these tRNAs to repair to the −1 frame, the position of the RNA pseudoknot relative to the slippery site, and the pseudoknot’s thermodynamic stability (5–7, 12, 15, 30, 35). The Ty1 +1 ribosomal frameshift also requires a ribosomal pause, but this occurs when an elongating ribosome encounters a rare AGG codon in a special context (a “hungry codon”). The elongating ribosome, having its P site occupied by a peptidyl-tRNA, is forced to pause with its A site unoccupied as a consequence of the low abundance of the cognate tRNACUUArg (22). If, during the course of the pause, the ribosome slips 1 base in the 3′ direction, this peptidyl-tRNA is capable of base pairing to the new P-site codon in the +1 reading frame. The new A-site codon corresponds to an abundant tRNAGCCGly. If this tRNA can be inserted into the +1 frame codon, then the ribosomal frameshift can become established.
The 29-kDa pokeweed antiviral protein (PAP) isolated from Phytolacca americana is a ribosome-inactivating protein (RIP). PAP catalytically removes a specific adenine base from a highly conserved, surface-exposed stem-loop structure in the large rRNA of eukaryotic and prokaryotic ribosomes (19, 26). PAP displays broad-spectrum antiviral activity against plant and animal viruses, including influenza virus (41), poliovirus (44), herpes simplex virus (1), and human immunodeficiency virus (47). PAP removes an adenine base by specific cleavage of the N-glycosidic bond at A4324 in rat 28S rRNA and at homologous sites on ribosomes from other organisms. Ribosomes depurinated in this manner are unable to bind the elongation factor 2 (EF-2)–GTP complex, and protein synthesis is blocked at the translocation step (34, 36). We previously expressed a PAP cDNA in S. cerevisiae under the control of the galactose-inducible GAL1 promoter and showed that the expression of PAP inhibits the growth of yeast cells (28). Mutants of PAP that lose this growth-inhibiting ability have been isolated. One of the PAP mutants, pNT123-2, had a point mutation at the active site (E176V). This mutation abolished enzymatic activity in vitro in rabbit reticulocyte lysates (28) and in vivo in yeasts (28) and in transgenic plants (43).
In this report, we demonstrate that the expression of PAP in S. cerevisiae leads to specific inhibition of ribosomal frameshifting in the +1 direction and interferes with the ability of Ty1 to retrotranspose. In contrast, PAP expression in yeast does not affect ribosomal frameshifting in the −1 direction, nor does it interfere with maintenance of the M1-dependent killer virus phenotype. Our results are explained in light of a “kinetic pause” model of programmed ribosomal frameshifting. This is the first demonstration of specific inhibition of ribosomal frameshifting and retrotransposition by PAP and suggests that this inhibition may be a general mechanism for inhibition of other viral or cellular mRNAs that use programmed +1 ribosomal frameshifting.
MATERIALS AND METHODS
Strains and media.
S. cerevisiae PSY1 (MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100) was used for all of the assays. Strain JD759 {MATα kar1-1 arg1 thr(1,X) [L-AHN M1]} was used as the cytoduction donor strain in order to introduce the L-A and M1 dsRNA viruses into PSY1. Strain 5X47 (a/α his1/+ trp1/+ ura3/+ K−R−) was the killer indicator strain used to score the killer phenotype as described previously (12). YPAD, YPG, SD, synthetic complete medium, and 4.7MB plates for testing the killer phenotype were prepared as previously reported (16). Synthetic complete medium (H−leu, H−ura, and H−ura,−leu) (e.g., synthetic complete medium H lacking leucine [H−leu]) with 2% dextrose, galactose, or raffinose was used for controlling the induction of the PAP gene from the pNT188 and pNT123 plasmid vectors.
Plasmids.
Plasmids pNT188 and pNT123 were constructed as described previously (28) by cloning the cDNAs encoding PAP 3′ of the GAL1 promoter in the yeast expression vector YEp351 containing the selectable marker LEU2 and the yeast expression vector pAC55 containing the selectable marker URA3, respectively. The expression of PAP in both pNT188 and pNT123 is under the control of the galactose-inducible GAL1 promoter. The PAP catalytic site mutant pNT123-2 was generated by ethyl methanesulfonate mutagenesis and characterized previously (28, 43). Plasmids pT125 (0 frame control), pF8 (L-A-derived −1 ribosomal frameshift test vector), and pJD104 (Ty1-derived +1 ribosomal frameshift test vector), used in the in vivo ribosomal frameshifting assay, were described previously (2, 12) (Fig. 1A). pTY1HIS3AI was used to measure Ty1 retrotransposition frequencies (10).
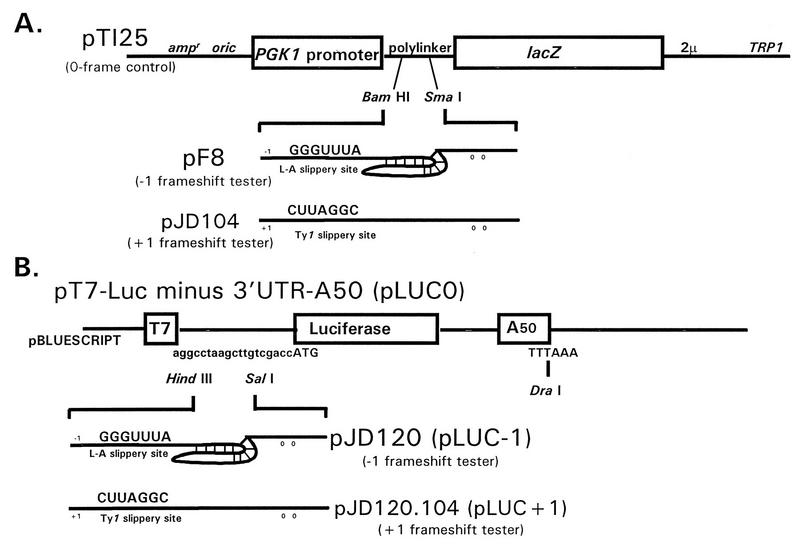
FIG. 1.
Vectors used to measure programmed ribosomal frameshifting efficiencies in vivo (A) and in vitro (B). The in vivo 0 frame control reporter plasmid pT125 and the −1 ribosomal frameshift test plasmid pF8 are described elsewhere (12). The +1 ribosomal frameshift test plasmid pJD104 is described elsewhere (2). In these constructs, transcription is driven from the constitutive phosphoglycerol kinase 1 (PGK1) promoter. The in vitro 0 frame control plasmid pLUC0 and the −1 ribosomal frameshift test plasmid pJD120 are described elsewhere (14). Construction of the in vitro +1 ribosomal frameshift plasmid pJD120.104 is described in Materials and Methods. The efficiencies of programmed ribosomal frameshifting were determined by dividing the enzymatic activities produced from the frameshift reporters (−1 or +1) by the enzymatic activities produced from the 0 frame controls and multiplying the resulting ratios by 100. The approximate locations of −1, +1, and 0 frame termination codons are indicated by numbers.
Plasmids pT7-LUC minus 3′UTR-A50 (23) (referred to as pLUC0) and pJD120 were used to produce synthetic 0 frame luciferase and synthetic L-A-derived −1 ribosomal frameshift test mRNAs, respectively, as described previously (14). The synthetic oligonucleotides 5′ CCCCCCATGGTAACCCCGGGCTG 3′ and 5′ CCCCAAGCTTATGACTTCTAGGATC 3′ were used to amplify the Ty1-derived +1 ribosomal frameshift signal from pJD104 by use of the PCR. The approximately 200-bp DNA fragment was digested with HindIII and NcoI and ligated into similarly digested pLUC0 to make pJD120.104. In this plasmid, the luciferase gene is 3′ of the Ty1 +1 ribosomal frameshift signal and in the +1 frame with respect to the AUG translational start site. These plasmids are shown in Fig. 1B.
Genetic methods. (i) Ty1 retrotransposition frequency.
PSY1 cells harboring either pNT188 or YEp351 were transformed with pTy1HIS3AI (10) and selected for on H−ura,−leu with 2% dextrose. Transformants were then grown on H−ura,−leu with 2% galactose at room temperature for 4 days. Patches of cells were subsequently replica plated back onto H−ura,−leu with 2% dextrose and incubated at 30°C for 2 days. After incubation, the cells were replica plated onto H−his or grown in H−ura,−leu liquid medium. The optical density at 550 nm was determined for cells grown in liquid medium, and 10-fold dilutions of cells from 104 to 108 CFU were seeded onto H−his medium. Retrotransposition frequencies were calculated by dividing the number of His+ colonies by the total number of CFU seeded onto the plate.
(ii) Killer assay.
PSY1 cells harboring pNT188 or YEp351 were cured of mitochondrial DNA ([rho0]) by growth on selective medium containing 33 μg of ethidium bromide per ml. L-A and M1 were introduced into [rho0] cells by cytoduction for 7 h at 30°C as described previously with JD759 as the donor cells (15). Cells were streaked for single colonies onto H−arg medium to select against the donor strain and subsequently replica plated to SD, YPG, and 4.7MB plates seeded with 5X47 killer indicator cells. Cytoductants were identified by growth on H−arg and YPG, no growth on SD, and killer phenotypes. To test for the effects of PAP on the maintenance of L-A and M1, cells were grown on H−leu containing 2% galactose for 4 days at 24°C. Cells were then incubated in H−leu liquid medium containing 2% dextrose overnight at 30°C, seeded onto H−leu containing 2% dextrose at densities of approximately 100 CFU/plate, grown for single colonies, and replica plated to killer indicator plates.
(iii) Measurement of ribosomal frameshifting in vivo.
Cells harboring pNT123, pNT123-2, or vector alone were transformed with either pT125, pF8, or pJD104, and transformants were selected on H−ura,−trp medium. A minimum of three independent transformants from each group were grown overnight in H−ura,−trp containing 2% raffinose at 30°C. The cultures were then split, centrifuged, resuspended in 2 ml of H−ura,−trp containing either 2% raffinose or 2% galactose, and grown for 5 h. This procedure will induce PAP expression, as shown earlier (28). β-Galactosidase activities were determined as described previously (12). All assays were performed in triplicate, and each assay was repeated at least three times. Percent inhibition by PAP of general translation (0 frame) was calculated by determining the ratio of β-galactosidase activities produced by induced and uninduced cells. Percent inhibition by PAP of ribosomal frameshifting was calculated by determining the ratio of ribosomal frameshifting efficiencies in induced and uninduced cells.
(iv) Luciferase assay.
Synthetic 7methyl-Gppp poly(A)-tailed mRNAs were prepared as T7 RNA polymerase runoff transcripts from DraI-digested pLUC0, pJD120, and pJD120.104 with a mMessagemMachine kit (Ambion). Translation-competent rabbit reticulocyte lysates were commercially obtained, and in vitro translation reactions were carried out as described by the manufacturer (Ambion). Lysates (20 μl) were preincubated at 28°C for 15 min with 1 to 4 pg of PAP, and then 20 ng of each of the mRNAs (Luc0, Luc−1, and Luc+1) was added to the PAP-lysate mixture and allowed to incubate at 28°C for 45 min. Each data point was assayed in triplicate. After incubation, the mixture was placed on dry ice to stop the reaction and subsequently was allowed to thaw on ice. Luciferase activities were determined with a Turner 20/20 luminometer. Ribosomal frameshift efficiencies were calculated by determining the ratio of luciferase activities produced by the test mRNAs (Luc−1 and Luc+1) and the 0 frame control mRNA (Luc0).
(v) Nuclease protection assays.
Total cellular RNA was isolated from induced and uninduced cells harboring PAP and frameshift test vectors by previously described methods (9) to measure the effects of PAP on mRNA abundances in vivo. To measure the effects of PAP on mRNA abundances in vitro, the synthetic luciferase test mRNAs were extracted from rabbit reticulocyte extracts by organic extraction after the incubation described above. RNase protection assays followed procedures described elsewhere (33). Briefly, RNA (10 μg) from each sample was resuspended to a total volume of 21 μl in hybridization buffer [40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.4), 1 mM EDTA, 0.4 M NaCl, 80% formamide]. The buffer contained excess probe. A 200-nucleotide (nt) [α-32P]CTP-labelled minus-strand lacZ probe generated from a T7 RNA polymerase runoff transcript of HincII-digested pJD86 (16) and a 241-nt [α-32P]CTP-labelled minus-strand CYH2 probe generated from an SP6 RNA polymerase runoff transcript of HincII-digested pGEM-42-CYH2 (25) were used as probes to measure the effects of PAP on mRNA abundances in vivo. The CYH2 gene encodes the constitutively expressed ribosomal protein L29 (39) and was used as a loading control. A 300-nt [α-32P]CTP-labelled minus-strand luciferase probe generated from a T7 RNA polymerase runoff transcript of pLUC0 digested with ClaI was used to measure the effects of PAP on mRNA abundances in vitro. Hybridization was carried out initially at 70°C for 15 min and then at 50°C for 5 h. RNase T1 and RNase A (200 μl of a 300 mM NaCl–10 mM Tris [pH 7.5]–5 mM EDTA buffer containing 286 U of RNase T1 and 0.72 μg of RNase A) were added to the annealed RNAs, and the mixture was incubated at room temperature for 15 min. Seventeen microliters of a proteinase K-sodium dodecyl sulfate solution (1:4 ratio of proteinase K at 10 mg/ml to 10% sodium dodecyl sulfate) was added, and the reaction mixtures were incubated for 15 min at 37°C. The reaction mixtures were extracted with an equal volume of phenol-chloroform equilibrated to pH 7.5 and were centrifuged for 5 min, the aqueous layers were removed, carrier tRNA (20 μg) was added, and nucleic acids were precipitated with 2.5 volumes of ethanol at −20°C for 15 min. Dried pellets were resuspended in 20 μl of loading dye and denatured at 100°C for 3 min, and RNA samples were electrophoretically separated through 6% polyacrylamide denaturing gels, which were dried and exposed for autoradiography. The protected RNA fragments were quantitated with a Bio-Rad model G-670 imaging densitometer. The relative lacZ mRNA abundances were calculated by determining the ratio of lacZ- to CYH2-protected RNA fragment band intensities.
RESULTS
In vivo frameshifting.
RIP-mediated depurination of the large ribosomal subunit RNA results in increased susceptibility of the RNA sugar-phosphate backbone to hydrolysis at the depurination site. Hence, when depurinated rRNA is treated with aniline, a small fragment which corresponds to the 3′-terminal end of the large rRNA is released (3, 26, 38). Previous results demonstrated that when ribosomes from wild-type yeast were incubated with PAP and the rRNA extracted from these ribosomes was treated with aniline, a 367-nt RNA fragment was released (43). When ribosomes were isolated from yeast expressing PAP (pNT123) or the active-site mutant PAP (pNT123-2) and subjected to the RNA depurination assay, the diagnostic 367-nt RNA fragment was generated from cells expressing PAP but not from cells expressing the active-site mutant PAP (43). These results indicated that yeast ribosomes are depurinated in vivo by expression of wild-type PAP but not by expression of the active-site mutant PAP (43).
The effects of PAP on programmed −1 and +1 ribosomal frameshifting were assayed with PSY1 cells cotransformed with a series of TRP1- and URA3-based vectors (Fig. 1A). The TRP1-based vectors were used to assay programmed ribosomal frameshifting. pF8 (12) and pJD104 (2) were used to measure β-galactosidase activities produced as a consequence of L-A sequence-directed −1 and of Ty1 sequence-directed +1 ribosomal frameshift events, respectively. pT125 (12) was used as the 0 frame β-galactosidase standard. The efficiency of programmed ribosomal frameshifting was determined by calculating the ratio of −1 frame or +1 frame to 0 frame β-galactosidase activities and multiplying by 100 (12). The URA3-based vectors harbored PAP (pNT123), the active-site mutant PAP (pNT123-2), or a control. In pNT123 and pNT123-2, transcription of the genes encoding PAP was under the control of the GAL1 promoter. To induce the production of PAP, overnight cultures were split into selective media containing 2% galactose as the carbon source. Cells grown in 2% raffinose were used as uninduced controls. After 5 h of growth, β-galactosidase activities were determined, and the efficiencies of ribosomal frameshifting were calculated.
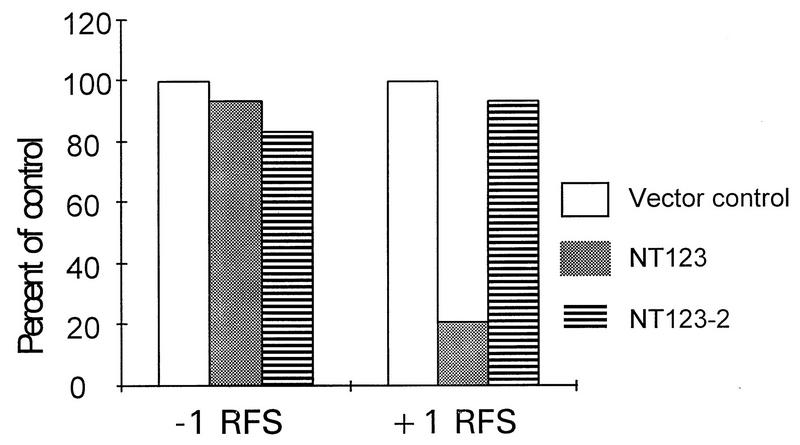
The results of these experiments are summarized in Table 1. Neither the presence of galactose alone nor the induction of PAP (pNT123) by galactose affected the overall translation of the 0 frame control reporter mRNA. Similarly, neither of these factors had any influence on translation of the −1 ribosomal frameshift reporter mRNA or on −1 ribosomal frameshifting efficiency. Interestingly, both growth in galactose and the induction of PAP affected the translation of the +1 ribosomal frameshift reporter mRNA (pJD104). Galactose alone increased the translation of this reporter 2.26-fold above the raffinose control. Consequently, the efficiency of +1 ribosomal frameshifting was stimulated 2.28-fold under these conditions. This trend was observed in both the absence of PAP (PSY1) and the presence of the active-site mutant PAP (pNT123-2). Induction of PAP reversed this trend, resulting in decreased +1 frame reporter β-galactosidase activity (Gal/Raf ratio, 0.46) and a corresponding decrease in +1 ribosomal frameshifting efficiency (Gal/Raf ratio, 0.47). As a true monitor of the overall effects of PAP on programmed ribosomal frameshifting, the ratios of frameshifting in PAP-induced and uninduced cells (Gal/Raf in pNT123 and pNT123-2) were normalized to those in control cells (Gal/Raf in PSY1). As indicated in Table 1 and as shown in Fig. 2, active PAP inhibited Ty1 sequence-directed +1 ribosomal frameshifting to approximately 21% of wild-type levels ([0.47/2.28] × 100). The expression of inactive PAP (pNT123-2) had no such effect ([2.13/2.28] × 100, or 93%). With respect to programmed −1 ribosomal frameshifting, there were no significant differences in the ratios between cells expressing the active and cells expressing the inactive forms of PAP.
TABLE 1.
In vivo effects of PAP
| Plasmid and growth conditions | β-Galactoside activity in the presence ofa:
|
% −1 RFSb | % +1 RFSc | ||
|---|---|---|---|---|---|
| pT125 (0) | pF8 (−1) | pJD104 (+1) | |||
| PSY1 (vector) | |||||
| Raffinosed | 38.8 ± 1.52 | 1.50 ± 0.28 | 0.69f ± 0.15 | 3.9 | 1.8f |
| Galactosed | 38.1 ± 2.37 | 1.61 ± 0.45 | 1.56f ± 0.33 | 4.2 | 4.1f |
| Gal/Raf (% vector control)e | 0.98 | 1.07 | 2.26 | 1.07 (100) | 2.28 (100) |
| pNT123 (active PAP) | |||||
| Raffinose (−PAP) | 43.35 ± 1.48 | 1.26 ± 0.15 | 0.84g ± 0.14 | 2.9 | 1.9g |
| Galactose (+PAP) | 41.54 ± 1.83 | 1.19 ± 0.16 | 0.37g ± 0.05 | 2.9 | 0.89g |
| Gal/Raf (% vector control) | 0.96 | 0.94 | 0.46 | 1.0 (93) | 0.47 (21) |
| pNT123-2 (inactive PAP) | |||||
| Raffinose (−PAP) | 33.60 ± 1.16 | 1.26 ± 0.25 | 0.54f ± 0.06 | 3.8 | 1.6f |
| Galactose (+PAP) | 34.06 ± 2.70 | 1.15 ± 0.29 | 1.16f ± 0.30 | 3.4 | 3.4f |
| Gal/Raf (% vector control) | 1.01 | 0.91 | 2.15 | 0.89 (83) | 2.13 (93) |
β-Galactosidase activities, in arbitrary units, were used as a measure of translational competence. In pT125, lacZ is in the 0 frame. pF8 and pJD104 are the −1 and +1 ribosomal frameshift reporter plasmids, respectively; the reading frames of the lacZ reporter genes are indicated in parentheses.
Percent −1 ribosomal frameshifting (RFS) was calculated by multiplying the ratio of pF8/pT125 β-galactosidase activities by 100.
Percent +1 ribosomal frameshifting was calculated by multiplying the ratio of pJD104/pT125 β-galactosidase activities by 100.
Cells were grown in selective media containing either 2% raffinose (PAP uninduced [−PAP]) or 2% galactose (PAP induced [+PAP]) for 5 h.
The Gal/Raf ratio measures the effect of PAP induction on β-galactosidase activities and programmed ribosomal frameshifting; in percent vector control, the PSY1 Gal/Raf ratio is the denominator and the Gal/Raf ratios of the test plasmids are the numerators.
t < 0.05 (Student’s t test).
t < 0.01 (Student’s t test).
FIG. 2.
Effects of PAP on programmed ribosomal frameshifting in vivo. Cultures of yeast cells harboring frameshift indicator vectors (0 frame control and −1 or +1 ribosomal frameshift test vectors) and either pNT123 (galactose-inducible wild-type PAP), pNT123-2 (galactose-inducible active-site mutant PAP), or no other vector were split into selective media containing either 2% galactose (induced) or 2% raffinose (uninduced) and incubated for 5 h. β-Galactosidase activities were then determined. The efficiencies of programmed ribosomal frameshifting are taken from Table 1. To obtain the percent of control, the ratios of frameshifting in PAP-induced and uninduced cells (Gal/Raf in pNT123 and pNT123-2; Table 1) were normalized to those in control cells (Gal/Raf in PSY1; Table 1). RFS, ribosomal frameshift.
In vitro frameshifting.
The efficiencies of −1 and +1 ribosomal frameshifting were examined in vitro with synthetic luciferase-based reporters (Fig. 1B) in translationally competent rabbit reticulocyte extracts. The efficiencies of ribosomal frameshifting were determined by measuring the ratios of light units produced by the frameshift reporter mRNAs (Luc−1 and Luc+1) and dividing them by those produced by the 0 frame control mRNA (Luc0). The effects of PAP on −1 and +1 ribosomal frameshifting were examined by the addition of purified PAP to the translation extracts. The results of these experiments are summarized in Table 2. Although the addition of purified PAP inhibited overall translation up to approximately 40% (compare 0 frame with no PAP to 0 frame with 4 pg of PAP), the luciferase activity of the −1 frame construct decreased in parallel with that of the 0 frame construct such that there were no changes in −1 ribosomal frameshifting efficiencies. However, the luciferase activity generated from the +1 frame construct decreased much more rapidly than that generated from the 0 frame construct such that there was an overall decrease in +1 ribosomal frameshifting efficiency, from 15.6% (no PAP) to only 1.1% (4 pg of PAP), an approximate 94% inhibition of +1 ribosomal frameshifting. Figure 3 shows a plot of the ratios of programmed ribosomal frameshifting as a percentage of the 0 frame control value. The addition of PAP specifically inhibited +1 ribosomal frameshifting. These data indicate that PAP has a specific and direct effect on the mechanisms that govern +1 ribosomal frameshifting.
TABLE 2.
In vitro effects of PAP
| PAP (pg) | Light units in the presence of the indicated constructa:
|
% −1 RFSb | % +1 RFSb | ||
|---|---|---|---|---|---|
| 0 frame | −1 frame | +1 frame | |||
| None | 481.1 ± 8.39 | 58.3 ± 0.95 | 72.1 ± 2.90 | 12.1 | 15.6 |
| 1 | 476.2 ± 2.42 | 56.3 ± 3.31 | 62.4 ± 1.18 | 11.8 | 13.1 |
| 2 | 395.2 ± 3.61 | 47.3 ± 4.79 | 31.8 ± 3.13 | 12.0 | 8.0 |
| 3 | 335.8 ± 16.0 | 41.5 ± 1.00 | 10.7 ± 2.19 | 12.4 | 3.2 |
| 4 | 290.7 ± 8.39 | 34.1 ± 2.19 | 3.2 ± 0.18 | 11.7 | 1.1 |
For 0 frame, −1 frame, and +1 frame constructs, 20 ng each of synthetic Luc0, Luc−1, and Luc+1 mRNAs, respectively, were used.
Percent ribosomal frameshifting (RFS) was calculated by multiplying the ratio of light units generated by Luc−1/Luc0 or Luc+1/Luc0 by 100.
FIG. 3.
Effects of PAP in vitro on −1 and +1 ribosomal frameshifting (RFS) efficiencies. Translation-competent rabbit reticulocyte lysates were preincubated at 28°C for 15 min with the indicated amounts of purified PAP. Then, 20 ng of each of the synthetic luciferase reporter mRNAs (0 frame control and −1 or +1 ribosomal frameshift test mRNAs) was added to the PAP-lysate mixture and incubated at 28°C for 45 min. Luciferase activities were then determined by luminometry. Ribosomal frameshifting efficiencies were calculated by determining the ratios of luciferase activities produced by the test mRNAs to the 0 frame control value. Percent of control represents the efficiency of ribosomal frameshifting plotted as a percentage of that in the no-PAP control.
Nuclease protection assays.
The −1 and +1 lacZ reporter mRNAs both contain in-frame nonsense codons approximately 200 nt from their 5′ ends, followed by over 3.1 kb of lacZ message, and these mRNAs are intrinsically unstable (9). In contrast, the 0 frame control mRNA produced from pT125 is very stable. Thus, changes in the stabilities of either of the reporter mRNAs might alter the amount of β-galactosidase present in the cell, which in turn might affect the apparent efficiency of programmed ribosomal frameshifting. In order to examine these possibilities, nuclease protection assays were performed. A 200-nt 32P-labelled minus-strand RNA corresponding to the 3′ end of the lacZ mRNA was transcribed and ybridized, in the presence of excess probe, with total RNA extracted from induced and uninduced cells harboring the frameshift vectors (pT125, pF8, or pJD104) and pNT123, pNT123-2, or vector alone. A 260-nt 32P-labelled minus-strand CYH2 mRNA, which encodes the constitutively expressed ribosomal protein L29 (39), served as the internal loading control for each sample. Samples were electrophoretically separated, and the intensities of the protected bands were determined by scanning densitometry. The ratios of the intensities of the 200-nt lacZ band to the 241-nt CYH2 band were determined, and these ratios were used as relative measures of steady-state lacZ mRNA abundance. No differences were noted in the ratios of CYH2 mRNA to 0 frame, −1 frame, or +1 frame lacZ mRNAs for cells grown with galactose versus raffinose (data not shown). The effects of PAP on the 0 frame, −1 frame, and +1 frame luciferase reporter mRNAs were also assayed in vitro. Increasing concentrations of PAP had no effect on luciferase mRNA stabilities (data not shown). Thus, the observed decreases in +1 ribosomal frameshifting efficiency in PAP-induced cells in vivo and upon the addition of PAP to translationally competent reticulocyte lysates in vitro are not due to the preferential destabilization of the +1 lacZ reporter mRNA in either of these systems. These data support the conclusion that PAP specifically inhibits +1 ribosomal frameshifting.
Retrotransposition and maintenance of the killer phenotype.
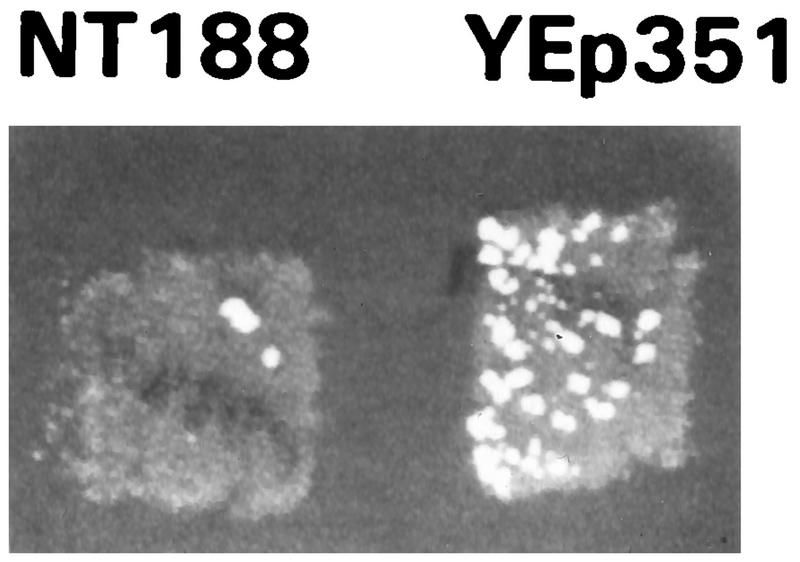
The effects of PAP on the ability of cells to propagate two different sets of ribosomal frameshift-dependent viruses were examined. To examine the effects of PAP on Ty1 retrotransposition, galactose was used to coinduce the transcription of PAP and of a HIS3-tagged Ty1 mRNA from LEU2-selectable pNT188 and URA3-selectable pTy1HIS3AI plasmids, respectively. Control cells harboring pTy1HIS3AI contained the LEU2-selectable YEp351 plasmid. In pTy1HIS3AI, transcription of a Ty1 cDNA is induced by galactose (10). This cDNA also contains the yeast HIS3 gene in the reverse orientation and into which an artificial intron has been introduced. The intron is spliced out of the galactose-induced transcript, and the spliced transcript is packaged and copied into Ty1 cDNA in Ty1 viruslike particles. These enter the nucleus, and the cDNA is integrated into a chromosomal locus. The endogenous HIS3 promoter can then transcribe this gene. Conversion of his3 cells from His− to His+ is used as an indicator of Ty1 retrotransposition from the plasmid to a host cell chromosome. The entire process is dependent on viral particle morphogenesis, which in turn is extremely sensitive to alterations in programmed +1 ribosomal frameshifting efficiencies (reviewed in reference 11). Thus, changes in Ty1 retrotransposition frequencies are indicative of the effect of PAP on programmed +1 ribosomal frameshifting efficiencies. Figure 4 shows, on a qualitative level, that Ty1 retrotransposition frequencies are substantially reduced in the presence of PAP. To quantitate the inhibitory effect of PAP on Ty1 retrotransposition frequencies, cells that had undergone galactose induction were grown in liquid medium and spread onto H−his medium at densities ranging from 104 to 108 CFU. Ty1 retrotransposition frequencies were directly calculated by determining the ratio of His+ CFU to the total number of CFU per plate. Retrotransposition frequencies in control cells (YEp351) were 1.31 × 10−2, whereas those in cells expressing PAP (pNT188) were 8.6 × 10−5. Thus, the inhibition of Ty1 retrotransposition by PAP is greater than 99%.
FIG. 4.
PAP inhibits Ty1 retrotransposition. PSY1 cells were transformed with galactose-inducible PAP (pNT188) or vector (YEp351) and pTy1HIS3AI. Cells were grown at 24°C on medium containing 2% galactose but lacking uracil and leucine for 4 days, replica plated at 30°C to medium containing 2% dextrose but lacking uracil and leucine, and subsequently replica plated to medium containing 2% dextrose but lacking histidine. Growth of the colonies was indicative of Ty1 retrotransposition.
L-A and M1 were introduced into PSY1 cells harboring either pNT188 or YEp351 as described in Materials and Methods. PAP was induced in these cells as described above for 4 days. Cells were subsequently grown in liquid cultures, and approximately 100 CFU of cells was spread onto H−leu medium (four plates each). Individual colonies were allowed to grow at 30°C and then were replica plated to 4.7MB killer indicator plates. No qualitative or quantitive differences were observed between the PAP-induced and vector control cells with regard to their killer phenotypes (data not shown). These results suggest that PAP may not have an effect on killer phenotype maintenance or, if there is an effect, that it is too subtle to be detected by these assays.
DISCUSSION
The translocation step of protein synthesis divides the population of ribosomes between those having both P and A sites occupied by a peptidyl-tRNA and an aminoacyl-tRNA, respectively (the substrate for L-A-promoted −1 ribosomal frameshifting), and those having only the P site occupied by a peptidyl-tRNA (the substrate for Ty1-promoted +1 ribosomal frameshifting). We interfered with translocation by using the 29-kDa PAP isolated from P. americana. PAP inhibits EF-2-mediated translocation by catalytically depurinating a specific adenine residue in the α-sarcin loop of eukaryotic 28S rRNA (19, 28). We previously demonstrated that yeast cells lack endogenous RIP activity but that recombinant PAP is enzymatically active when expressed in intact yeast cells and purified PAP is active in yeast cell extracts (43). Here, we observed that the in vivo induction of PAP had a cytostatic rather than a cytotoxic effect. Cell growth was strongly inhibited when pNT123- or pNT188-harboring cells were grown in the presence of galactose. However, these cells resumed normal growth phenotypes upon transfer to glucose. Thus, PAP did not kill the cells per se but rather arrested cell growth and protein synthesis.
We investigated the effects of plasmid-borne, galactose-inducible PAP and an active-site mutant PAP (43) on programmed ribosomal frameshifting efficiencies and virus propagation by using both intact cells and translationally competent rabbit reticulocyte extracts. PAP specifically decreased +1 ribosomal frameshifting efficiencies to approximately 20% of wild-type levels in vivo (Table 1 and Fig. 2). PAP also strongly and specifically inhibited +1 ribosomal frameshifting in an in vitro translation system (Table 2 and Fig. 3). There were no corresponding changes in −1 ribosomal frameshifting efficiencies or in the relative steady-state abundances of +1 ribosomal frameshift reporter mRNAs in the presence of PAP in either the in vivo or the in vitro system. These data demonstrated that PAP acts directly to specifically inhibit Ty1-directed programmed +1 ribosomal frameshifting.
Our research has focused on characterizing the molecular mechanisms underlying programmed ribosomal frameshifting. We previously used genetic analyses to identify nine complementation groups of yeast mof (maintenance of frame) mutants having increased efficiencies of −1 ribosomal frameshifting (9, 15–17). Programmed ribosomal frameshifting is a kinetic phenomenon in which ribosomal pauses at the Ty1 and L-A slippery sites drive the +1 and −1 frameshifts, respectively. A kinetic pause model of programmed ribosomal frameshifting predicts that changes in the length of time that ribosomes are paused at these frameshift signals should result in changes in the probability of ribosomal slippage. Since the ribosomal pauses occur during the course of translational elongation, the kinetic parameters that might influence these processes must be defined in the context of the three limiting steps of the translational elongation cycle (reviewed in reference 27). These are (i) recognition and insertion into the ribosomal A site of cognate aminoacyl-tRNA by EF-1; (ii) peptide transfer (mediated by the ribosomal peptidyl transferase center); and (iii) translocation mediated by EF-2. We previously targeted the first step by using mutants of TEF2 (encoding elongation factor 1α), demonstrating that specific TEF2 mutants affect either −1 or +1 ribosomal frameshifting efficiencies (13). The peptidyl transferase inhibitors anisomycin and sparsomycin were used to target the second step; these inhibitors specifically altered −1 ribosomal frameshifting efficiencies and promoted the loss of both L-A and M1 (14). In this report, we used PAP to target the translocation step. The model predicts that translocation defects should decrease the ratio of ribosomes with an unoccupied A site (substrate for Ty1-directed +1 ribosomal frameshifting) to an EF-1–aminoacyl-tRNA–GTP ternary complex. Thus, the A sites of ribosomes that have passed through the translocation step should be rapidly occupied by the ternary complex, effectively decreasing the length of time that these ribosomes are paused at the Ty1 +1 frameshift signal. This process should lead to a decrease in the overall probability of ribosomal slippage. As predicted by the model, PAP decreased the efficiencies of programmed +1 ribosomal frameshifting in both intact cells and an in vitro assay system. Further, a mutant PAP with an inactivated active site (pNT123-2), which does not inhibit translocation, had no such effect. Notably, PAP had no effect on −1 ribosomal frameshifting efficiencies. PAP-inhibited ribosomes should have both their A and P sites occupied by tRNAs and, as such, could theoretically be substrates for programmed −1 ribosomal frameshifting. The inability of PAP to affect this process suggests that ribosomes that have passed through the peptidyl transfer step but that have not translocated are no longer capable of shifting in the −1 direction. Thus, the results presented here provide the first evidence that programmed −1 ribosomal frameshifting does not occur after the peptidyl transferase reaction and narrow the window during which −1 ribosomal frameshifting can occur.
It has been demonstrated that increasing the efficiency of +1 ribosomal frameshifting by starving cells for polyamines (2) or by disrupting the gene encoding tRNACUUArg (HSX1) (32) strongly inhibits Ty1 retrotransposition. Similarly, decreasing the efficiency of +1 ribosomal frameshifting by overexpressing tRNACUUArg also inhibits Ty1 retrotransposition (46). Here we demonstrate that PAP-promoted inhibition of +1 ribosomal frameshifting also has a strong inhibitory effect on this process (Table 2 and Fig. 3). Although it has been reported that PAP can inhibit the replication of human immunodeficiency virus, a virus that requires a −1 ribosomal frameshift, we did not observe any effect on either L-A sequence-directed −1 ribosomal frameshifting or the ability of cells to maintain the M1-dependent killer virus phenotype. A critical difference between assays for Ty1 retrotransposition and those for killer phenotype maintenance is that in the former, the cells start out with no Ty1 viral particles, whereas in the latter, the cells contain approximately 104 L-A and M1 viral particles. Thus, with regard to the killer assay, the cells are already superinfected, whereas the Ty1 retrotransposition assay requires a de novo infection by inducing transcription and translation of the Ty1 virus. It is possible that PAP exerts a general effect on translation that interferes with the establishment of an infection and that the killer assay is not sensitive enough to observe this effect. Alternatively, PAP may specifically recognize and promote the destabilization of 7-methyl-Gppp-capped or polyadenylated viral RNAs. The Ty1 mRNA used in our study was capped. In contrast, L-A and M1 mRNAs may not be suitable substrates for PAP because they are not capped or polyadenylated (reviewed in reference 45). Recently, we reported that a nontoxic C-terminal deletion mutant of PAP which does not depurinate host ribosomes inhibits viral infection, suggesting that the antiviral activity of PAP is not solely due to the depurination of host ribosomes (43). The results reported here indicate that PAP specifically inhibits Ty1 but not L-A and M1, providing further evidence that the antiviral activity of PAP is not due solely to a general inhibition of host protein synthesis.
ACKNOWLEDGMENTS
We thank Peter Day and Terri Goss Kinzy for critical reading of the manuscript and Annette Chiang for constructing pNT188.
This work was supported in part by grants to J.D.D. from the Foundation of the University of Medicine and Dentistry of New Jersey (grant 8-97) and the State of New Jersey Commission on Cancer Research (grant 96-62-CC2-00) and by National Science Foundation (NSF) grant NSFMCB96-31308 to N.E.T. B.P. was supported by the NSF Research Experience for the Undergraduates (REU) Program.
REFERENCES
- 1.Aron G M, Irvin J D. Inhibition of herpes simplex virus multiplication by the pokeweed antiviral protein. Antimicrob Agents Chemother. 1980;17:1032–1033. doi: 10.1128/aac.17.6.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasundaram D, Dinman J D, Wickner R B, Tabor C W, Tabor H. Spermidine deficiency increases +1 ribosomal frameshifting efficiency and inhibits Ty1 retrotransposition in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:172–176. doi: 10.1073/pnas.91.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass H W, Webster C, O’Brian G R, Roberts J K, Boston R S. A maize ribosome-inactivating protein is controlled by the transcriptional activator Opaque-2. Plant Cell. 1992;4:225–234. doi: 10.1105/tpc.4.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I. Ribosomal frameshifting on viral RNAs. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 5.Brierley I A, Dingard P, Inglis S C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brierley I A, Jenner A J, Inglis S C. Mutational analysis of the “slippery sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brierley I A, Rolley N J, Jenner A J, Inglis S C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bussey H. K1 killer toxin, a pore-forming protein from yeast. Mol Microbiol. 1991;5:2339–2343. doi: 10.1111/j.1365-2958.1991.tb02079.x. [DOI] [PubMed] [Google Scholar]
- 9.Cui Y, Dinman J D, Peltz S W. mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curcio M J, Garfinkel D J. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinman J D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinman J D, Icho T, Wickner R B. A −1 ribosomal frameshift in a double-stranded RNA virus forms a Gag-Pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinman J D, Kinzy T G. Translational misreading: mutations in translation elongation factor 1α differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA. 1997;3:870–881. [PMC free article] [PubMed] [Google Scholar]
- 14.Dinman J D, Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Peptidyl transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficiencies: development of model systems. Proc Natl Acad Sci USA. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinman J D, Wickner R B. Ribosomal frameshifting efficiency and Gag/Gag-Pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinman J D, Wickner R B. Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered −1 ribosomal frameshifting efficiencies. Genetics. 1994;136:75–86. doi: 10.1093/genetics/136.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinman J D, Wickner R B. 5S rRNA is involved in fidelity of translational reading frame. Genetics. 1995;141:95–105. doi: 10.1093/genetics/141.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donahue T F, Cigan A M. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol. 1988;8:2955–2963. doi: 10.1128/mcb.8.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endo Y, Tsurugi K, Lambert J M. The site of action of six different ribosome-inactivating proteins from plants on eukaryotic ribosomes: the RNA N-glycosidase activity of the proteins. Biochem Biophys Res Commun. 1988;150:1032–1036. doi: 10.1016/0006-291x(88)90733-4. [DOI] [PubMed] [Google Scholar]
- 20.Farabaugh P J. Post-transcriptional regulation of transposition by Ty retrotransposons of Saccharomyces cerevisiae. J Biol Chem. 1995;270:10361–10364. doi: 10.1074/jbc.270.18.10361. [DOI] [PubMed] [Google Scholar]
- 21.Fujimura T, Wickner R B. Gene overlap results in a viral protein having an RNA binding domain and a major coat protein domain. Cell. 1988;55:663–671. doi: 10.1016/0092-8674(88)90225-5. [DOI] [PubMed] [Google Scholar]
- 22.Gafner J, De Robertis E M, Phillippsen P. Delta sequences in the 5′ non-coding region of yeast tRNA genes. EMBO J. 1983;2:583–591. doi: 10.1002/j.1460-2075.1983.tb01467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallie D R, Feder J N, Schimke R T, Walbot V. Post-transcriptional regulation in higher eukaryotes: the role of the reporter gene in controlling expression. Mol Gen Genet. 1991;288:258–265. doi: 10.1007/BF00282474. [DOI] [PubMed] [Google Scholar]
- 24.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 25.Hagan K W, Ruiz-Echevarria M J, Quan Y, Peltz S W. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartley M R, Legname G, Osborn R, Chen Z, Lord J M. Single-chain ribosome inactivating proteins from plants depurinate Escherichia coli 23S ribosomal RNA. FEBS Lett. 1991;290:65–68. doi: 10.1016/0014-5793(91)81227-y. [DOI] [PubMed] [Google Scholar]
- 27.Hershey J W B. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 28.Hur Y, Hwang D-J, Zoubenko O, Coetzer C, Uckun R M, Tumer N E. Isolation and characterization of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: identification of residues important for toxicity. Proc Natl Acad Sci USA. 1995;92:8448–8452. doi: 10.1073/pnas.92.18.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Icho T, Wickner R B. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem. 1989;264:6716–6723. [PubMed] [Google Scholar]
- 30.Jacks T, Madhani H D, Masiraz F R, Varmus H E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacks T, Varmus H E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 32.Kawakami K, Paned S, Faioa B, Moore D P, Boeke J D, Farabaugh P J, Strathern J N, Nakamura Y, Garfinkel D J. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993;135:309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiledjian M, Kadesch T. Post-transcriptional regulation of the human liver/bone/kidney alkaline phosphatase gene. J Biol Chem. 1991;266:4207–4213. [PubMed] [Google Scholar]
- 34.Montanaro L, Sperti S, Mattioli A, Testoni G, Stirpe F. Inhibition by ricin of protein synthesis in vitro. Inhibition of the binding of elongation factor 2 and of adenosine diphosphate-ribosylated elongation factor 2 to ribosomes. Biochem J. 1975;146:127–131. doi: 10.1042/bj1460127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morikawa S, Bishop D H L. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osborn R W, Hartley M R. Dual effects of the ricin A chain on protein synthesis in rabbit reticulocyte lysate. Inhibition of initiation and translocation. Eur J Biochem. 1990;193:401–407. doi: 10.1111/j.1432-1033.1990.tb19353.x. [DOI] [PubMed] [Google Scholar]
- 37.Somogyi P, Jenner A J, Brierley I A, Inglis S C. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stirpe F, Bailey S, Miller S P, Bodley J W. Modification of ribosomal RNA by ribosome-inactivating proteins from plants. Nucleic Acids Res. 1988;16:1349–1357. doi: 10.1093/nar/16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stocklein W, Piepersberg W. Altered ribosomal protein L29 in a cycloheximide-resistant strain of S. cerevisiae. Curr Genet. 1980;1:177–183. doi: 10.1007/BF00390941. [DOI] [PubMed] [Google Scholar]
- 40.TenDam E, Pleij K, Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992;31:11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- 41.Tomlinson J A, Walker V M, Flewett T H, Barclay G R. The inhibition of infection by cucumber mosaic virus and influenza virus by extracts from Phytolacca americana. J Gen Virol. 1974;22:225–232. doi: 10.1099/0022-1317-22-2-225. [DOI] [PubMed] [Google Scholar]
- 42.Tu C, Tzeng T-H, Bruenn J A. Ribosomal movement impeded at a pseudoknot required for ribosomal frameshifting. Proc Natl Acad Sci USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tumer N E, Hwang D-J, Bonness M. C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc Natl Acad Sci USA. 1997;94:3866–3871. doi: 10.1073/pnas.94.8.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ussery M A, Irvin J D, Hardesty B. Inhibition of poliovirus replication by a plant antiviral peptide. Ann N Y Acad Sci. 1977;284:431–440. doi: 10.1111/j.1749-6632.1977.tb21979.x. [DOI] [PubMed] [Google Scholar]
- 45.Wickner R B. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Boeke J D. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc Natl Acad Sci USA. 1990;87:8360–8364. doi: 10.1073/pnas.87.21.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarling J M, Moran P A, Haffar O, Sias J, Richman D D, Spina C A, Myers D E, Kuelbeck V, Ledbetter J A, Uckun F M. Inhibition of HIV replication by pokeweed antiviral protein targeted to CD4+ cells by monoclonal antibodies. Nature. 1990;347:92–95. doi: 10.1038/347092a0. [DOI] [PubMed] [Google Scholar]