Abstract
Colorectal cancer (CRC) remains a major global health burden with high mortality rates, underscoring the need for effective therapies. This study explores the acetylation characteristics in CRC using single-cell RNA sequencing (scRNA-seq) and weighted gene co-expression network analysis (WGCNA), assessing their relationship with prognosis and the immune microenvironment. We analyzed two scRNA-seq datasets from the GEO database to identify distinct cell subtypes. Acetylation activity scores were calculated using the ssGSEA method. A WGCNA was constructed to identify gene modules associated with acetylation. An acetylation-related prognostic signature (ARPS) was developed, and its clinical significance was evaluated through survival analysis and immune landscape characterization. Acetylation activity was significantly elevated in epithelial, endothelial, and stromal cells. Based on the results of scRNA-seq, WGCNA identified 169 acetylation-related genes. Intersection with 1,691 acetylation-related differentially expressed genes (DEGs) yielded 131 common genes. Combining clinical data with the expression profiles of these genes, we employed 101 machine learning algorithms to develop an ARPS that accurately predicts the prognosis of CRC patients. Low-risk patients showed increased infiltration of immune cells, enhanced immune function, and better responses to immunotherapy. These findings underscore the clinical significance of acetylation features in CRC prognosis and immune response, highlighting their potential as biomarkers and therapeutic targets.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-21081-8.
Keywords: Colorectal cancer, Acetylation, Tumor microenvironment, Prognostic model, Immunotherapy
Subject terms: Colorectal cancer, Immunotherapy, Tumour immunology, Risk factors
Introduction
Colorectal cancer (CRC) remains a significant global health burden, ranking as the third most common cause of cancer mortality worldwide1. Despite advancements in early detection and therapies, including surgical resection, chemotherapy, and targeted therapies, the prognosis for advanced CRC remains poor due to tumor heterogeneity and the complex tumor microenvironment (TME)2. Therefore, understanding CRC’s molecular biology is crucial for developing more effective treatment modalities and improving patient outcomes.
Acetylation-related processes in the TME have gained significant attention in the context of CRC3–6. Acetylation, a critical post-translational modification, regulates gene expression, protein stability, and cellular signaling pathways central to tumor biology7. Altered acetylation patterns influence the interactions between tumor cells and the TME, modulate immune responses and impactcancer progression7–10. These modifications may also affect the efficacy of immunotherapy, suggesting that mapping the acetylation landscape within the TME could provide insights into tumor behavior and therapeutic responses11–13. The tumor microenvironment (TME) is a dynamic ecosystem where acetylation modifications critically regulate stromal-immune crosstalk14–16. Recent studies reveal that acetyltransferases reprogram cancer-associated fibroblasts (CAFs) to promote extracellular matrix remodeling through metabolic alterations, facilitating immune exclusion in lung cancer15. Histone acetylation in dendritic cells dictates antigen presentation capacity and T-cell priming efficiency in pan-cancer models14. Post-translational modifications (PTMs) like B4GALT2-mediated glycosylation create physical barriers to lymphocyte infiltration, a mechanism conserved in immune-cold tumors16. In CRC specifically, acetylation of ELMO1 modulates Rac1-dependent stromal invasion3, while KAT7-mediated crotonylation competes with acetylation to drive tumorigenesis4. This suggests acetyl-CoA flux acts as a metabolic switch coordinating TME immunosuppression. Our study leverages single-cell resolution to decode this axis in CRC. The exploration of acetylation-related biomarkers may identify novel therapeutic targets and prognostic indicators, enabling personalized CRC management. By delving into the role of acetylation in CRC, researchers not only advance our understanding of the disease but also open new avenues for developing innovative therapeutic strategies aimed at improving patient prognosis and survival rates.
This study aims to characterize acetylation features in CRC and their association with the prognosis and the immune microenvironment through a stepwise framework: (1) Single-cell mapping of acetylation heterogeneity across TME compartments using scRNA-seq to resolve cellular diversity and identify context-specific acetylation patterns; (2) Network-based identification of conserved acetylation modules through WGCNA, exploring gene co-expression patterns and acetylation-associated functional units; (3) Machine learning integration for clinical translation, constructing a prognostic model to derive novel biomarkers and therapeutic targets for personalized strategies; and (4) Experimental validation of therapeutic implications. This integrated approach bridges molecular features with clinical outcomes to advance personalized CRC management, enhancing understanding of molecular mechanisms and potentially improving patient outcomes through tailored therapeutics.
Materials and methods
Acquisition of patients’ datasets
The transcriptomic data and clinical information of CRC were collected from multiple databases. Two scRNA-seq datasets (GSE132465 and GSE144735) were downloaded from the Gene Expression Omnibus (GEO) database, comprising 23 and 12 CRC samples, respectively. Bulk RNA-seq data and clinical information from The Cancer Genome Atlas (TCGA) included 51 normal and 647 CRC samples. During processing, we excluded samples with incomplete clinical information or a survival time of less than 30 days. To further validate our findings, we used the GSE39582 dataset from the GEO, which includes 562 tumor samples.
Single-cell quality control and cell type annotation
scRNA-seq data was processed using the “Seurat” package in R. Preliminary quality control protocols were established to exclude subpar cells, ensuring the retention of those exhibiting gene expression levels ranging from 300 to 7,000 genes, while also maintaining mitochondrial gene composition under 10%. Data were normalized using the LogNormalize method, and the top highly variable genes were identified using the FindVariableFeatures function, retaining 3,000 genes for subsequent analysis. The Harmony algorithm was employed to address batch effects among the samples. Dimensionality reduction was achieved through principal component analysis (PCA) and t-distributed Stochastic Neighbor Embedding (tSNE). Clusters were delineated using FindNeighbors and FindClusters functions. Cells were then annotated according to the “SingleR” package and manual curation. Marker genes for each cluster was determined via FindAllMarkers function.
Single sample gene set enrichment analysis (ssGSEA)
To explore the significance of acetylation in the TME, this study utilized ssGSEA, a sophisticated analytical tool designed to evaluate the enrichment of specified gene sets in individual samples based on gene expression data. The list of acetylation-related genes (ARGs) allowed for the computation of acetylation scores for each cell in the TCGA cohort. Samples were categorized into two groups based on acetylation scores, specifically high and low, utilizing the optimal score thresholds. To evaluate the differential expression between these groups, the FindAllMarkers function was employed, applying a log2 fold change threshold of 0.35 alongside a minimum detection rate of 35%. The FindAllMarkers function was utilized to select marker genes of high and low acetylation score groups. The resulting p-values were adjusted for multiple testing using the Benjamini-Hochberg false discovery rate (FDR) method, with an FDR < 0.05 considered significant. This comprehensive approach highlights the multifaceted role of acetylation and immune cell interactions in the TME of CRC.
Weighted co‑expression network analysis (WGCNA)
WGCNA is a method of unsupervised learning that emphasizes the patterns of gene co-expression. This method constructs a weighted network of gene co-expressions to ascertain the interrelationships among genes and categorizes these interconnections into distinct modules. To identify gene sets related to acetylation, we performed WGCNA analysis on TCGA data using the “WGCNA” R package. First, we calculated the correlation between gene pairs using gene expression profiles and converted this into a co-expression matrix. P-values were adjusted via Bonferroni correction to account for multiple comparisons across modules, with adjusted p < 0.05 deemed significant. Next, we set a soft threshold to construct a scale-free network among genes and convert the adjacency matrix into a Topological Overlap Matrix (TOM). Following this, we utilize a dynamic tree-cutting algorithm to group genes and delineate modules. Ultimately, we identify the module that exhibits the highest correlation with the acetylation score for subsequent examination.
Integrating machine learning methods to generate prognostic signature
To identify a set of genes associated with acetylation, an intersection analysis was performed between the acetylation-related DEGs and the genes derived from the WGCNA modules. Prognostic genes were generated using univariate Cox regression analysis within the TCGA dataset (P < 0.05). To develop a consensus prognostic signature for CRC, we integrated ten machine learning algorithms: RSF, Enet, Lasso, stepwise Cox regression, Ridge regression, CoxBoost, plsRcox, SuperPC, GBM, and survival SVM. Notably, algorithms such as LASSO, RSF, stepwise Cox, and CoxBoost offer intrinsic feature selection. A total of 101 algorithm combinations were constructed for prediction models in the TCGA dataset, applying a leave-one-out cross-validation (LOOCV) framework alongside 10-fold cross-validation. These combinations were employed to independently generate an acetylation-related prognostic signature (ARPS) leveraging acetylation-specific genes. Subsequently, each model’s performance was assessed across TCGA and GSE39582 validation datasets by calculating Harrell’s concordance index (C-index). Ultimately, the consensus acetylation-related prognostic signature (ARPS) for CRC was determined by averaging the C-indices across the two cohorts, identifying the model with the highest average C-index as the optimal ARPS. For each cohort, the ARPS score was computed using the model derived from the training cohort.
According to the optimal ARPS score, the CRC samples were stratified into groups of low and high risk. The Kaplan-Meier survival analysis was conducted to compare OS between the high- and low-risk groups. The ARPS’s prognostic ability was evaluated by generating a receiver operating characteristic (ROC) curve using the “timeROC” package. Then, the predictive accuracy of the ARPS score was further confirmed by the samples of the GSE39582 cohort.
Clinical value of ARPS
Relevant clinical data CRC patients were obtained from TCGA. The training cohort consisted of patients with various clinical variables such as age, gender, tumor site, tumor status, and TNM stage. To validate the clinical significance of the ARPS score, we examined their correlation with clinicopathological factors. Next, univariate and multivariate Cox analyses were performed to evaluate the independent prognostic value of ARPS when combined with clinical parameters. To correct for multiple testing, the Benjamini-Hochberg FDR method was applied, retaining genes with FDR < 0.05.
Analysis of tumor microenvironment
The “ESTIMATE” R package was utilized to calculate stromal scores, immune scores, and tumor purity across different risk groups. To further evaluate immune cell infiltration, we employed the CIBERSORT algorithm to assess the abundance of 22 immune cell types within different risk groups. Additionally, single-sample gene set enrichment analysis (ssGSEA) was conducted to quantify the infiltration of these immune cells, as well as to examine the overall immune function pathways between the high-risk and low-risk groups. Results from ssGSEA were visualized using boxplots to illustrate differences in immune cell abundance.
The potential role of ARPS in immunotherapy
To assess the effectiveness of immunotherapy responses, we analyzed immune checkpoint expression differences between high- and low-risk groups. Next, we utilized the Tumor Immune Dysfunction and Exclusion (TIDE) algorithm (http://tide.dfci.harvard.edu/) to predict TIDE scores, which helped us evaluate immunotherapy sensitivity across these risk categories. TIDE data for CRC were sourced from the TIDE database to compare dysfunction and exclusion, alongside overall TIDE scores. The immunophenoscore (IPS) data for CRC patients were obtained from The Cancer Immunome Atlas (TCIA) (https://www.tcia.at/home) to predict responses to biological therapies such as CTLA-4 and PD-1 inhibitors. Similarly, IPS data were analyzed to determine the differential efficacy of monotherapies and combination therapies involving anti-CTLA-4 and anti-PD-1, comparing the outcomes among two risk groups. Furthermore, clinical and survival data related to different immunotherapy regimens were extracted from the IMvigor 210 database, which included 348 patients treated with a PD-L1-targeting antibody. This allowed us to validate the prognostic capabilities of the ARPS within the IMvigor210 cohort. Additionally, the tumor mutation burden (TMB), a critical indicator of response to immune checkpoint inhibitors, was calculated for each CRC sample. This quantification of tumor immunogenicity, alongside a comparative analysis among two risk groups, provided deeper insights into the relationship between mutational load, risk scores, and the overall prognostic outcomes in CRC patients receiving immunotherapy.
Screening of sensitive drugs
The “oncoPredict” package was employed to assess drug sensitivity, specifically focusing on the half-maximal inhibitory concentration (IC50) values for therapeutic drugs. Potential therapeutics for risk groups were identified by comparing sensitivity profiles of 198 drugs using transcriptomic data.
Gene set enrichment analysis (GSEA)
GSEA was conducted utilizing the “clusterProfiler” package to investigate the variations in Gene Ontology (GO) terms and KEGG signaling pathways associated with the ARPS in CRC across two distinct risk groups. The analysis focused on comparing Hallmark, GO terms, and KEGG pathways17–19. Significance thresholds were set as |NES| > 1, nominal p < 0.05, and FDR q-value < 0.25 to control for false positives arising from multiple hypothesis testing. Background gene sets, including c5.go.Hs.symbols.gmt for (GO) and c2.cp.kegg.Hs.symbols.gmt for KEGG pathways, were sourced from the MSigDB database (http://software.broadinstitute.org/gsea/msigdb).
Statistical analysis
All statistical analyses were conducted using R software version 4.3.1, with a significance level set at p < 0.05 to determine statistical significance. Differences in continuous variables were evaluated using either the Wilcoxon rank-sum test or Student’s t-test. Survival differences were analyzed by Kaplan-Meier analysis alongside the log-rank test. P < 0.05 was considered statistically significant.
Results
Acetylation characteristics in single-cell transcriptome
To explore the acetylation features and immune cell dynamics in CRC, we analyzed single-cell transcriptomic data from two GEO datasets (GSE132465 and GSE144735). Following rigorous quality control, 35 CRC samples were retained. To address potential batch effects, we utilized the Harmony algorithm, which facilitated the dimensionality reduction and allowed for the identification of 9 distinct cell subtypes within the CRC samples (Fig. 1A). Using SingleR and published cell markers, we annotated 54,069 cells into nine major clusters,: B Cells, dendritic cells, endothelial cells, epithelial cells, mast cells, myeloid cells, plasma cells, stromal cells, and T cells (Figs. 1B, C). To quantify the acetylation features in each cell type, we calculated the acetylation activity score across different cell types using ssGSEA methods (Fig. 1D). Epithelial cells exhibited the highest acetylation scores, followed by endothelial and stromal cells, while immune cells showed significantly lower activity (Fig. 1E). Cells were stratified into high and low acetylation groups, yielding 1,815 DEGs (FDR < 0.05) for further investigation.
Fig. 1.
Acetylation features in single-cell transcriptomics. (A) t-SNE plot shows the regrouping of CRC single cells into 9 separate clusters. (B) t-SNE plot shows the cell types identified by marker genes. (C) The expression patterns of the cell type-specific marker genes across the cell clusters. (D) The activity score of acetylation in each cell. (E) The distribution of the acetylation scores in different cell types.
Network analysis identifies core acetylation modules
To explore the genes linked to acetylation, we employed the ssGSEA technique to measure the acetylation scores for each sample from TCGA. Subsequently, we developed a WGCNA to pinpoint modules that exhibit a significant correlation with acetylation, utilizing differentially expressed genes related to acetylation at the single-cell level (Fig. 2A). We illustrated the clustering relationships among tumor samples by creating a hierarchical clustering dendrogram of the samples (Fig. 2B). The heatmap at the bottom displays the acetylation scores for each sample, highlighting the relative activity of acetylation features in the samples. The optimal soft threshold chosen is 8 (R2 = 0.85), which ensures that the network adheres to the scale-free topology standard (Fig. 2C). Furthermore, the minimum gene count required for each module is established at 50, leading to the discovery of eight unique modules (Fig. 2D). The green module demonstrates a significant association with the acetylation score, exhibiting a correlation coefficient (R) of -0.75 and a adjusted p-value lower than 0.05. Consequently, we obtained a total of 169 genes within the green module. By intersecting these with the 1,691 DEGs related to acetylation (Fig. 2E), we obtained 131 genes (Fig. 2F). These genes are regarded as playing a crucial role in acetylation processes at both the transcriptomic and single-cell transcriptomic levels. We performed GO enrichment analysis to investigate the distribution of the identified genes across biological processes (BP), cellular components (CC), and molecular functions (MF). The findings indicated that these genes are significantly enriched in energy metabolism, protein synthesis, and membrane transport (Fig. 2G).
Fig. 2.
Weight co-expression network and gene enrichment analysis. (A) Dendrogram showing the hierarchical clustering of TCGA samples. The heatmap at the bottom represents the acetylation scores of each sample. (B) Cluster dendrogram of the WGCNA analysis. (C) Selection of the optimal soft threshold power. (D) Module-trait heatmap showing that the green modules were closely related to the acetylation trait. (E) Volcano plot showing differential analysis results between TCGA samples and normal samples. (F) Venn plot showing the intersecting genes between the green modules and DEGs in bulk RNA-seq. (G) GO enrichment of the overlapping genes. (H) Univariate Cox regression analysis of 30 acetylation-related genes.
Machine learning integrates acetylation features into clinical risk prediction
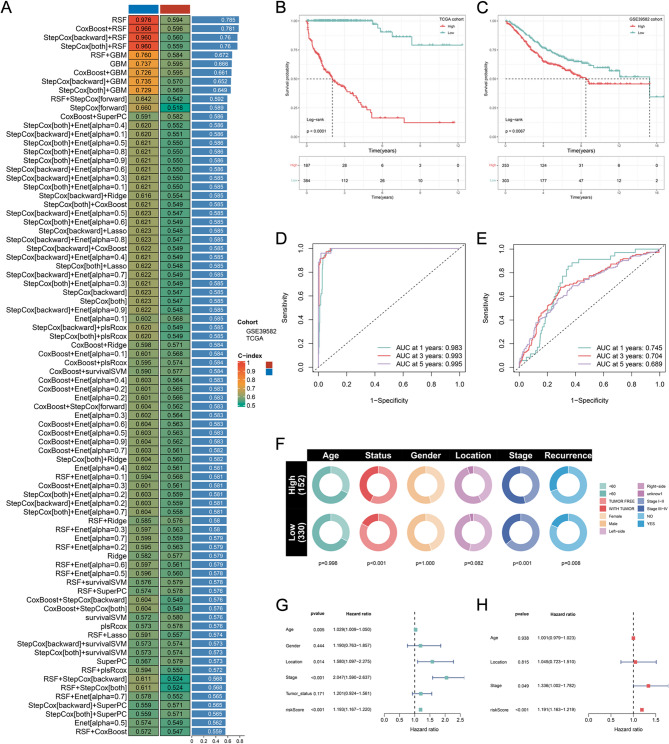
Leveraging these acetylation-related genes, we next asked whether machine learning could integrate them into a clinically actionable prognostic tool. Following univariate Cox regression analysis of the 131 acetylation-related genes and FDR correction (q < 0.05), 30 were significantly correlated with the OS of CRC patients (Fig. 2H). To establish a robust prognostic signature centered on acetylation-related molecular features, we integrated a total of 30 prognostic genes into our analytical framework, utilizing a LOOCV approach. We developed predictive models leveraging 101 algorithmic combinations and implemented 10-fold cross-validation within the TCGA training cohort. For both training and validation cohorts, we computed the average C-index for each model. The resultant optimal model was identified as the CoxBoost + RSF combination, achieving the highest average C-index of 0.781 (Fig. 3A). Consequently, we formulated an ARPS, which comprises ten pivotal acetylation-related genes: DCTPP1, TXNDC12, RPS24, TCEAL4, RAB5C, NME1, VOPP1, SMAGP, MRPL22, and HINT1. ARPS scores were subsequently calculated for all samples across the two cohorts according to the expression profiles of these ten genes.
Fig. 3.
Construction and evaluation of a ARPS. (A) The C-index of 101 kinds prognostic models of TCGA and GEO datasets. (B,C) The Kaplan-Meier survival curves for the two ARPS groups in the TCGA dataset (B) and GEO (C) dataset. (D,E) Time-dependent ROC curve of the ARPS in TCGA training dataset (D) and GEO (E) datasets. (F) Correlations of two ARPS groups with clinical characteristics in the TCGA dataset. (G,H) Univariate (G) and multivariate (H) Cox analysis of ARPS score and clinicopathological parameters.
In both training and validation cohorts, the optimal ARPS score was used to divide CRC samples into two risk subgroups. The Kaplan-Meier analysis indicated that individuals classified as high-risk experienced a reduced duration of survival compared to those classified as low-risk in the TCGA and GEO datasets (P < 0.05; Figs. 3B, C). Further analysis using the ROC curve demonstrated that in the TCGA dataset, the AUC values were 0.983, 0.993, and 0.995 for 1, 3, and 5 years (Fig. 3D), while in the GSE39582 dataset, the values were 0.745, 0.704, and 0.689 (Fig. 3E). These findings affirm the effectiveness of the ARPS in providing precise prognostic predictions.
Clinical significance of the ARPS
We compared the distribution of clinical characteristics across the two ARPS score groups. Significant differences in TNM stage, tumor status, and recurrence status were observed between risk groups (Fig. 3F). Specifically, when contrasting the low-risk subgroup with the high-risk subgroup, there was a notable rise in the percentage of patients classified as stage III and IV within the high-risk group (Fig. 3F). Likewise, the proportion of patients with tumor status and recurrence status was evidently higher in the high-risk subgroup (Fig. 3F). Univariate and multivariate analyses demonstrated that the ARPS score remained unaffected by other clinicopathological variables (Figs. 3G, H). In multivariate regression analysis (Fig. 3H), the ARPS score and TNM stage were identified as separate prognostic factors.
Association of risk scores and tumor immune microenvironment
Given the TME’s clinical significance, we investigated how ARPS risk groups shape immune landscapes.We utilized the ESTIMATE algorithm to calculate immune scores, stromal scores, estimate scores, and tumor purity within the TME. The Wilcoxon test revealed that low-risk CRC patients exhibited markedly higher immune, stromal, and estimate scores, alongside reduced tumor purity scores (Fig. 4A-D). The CIBERSORT analysis showed high-risk tumors enriched for M0 and M2 subtypes (Fig. 4E), while, the low-risk tumors had increased CD8 + T cells, resting and activated dendritic cells, mast cells, follicular helper T cells, and Tregs. To validate the association between ARPS and tumor immune microenvironment, we analyzed an independent cohort (GSE39582). Consistent with our initial findings, low-risk CRC patients exhibited markedly higher immune, estimate scores, and reduced tumor purity scores (Figures S1A-D). Similarly, low-risk patients exhibited significantly higher abundance of CD8 + T cell and activated dendritic cells, while high-risk patients enriched for M2 macrophages (Figure S1E). Furthermore, ssGSEA analysis revealed a notably elevated presence of B cells, CD8 + T lymphocytes, and dendritic cells within the low-risk group (Fig. 4F). Additionally, ssGSEA analysis highlighted significant differences in immune function scores between the two risk groups, indicating that low-risk CRC samples had greater enrichment in APC_co-inhibition, checkpoint, MHC class I, CCR, parainflammation, and T cell co-inhibition (Fig. 4G).
Fig. 4.
The immune landscape associated with the ARPS in the TCGA cohort. (A–D) The immune score, stromal score, estimate score, and tumor purity were applied to quantify the different immune statuses between the high- and low-risk groups. (E) Box plot displays the differential abundance of 22 infiltrative immune cells by CIBERSORT database between high-risk and low-risk groups. (F) The level of immune cells in different ARPS score groups. (G) The level of immune-related function in different ARPS score groups. ***p < 0.001, **p < 0.01, *p < 0.05; Benjamini-Hochberg adjusted.
Role of ARPS in immunotherapy
Recent studies have highlighted the complex relationship between antigen presentation, tumor mutation burden (TMB), and the efficacy of immunotherapy in cancer patients20. An increased diversity in antigen presentation, characterized by high expression of immunological checkpoints such as CTLA4, PD-L1, and LAG3, suggests a greater potential for positive responses to immunotherapy, particularly in CRC patients with low ARPS scores. Boxplot analyses indicated that these patients exhibited significantly higher expressions of these checkpoints (Fig. 5A; all p < 0.05), correlating with better therapeutic outcomes. Furthermore, TMB has been recognized as an essential determinant affecting the efficacy of immunotherapy. In CRC patients with low ARPS scores, a higher TMB was observed (Fig. 5B), reinforcing the notion that elevated TMB levels are associated with improved responses to immunotherapy. Conversely, a low TIDE score indicates a better response to treatment and a reduced risk of immune escape, as evidenced by higher TIDE scores in patients with higher IRS scores (Fig. 5C; all p < 0.05). The Immunophenoscore (IPS), which reflects the immune profile of tumors, also plays a crucial role in predicting immunotherapy outcomes. A higher IPS is linked to better therapeutic benefits, and its application in conjunction with immunological checkpoint blockers such as CTLA4 and PD1 has shown promising potential in CRC treatment. Low-risk patients consistently displayed more favorable responses to immunotherapy treatments across various combinations of CTLA4 and PD1 expressions (Figs. 5D-G). Consequently, CRC patients with low ARPS scores may derive greater benefits from immunotherapy. To further confirm this finding, we calculated the ARPS score in patients undergoing immunotherapy. In the IMvigor210 cohort, non-responders had significantly higher ARPS scores than responders (p < 0.01), as illustrated in Fig. 5H. Furthermore, patients with higher ARPS scores demonstrated a lower overall survival (OS) and a reduced response rate (Fig. 5I, J), which further substantiates our earlier observations.
Fig. 5.
ARPS acted as an indicator for immunotherapy benefits in CRC. (A) The difference in the expression of immune checkpoint-related genes between high-risk and low-risk groups. (B) The TMB difference between the two groups. (C) The difference in TIDE score between the different groups. (D–G) Differences in IPS among different risk groups in different situations. (H) Comparison of ARPS score between progressive disease (PD)/stable disease (SD) and complete response (CR)/partial response (PR) groups. (I) The distribution of immunotherapeutic response in two groups stratified by ARPS in IMvigor210 cohort. (J) Kaplan-Meier survival curves with log-rank test for different ARPS groups to compare the OS differences in IMvigor210 cohort. *P < 0.05; **P < 0.01; ***P < 0.001.
Predictive analysis for drug therapy
Drug resistance poses a significant obstacle in the management of cancer, frequently leading to diminished therapeutic effectiveness and adverse clinical results for CRC patients. To improve treatment effectiveness, we investigated whether ARPS characteristics can accurately predict the sensitivity to therapeutic drugs. In our analysis, we utilized the “oncoPredict” package to evaluate the IC50 values of 198 drugs. In low-risk patients, the IC50 values for drugs such as 5-Fluorouracil, Gemcitabine, Bortezomib, Cediranib, Crizotinib, and Gefitinib were significantly lower (Fig. 6A–F), indicating that individuals with low-risk scores may respond better to these treatments. Conversely, Sepantronium bromide and Trametinib showed a trend of decreased IC50 values in the high-risk group (Fig. 6G, H), suggesting that these drugs may be more effective in this population.
Fig. 6.
Prediction of drug sensitivity by OncoPredict in high-risk and low-risk groups. (A) 5-Fluorouracil. (B) Gemcitabine. (C) Bortezomib. (D) Cediranib. (E) Crizotinib. (F) Gefitinib. (G) Sepantronium bromide. (H) Trametinib. *P < 0.05; **P < 0.01; ***P < 0.001.
Potential biological functions and pathway analyses in two risk groups
To investigate the molecular mechanisms underlying ARPS, we subsequently performed GSEA on both GO and KEGG gene sets to elucidate the differences in biological functions and pathways between the high-risk and low-risk groups (Figs. 7A-D). For GO terms, the high-risk cohort exhibited significant enrichment in categories such as “BP cell morphogenesis involved in neuron differentiation”, “BP cell part morphogenesis”, and “BP developmental growth”. Conversely, the low-risk group demonstrated notable enrichment in “BP mitochondrial translation”, “BP purine containing compound metabolic process”, and “CC mitochondrial matrix” (Fig. 7A, B). Regarding KEGG terms, the GSEA findings indicated that the high-risk group was predominantly enriched in pathways such as the “MAPK signaling pathway”, “cell cycle”, and “focal adhesion” (Fig. 7C). In contrast, the low-risk group showed significant enrichment in pathways related to “chemokine signaling pathway”, “cytokine-cytokine receptor interaction”, and “intestinal immune network for IgA production” (Fig. 7D).
Fig. 7.
Prediction of drug sensitivity by OncoPredict in high-risk and low-risk groups. (A,B) GO analysis between high-risk group (A) and low-risk group (B) utilizing GSEA method. (C,D) KEGG analysis between high-risk group (C) and low-risk group (D) utilizing GSEA method. Significance thresholds: |NES|>1, NOM p < 0.05, FDR q < 0.25. Bar color: NES value; bar length: -log₁₀(FDR).
Discussion
Colorectal cancer (CRC) remains a leading cause of cancer-related mortality worldwide, with current treatment strategies often limited by the heterogeneity of tumor biology and patient responses. This complexity necessitates innovative approaches integrating phenotypic characteristics to enhance therapeutic decision-making and patient outcomes. Our study highlights the critical role of acetylation features in CRC, suggesting that a deeper understanding of acetylation within the TME could significantly improve prognostic assessments and treatment strategies.
Our findings demonstrate that compartment-specific acetylation dynamics (epithelial/stromal; Fig. 1E) stratify CRC into clinically distinct subtypes through a machine learning-derived ARPS. This robust model stratifies patients by: (i) prognosis (increased mortality in high-risk patients; Fig. 3B-C), (ii) treatment response (enhanced immunotherapy efficacy in low-risk cohorts; Fig. 5H-J), and (iii) TME reprogramming (acetylome linkage of metabolic pathways to immune checkpoint regulation; Figs. 5A, 7B). Collectively, our findings emphasize the need to explore acetylation dynamics for developing precision interventions in CRC and improving patient management. Our machine learning-derived ARPS aligns with emerging multi-omics frameworks for TME decoding. The iMLGAM model14 similarly integrates genetic and epigenetic features to predict immunotherapy response, validating our combinatorial approach. Crucially, the immune-excluded phenotype in high-risk patients mirrors B4GALT2-mediated barriers in LUAD16, suggesting conserved PTM-driven immune evasion. This synergy implies acetyltransferase inhibitors could overcome stromal exclusion-a strategy enabled by nanotechnology-based delivery systems15.
The ten-gene ARPS (DCTPP1, TXNDC12, RPS24, TCEAL4, RAB5C, NME1, VOPP1, SMAGP, MRPL22, HINT1) may influence CRC progression through acetylation-mediated pathways. HINT1, a tumor suppressor regulating AP-1 transcription and apoptosis21–23, requires acetylation for protein stabilization. Its downregulation in high-risk patients suggests impaired acetylation may promote immune evasion by reducing tumor immunogenicity. Conversely, RAB5C—a regulator of EGFR endocytosis—may undergo acetylation-induced dysregulation (as observed in related GTPases), potentially amplifying oncogenic MAPK signaling through enhanced receptor recycling. This aligns with MAPK pathway enrichment in high-risk patients and could drive M2 macrophage polarization via cytokine overproduction. NME1, which modulates purine metabolism and metastasis suppression, gains functional enhancement through K12 acetylation24. Its association with low-risk signatures may explain purine metabolism enrichment, potentially supporting anti-tumor immune responses. TXNDC12, an ER stress mediator, may experience acetylation-dependent alterations in protein folding efficiency, impairing antigen presentation via MHC-I in high-risk tumors. These mechanisms collectively shape the immunosuppressive TME observed in high-risk patients—characterized by reduced CD8 + T cells and elevated TIDE scores.GSEA revealed significant differences in pathway enrichment between high-risk and low-risk groups in CRC. The high-risk group was predominantly enriched in the MAPK signaling pathway and cell cycle-related processes, which are critical for cell proliferation and survival, suggesting that these pathways may contribute to tumor aggressiveness and poor prognosis in CRC patients. The MAPK pathway, known for its role in regulating various cellular activities including growth, differentiation, and apoptosis, has been implicated in cancer progression and therapeutic resistance25–27. Conversely, the low-risk group showed enrichment in pathways related to cytokine-cytokine receptor interactions, chemokine signaling, and the intestinal immune network for IgA production, which are essential for immune response modulation and tumor microenvironment regulation. This indicates that the low-risk group may have a more advantageous immune landscape, potentially enhancing their response to immunotherapies. These divergent pathways highlight the distinct biological behaviors of CRC subtypes, emphasizing the importance of understanding these pathways for developing targeted therapies. The findings from this study not only provide insights into the molecular mechanisms underlying CRC progression but also suggest potential therapeutic targets that could improve patient outcomes through personalized treatment strategies. Understanding the interplay between these pathways and their impact on tumor biology is crucial for advancing CRC management and improving prognostic predictions.
The immune landscape in CRC reveals significant differences between low-risk and high-risk patient groups, which may have profound implications for treatment strategies. In our study, low-risk patients exhibited elevated levels of immune cell infiltration and enhanced immune functionality, suggesting a more favorable environment for immunotherapy responses. Conversely, high-risk patients demonstrated characteristics associated with immune evasion, including a higher TIDE score and a distinct immune profile that may hinder effective immune responses. These findings align with previous research indicating that patients with higher immune infiltration often experience better outcomes following immunotherapy, as their tumors are more likely to be recognized and attacked by the immune system. The identification of specific immune cell types, such as CD8 + T cells and dendritic cells, which were more abundant in the low-risk group, further supports the notion that a robust immune response is crucial for effective cancer treatment28–32. The elevated immune scores and lower tumor purity observed in low-risk patients further support the notion that a favorable immune landscape is associated with better prognosis and therapeutic responses. The implications of these results are significant, as they suggest that stratifying patients based on their immune profiles could guide therapeutic decisions, particularly in the context of immunotherapy. By understanding the dynamics of immune cell infiltration and the TME, clinicians may be better equipped to predict patient responses to treatment and tailor interventions accordingly. This research underscores the importance of integrating immune analysis into prognostic models, as it may enhance our ability to pinpoint patients who are most likely to benefit from immunotherapeutic approaches, ultimately improving clinical outcomes in CRC management.
Our IC50 predictions align with established clinical responses: First, 5-FU efficacy in low-risk patients: The inverse correlation between ARPS scores and 5-FU sensitivity mirrors clinical data where high TIL (tumor-infiltrating lymphocyte) CRC shows 38% ORR vs. 18% in TIL-low tumors33. This synergy likely arises from immune-mediated tumor killing enhancing chemotherapeutic effects34. Second, argeted therapy for high-risk subgroup: The predicted Trametinib sensitivity in high-risk patients is grounded in their MAPK pathway activation (FDR < 0.001), consistent with MEK inhibitor trials reporting prolonged PFS (5.2 vs. 2.1 months) in MAPK-high CRC35. Third, novel therapeutic opportunities: Sepantronium bromide’s predicted efficacy in high-risk tumors suggests survivin inhibition as a strategy for aggressive, cell cycle-driven CRC-a hypothesis supported by phase I trials showing 22% disease control in refractory CRC36.
While our results offer valuable insights into the role of acetylation in shaping the TME, it is essential to acknowledge the limitations of this study. First, the reliance on single-cell RNA sequencing, although powerful, may not capture the full spectrum of cellular interactions and the spatial context of the tumor microenvironment. Future work should integrate spatial transcriptomics and proteomics to provide a more holistic view of the tumor ecosystem. Second, the reliance on acetylation activity as a sole marker may not capture the full spectrum of epigenetic modifications influencing gene expression. Future research should aim to elucidate the interplay between acetylation and other epigenetic mechanisms, as well as explore the functional consequences of the identified DEGs in the context of CRC. Third, while the ARPS showed strong prognostic capability in the discovery cohort (TCGA), its reduced AUC in the validation cohort (GSE39582) warrants consideration. This may arise from: (i) Technical biases (e.g., batch effects between RNA-seq platforms), (ii) Demographic differences (e.g., TCGA’s multinational vs. GSE39582’s European population), or (iii) Overfitting risk from high-dimensional feature selection. Though we mitigated overfitting through LOOCV and 10-fold cross-validation, future studies should validate ARPS in prospective, multi-center cohorts with standardized protocols. Notably, even with reduced AUC, the model significantly stratified survival (log-rank p < 0.05), supporting its clinical utility.
Conclusions
Our findings contribute to the growing body of evidence that highlights the significance of acetylation in cancer biology, suggesting that targeting acetylation pathways could enhance therapeutic strategies and improve patient outcomes in the context of tumor immunology and treatment resistance. The identification of specific DEGs associated with acetylation activity opens avenues for further research into their functional roles and potential as biomarkers for therapeutic response in cancer patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
KL and JFL contributed to study design. KL, WS, and YFZ extracted and analysed the data. All authors drafted the manuscript. KL, WS, and YFZ contributed to a critical revision of the manuscript. All authors approved the submitted version.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kai Li, Wei Song and Yuefeng Zhang.
References
- 1.Biller, L. H. & Schrag, D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA325 (7), 669–685 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Shin, A. E., Giancotti, F. G. & Rustgi, A. K. Metastatic colorectal cancer: mechanisms and emerging therapeutics. Trends Pharmacol. Sci.44 (4), 222–236 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, C. et al. Acetylation of ELMO1 correlates with Rac1 activity and colorectal cancer progress. Exp. Cell. Res.439 (1), 114068 (2024). [DOI] [PubMed] [Google Scholar]
- 4.Wang, M. et al. Competitive antagonism of KAT7 crotonylation against acetylation affects procentriole formation and colorectal tumorigenesis. Nat. Commun.16 (1), 2379 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou, X. et al. Deciphering the role of acetylation-related gene NAT10 in colon cancer progression and immune evasion: implications for overcoming drug resistance. Discover Oncol.16 (1), 774 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, X. et al. SNORA28 Promotes Proliferation and Radioresistance in Colorectal Cancer Cells through the STAT3 Pathway by Increasing H3K9 Acetylation in the LIFR Promoter. Adv. Sci.11(32), e2405332 (2024). [DOI] [PMC free article] [PubMed]
- 7.Crawford, C. E. W. & Burslem, G. M. Acetylation: a new target for protein degradation in cancer. Trends Cancer. 11 (4), 403–420 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, X. et al. Acetylation suppresses breast cancer progression by sustaining CLYBL stability. J. Translational Med.23 (1), 415 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miziak, P., Baran, M., Borkiewicz, L., Trombik, T. & Stepulak, A. Acetylation of histone H3 in cancer progression and prognosis. Int. J. Mol. Sci. ;25(20). (2024). [DOI] [PMC free article] [PubMed]
- 10.Wang, C. & Ma, X. The role of acetylation and deacetylation in cancer metabolism. Clin. Translational Med.15 (1), e70145 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu, Y., Zhang, H. & Nie, D. Histone modifications and metabolic reprogramming in tumor-associated macrophages: a potential target of tumor immunotherapy. Front. Immunol.16, 1521550 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu, Z., Nie, G., Peng, X., Zhan, X. & Ding, D. KAT8 catalyzes the acetylation of SEPP1 at lysine 247/249 and modulates the activity of CD8(+) T cells via LRP8 to promote anti-tumor immunity in pancreatic cancer. Cell. Bioscience. 15 (1), 24 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Malsi, K. et al. The role of lactylation in tumor growth and cancer progression. Front. Oncol.15, 1516785 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye, B. et al. iMLGAM: Integrated Machine Learning and Genetic Algorithm-driven Multiomics analysis for pan-cancer immunotherapy response prediction. Imeta.4(2), e70011 (2025). [DOI] [PMC free article] [PubMed]
- 15.Feng, J., Zhang, P., Wang, D., Li, Y. & Tan, J. New strategies for lung cancer diagnosis and treatment: applications and advances in nanotechnology. Biomark. Res.12 (1), 136 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, P. et al. Novel post-translational modification learning signature reveals B4GALT2 as an immune exclusion regulator in lung adenocarcinoma. J. Immunother. Cancer ;13(2). (2025). [DOI] [PMC free article] [PubMed]
- 17.Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28 (1), 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci.28 (11), 1947–1951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51 (D1), D587–d92 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmeri, M. et al. Real-world application of tumor mutational burden-high (TMB-high) and microsatellite instability (MSI) confirms their utility as immunotherapy biomarkers. ESMO open.7 (1), 100336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung, T. Y. et al. Deacetylation by SIRT1 promotes the tumor-suppressive activity of HINT1 by enhancing its binding capacity for β-catenin or MITF in colon cancer and melanoma cells. Exp. Mol. Med.52 (7), 1075–1089 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, H. et al. The HINT1 tumor suppressor regulates both gamma-H2AX and ATM in response to DNA damage. J. Cell. Biol.183 (2), 253–265 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motzik, A. et al. Post-translational modification of HINT1 mediates activation of MITF transcriptional activity in human melanoma cells. Oncogene36 (33), 4732–4738 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Iuso, D., Guilliaumet, J., Schlattner, U. & Khochbin, S. Nucleoside diphosphate kinases are ATP-Regulated carriers of Short-Chain Acyl-CoAs. Int. J. Mol. Sci. ;25(14). (2024). [DOI] [PMC free article] [PubMed]
- 25.Ma, Y. T. et al. Mechanisms of the JNK/p38 MAPK signaling pathway in drug resistance in ovarian cancer. Front. Oncol.15, 1533352 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng, Y., Li, Y. & Cao, H. BRD9 promotes the malignant phenotype of thyroid cancer by activating the MAPK/ERK pathway. Anticancer Drugs. 36 (5), 359–373 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hua, X., Wang, Y., Wang, C., Yang, H. & Liu, A. Hypoxia-related gene DDIT4 as a therapeutic biomarker promotes epithelial-mesenchymal transition in lung adenocarcinoma via the MAPK/ERK signaling pathway. Int. Immunopharmacol.157, 114739 (2025). [DOI] [PubMed] [Google Scholar]
- 28.Mazzoccoli, L. & Liu, B. Dendritic cells in shaping Anti-Tumor T cell response. Cancers (Basel)16(12). (2024). [DOI] [PMC free article] [PubMed]
- 29.Xiao, Z. et al. Impaired function of dendritic cells within the tumor microenvironment. Front. Immunol.14, 1213629 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reste, M. et al. The role of dendritic cells in tertiary lymphoid structures: implications in cancer and autoimmune diseases. Front. Immunol.15, 1439413 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, C., Liu, Y., Ali, N. M., Zhang, B. & Cui, X. The role of innate immune cells in the tumor microenvironment and research progress in anti-tumor therapy. Front. Immunol.13, 1039260 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, Y. et al. New insights on anti-tumor immunity of CD8(+) T cells: cancer stem cells, tumor immune microenvironment and immunotherapy. J. Translational Med.23 (1), 341 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganesh, K. et al. Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol.16 (6), 361–375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galluzzi, L., Buqué, A., Kepp, O., Zitvogel, L. & Kroemer, G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol.17 (2), 97–111 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Kopetz, S. et al. Phase II pilot study of Vemurafenib in patients with metastatic BRAF-Mutated colorectal cancer. J. Clin. Oncology: Official J. Am. Soc. Clin. Oncol.33 (34), 4032–4038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ambrosini, G., Adida, C. & Altieri, D. C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med.3 (8), 917–921 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.