Abstract
Human papillomavirus (HPV) E2 proteins regulate viral replication by binding to sites in the upstream regulatory region (URR) and by complex formation with the E1 origin recognition protein. In the genital HPV types, the distribution and location of four E2 binding sites (BS1 to BS4) which flank a single E1 binding site are highly conserved. We have examined the roles of these four E2 sites in the viral life cycle of HPV type 31 (HPV31) by using recently developed methods for the biosynthesis of papillomaviruses from transfected DNA templates (M. G. Frattini et al., Proc. Natl. Acad. Sci. USA 93:3062–3067, 1996). In transient assays, no single site was found to be necessary for replication, and mutation of the early promoter-proximal site (BS4) led to a fourfold increase in replication. Cotransfection of the HPV31 wild-type (HPV-wt) and mutant genomes with expression vectors revealed that E1 stimulated replication of HPV31-wt as well as the HPV31-BS1, -BS2, and -BS3 mutants. In contrast, increased expression of E2 decreased replication of these genomes. Replication of the HPV31-BS4 mutant genome was not further increased by cotransfection of E1 expression vectors but was stimulated by E2 coexpression. In stably transfected normal human keratinocytes, mutation of either BS1, BS3, or BS4 resulted in integration of viral genomes into host chromosomes. In contrast, mutation of BS2 had no effect on stable maintenance of episomes or copy number. Following growth of stably transfected lines in organotypic raft cultures, the differentiation-dependent induction of late gene expression and amplification of viral DNA of the BS2 mutant was found to be similar to that of HPV31-wt. We were unable to find a role for BS2 in our assays for viral functions. We conclude that at least three of the four E2 binding sites in the URRs of HPVs are essential for the productive viral life cycle. The specific arrangement of E2 binding sites within the URR appears to be more important for viral replication than merely the number of sites.
Human papillomaviruses (HPVs) infect squamous epithelial cells and induce hyperproliferative lesions (22, 29). The replication and transcription of HPVs is tightly linked to the differentiation state of the infected keratinocyte. After infection of basal cells, the viral genome, consisting of a circular double-stranded DNA of 7.9 kb, is replicated to 50 to 100 copies. The copy number of these episomes is then stably maintained through subsequent cell divisions of infected basal cells. Upon differentiation, the viral DNA is amplified to several thousand copies in suprabasal layers and differentiation-specific viral promoters are activated (22, 29).
In transient replication assays with bovine papillomavirus type 1 (BPV1), the viral E1 and E2 proteins are required to initiate replication at the viral origin which contains binding sites for these proteins (58, 59). Similarly, replication of plasmids containing HPV origins also require the action of both E1 and E2 proteins (5–8, 14, 31, 40, 43, 52). E1 has features characteristic of replication initiator proteins, including sequence-specific DNA binding, ATPase, and helicase activities. In addition, E1 interacts with cellular replication proteins such as the DNA polymerase α-primase complex (51). The E2 protein also exhibits sequence-specific DNA binding and was first described as a transcriptional regulator that either stimulates or represses promoter activity, dependent on the promoter context (32). E2 is also directly involved in viral DNA replication through complex formation with E1, which dramatically increases the binding of E1 to the origin. This stimulation is dependent on the relative affinity of the neighboring E2 binding sites (E2BS) and their distance to the E1 binding site (4, 13, 35, 46, 47, 57, 61).
Many studies on papillomavirus replication have been carried out with BPV1 as a model. However, differences exist between BPV1 and the genital HPVs with respect to the role of E2. All major early promoters of BPV1 are activated by and are highly dependent on E2, whereas the major early promoters of genital HPVs appear to be repressed by E2 (3, 9–11, 16, 18, 39, 42, 48, 50, 53–56). Second, the regulatory region of BPV1 has 12 E2BS with relative affinities varying 100-fold (30), whereas a large group of HPVs (including types 11, 16, 18, and 31) contain four high-affinity sites whose locations in the upstream regulatory region are highly conserved (2, 20, 44, 50). The role of E2BS has been examined in the genomic context of BPV1 by deletion analysis, which has revealed a high degree of redundancy in terms of transcriptional regulation (53). Also, in studies that addressed the importance of E2BS during the vegetative life cycle of BPV1, mutations in E2BS surrounding the origin showed no effect on transient or stable replication or the copy number of episomes (49). This finding indicated that no particular E2BS is essential for replication. However, at least seven E2BS are required for stable maintenance of the BPV1 origin in a cell line that constitutively expresses both E1 and E2. This finding suggested a requirement for a certain number but not arrangement of E2BS in this process (38). Since there are only four high-affinity sites in the genital HPVs, it is unclear what the requirements are for HPV E2 sequences during replication, transcriptional regulation, and/or partitioning.
The contribution of the individual E2BS of HPVs to transcriptional regulation has been assayed by using reporter constructs with E2 expression vectors (see Fig. 1 for numbering of E2BS). In these studies, E2 repressed the major early promoter by binding to binding site 4 (BS4), which is adjacent to the E6 open reading frame (9, 10, 42, 50, 54, 55). Transient replication analysis of HPV type 11 (HPV11) and HPV18 origin fragments also revealed a requirement of at least one E2BS for replication (5, 8, 31, 40, 43, 52). In this study, we investigated the role of the E2BS of HPV31 in the viral life cycle by a mutational analysis of each of the four sites in the upstream regulatory region in their genomic context. Three of the four sites were found to be essential for stable maintenance of viral episomes. Mutation of the fourth site did not alter any of the activities measured by our assays.
FIG. 1.
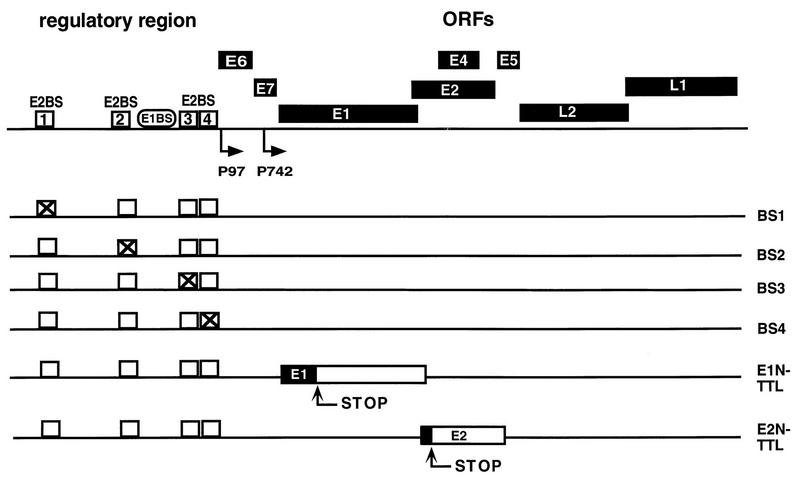
Schematic depiction of the linearized HPV31 genome. (Top) Upstream regulatory region located between the late and the early regions. Numbered boxes represent BS1 through -4, and the binding site for E1 is shown as an oval. The start sites for the early promoter P97 and the late P742 promoter are indicated by arrows. Open reading frames (ORFs) are shown in black. (Bottom) Mutated E2BS in mutant genomes are shown as crossed squares. The positions of translational stop codons introduced into the E1 and E2 genes are indicated by arrows.
MATERIALS AND METHODS
Recombinant plasmids.
Plasmid pUK-HPV31 contains the complete genome of HPV31 cloned into the EcoRI site of a modified pUC18 plasmid (26). Mutations were introduced by overlap extension PCR (19). The wild-type (wt) BS1 sequence was changed from ACCGTTTTCGGT to AAGCTTTTCATA in plasmid pHPV31-BS1 by replacing the RsrII-SpeI fragment (nucleotides [nt] 7415 to 7568) after PCR. The mutation creates a HindIII restriction site (underlined nucleotides). Mutations in BS2 (from ACCGTTTTCGGT to GTAGTTTTCGAA, creating a BsaAI site), BS3 (ACCGAAAGTGGT to CACGAAAGTACT, creating an ScaI site), and BS4 (ACCGAAAACGGT to TTCGAAAACCCA, creating an NcoI site) were introduced by PCR and used to replace the SpeI-HpaI fragment (nt 7568 to 212) in pUK-HPV31, resulting in pHPV31-BS2, -BS3, and -BS4. Plasmid pHPV31-E1N-TTL was also constructed by overlap extension PCR and creates a stop codon at nt 1039 in the E1 open reading frame. The mutated fragment was used to replace a BanII-SwaI fragment (nt 815 to 1645) in pmodBR-HPV31 (42a). Plasmid pHPV31-E2N-TTL was constructed by inserting a translation termination linker into the BclI site in E2, generating a stop codon at nt 2773 (22a). All PCR-generated mutations were confirmed by sequence analysis of the replaced fragment. Plasmid pRP742 was generated by PCR amplification of HPV31 nt 678 to 919, and the resulting fragment was then cloned into the BamHI site of pcDNAII (Invitrogen, San Diego, Calif.). Plasmid pRPA31L1 was generated by PCR amplification of HPV31 nt 5521 to 5703, and the resulting fragment was cloned into SalI/BamHI-digested pSP72 (Promega, Madison, Wis.). The HPV31 E1 and E2 expression vectors are based on pSG5 (Stratagene, La Jolla, Calif.) and have been described elsewhere (14).
Cells and transfections.
SCC-13 cells are derived from a squamous cell carcinoma of the cheek and were maintained in E medium in the presence of mitomycin-treated NIH 3T3 fibroblast feeder cells (41, 60). Normal human keratinocytes (NHKs) purchased from Clonetics (San Diego, Calif.) were grown in KGM (Clonetics) and were transfected at passages 2 to 4 with religated HPV31 DNA or mutated viral DNA and pSV2neo DNA as described previously (15). Cells were selected with geneticin (200 mg/ml; Gibco BRL, Gaithersburg, Md.) for 7 to 10 days in E medium supplemented with epidermal growth factor (5 ng/ml) on feeder cells, and resistant clones were expanded. The generation of stable cell lines was repeated four times with cells from two different donors to ensure reproducibility. Organotypic raft cultures were grown without protein kinase C activators as described previously (15, 33, 34).
Transient replication assay.
Plasmids containing the various HPV genomes (2.5 μg) were digested with EcoRI to excise the genomes from the vector sequences. Religated HPV genomes were then transfected into 60-mm-diameter dishes of SCC-13 cells (ca. 30% confluent), using 15 μl of Lipofectamine in OptiMEM (Gibco BRL) according to the manufacturer’s recommendations. The following day, the transfected cells were split onto 100-mm-diameter dishes and grown for an additional 3 to 4 days. Low-molecular-weight DNA was purified by the Hirt procedure (21), with the following modifications: cell pellets were digested with 50 μg of proteinase K per ml in 400 mM NaCl–10 mM EDTA–10 mM Tris HCl (pH 7.5)–0.2% sodium dodecyl sulfate (SDS) at 55°C for 3 h; NaCl was added to 1 M, and DNAs were precipitated at 4°C overnight followed by centrifugation (60 min, 4°C, 16,000 × g). Supernatants were extracted once with phenol-chloroform-isoamyl alcohol and once with chloroform before precipitation with isopropanol. Each sample was digested with 5 U of DpnI, 15 U of BanII, and 50 μg of RNase A per ml for 5 h prior to Southern analysis. Transfections were repeated at least three times with different DNA preparations.
Southern blot analysis.
Total cellular DNA from cell lines was isolated by proteinase K and RNase A digestion followed by phenol-chloroform extractions and ethanol precipitation. Digested DNAs were separated in 0.8% agarose gels and transferred onto GeneScreen Plus membranes (Dupont, Boston, Mass.). HPV31 fragments were detected with random-primed 32P-labeled HPV31 DNA (HiPrime kit; Boehringer Mannheim, Indianapolis, Ind.). Hybridizations were carried out in 50% formamide, 4× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–5× Denhardt’s–1% SDS–20 μg of salmon sperm DNA per ml at 42°C overnight. Blots were washed at room temperature twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS, twice in 0.1× SSC–0.1% SDS, and then twice in 0.1× SSC–1% SDS at 50°C. Blots were visualized by autoradiography and quantitated by phosphorimager (Fuji, Tokyo, Japan) analysis.
RNase protection analysis.
Total cellular RNA was isolated with Trizol reagent (Gibco BRL) from HPV31-containing keratinocytes. Precipitated RNA pellets were hybridized to 250,000 cpm of antisense 32P-labeled riboprobe transcribed from linearized pRP742 or pRPA31L1 in 10 μl of 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.4)–400 mM NaCl–1 mM EDTA–80% formamide overnight at 37°C. RNase digestion was performed by the addition of 300 μl of 10 mM Tris HCl (pH 7.5)–300 mM NaCl–5 mM EDTA containing RNase A (10 μg/ml) and RNase T1 (30 U/ml); digestion proceeded for 60 min at 37°C. The RNase reaction was stopped by adding SDS to 0.2% and 400 μg of proteinase K per ml, followed by digestion for 15 min at 37°C. Samples were then extracted with phenol-chloroform and precipitated with ethanol prior to resuspension in formamide loading buffer and resolution on a 5% denaturing polyacrylamide gel. Dried gels were visualized by autoradiography and quantitated by phosphorimaging.
In situ hybridization.
DNA in situ hybridization analysis was performed on 5-μm sections of paraffin-embedded raft tissue cross sections that had been fixed in 4% paraformaldehyde. Hybridization analysis was carried out with the Pathogene HPV in situ screening assay (Enzo Diagnostics, Farmingdale, N.Y.) according to the manufacturer’s instructions.
RESULTS
To evaluate the role of the four E2BS (BS1 to BS4) of HPV-31 during the viral life cycle, we generated HPV31 genomes in which individual E2BS were mutated. In addition, genomes which contain translation termination codons in the N terminus of either the E1 or the E2 gene were constructed (Fig. 1). The E2BS were mutated in both palindromic half sites of the recognition sequence (ACCGN4CGGT), which are specifically contacted by the E2 dimer (17). The binding capacity of each mutated site was analyzed in gel retardation assays, and all mutants were found to be defective for E2 binding (data not shown).
Mutation of E2BS4 increases transient replication of HPV-31.
To investigate whether the inactivation of a single E2BS influences the replication properties of HPV31, we first used a transient replication assay as described by Ustav and Stenlund (58) and Del Vecchio et al. (7). Wild-type HPV31 (HPV31-wt) DNA and HPV31 DNAs that contained mutations in each of BS1 through -4 (HPV31-BS1, -BS2, -BS3, and -BS4) or mutations in the E1 or the E2 gene (HPV31-E1N-TTL and HPV31-E2N-TTL) were released from the vector and religated prior to transfection. SCC-13 cells were transfected with the religated genomes, and low-molecular-weight DNA was isolated at 120 h posttransfection followed by Southern analysis (Fig. 2). To distinguish replicated from nonreplicated input DNA, the isolated DNA was digested with the restriction enzyme DpnI, which recognizes only DNA methylated in bacteria (37). The DNAs were also digested with an enzyme which cuts the HPV31 genome once to facilitate quantitation of copy number. The HPV31-E1N-TTL mutant genome did not replicate to significant levels, and the HPV31-E2N-TTL genome replicated on average to less than 10% of the wt genome level as measured by phosphorimaging analysis. These experiments were repeated three times with identical results. This analysis indicates that both E1 and E2 are required in the context of the viral genome for transient replication. Similar effects have been observed for BPV1 (58). While the HPV31-BS2 mutant replicated consistently to levels comparable to wt levels, replication of either the HPV31-BS1 or the HPV31-BS3 mutant was reduced to levels ranging from 30 to 50% of the wt level, as determined by phosphorimager analysis. Surprisingly, inactivation of the promoter-proximal BS4 reproducibly increased replication levels fourfold, indicating that this site acts as a negative regulator of replication (Fig. 2). Similar levels of replication were observed with the HPV31-BS4 mutant in NHKs, providing evidence that these effects are not specific for immortalized SCC-13 cells (data not shown).
FIG. 2.
Representative Southern analysis of transiently replicating HPV31 in SCC-13 cells. SCC-13 cells were transfected with religated HPV31-wt, -BS1, -BS2, -BS3, or -BS4 or genomes that contained a disrupted E1 (E1N-TTL) or E2 (E2N-TTL) gene. Low-molecular weight DNA was isolated 120 h posttransfection, digested with BanII and DpnI, and analyzed by Southern analysis using a full-length HPV31 probe. The marker lane (M) contains 100 pg of EcoRI-linearized HPV31 DNA.
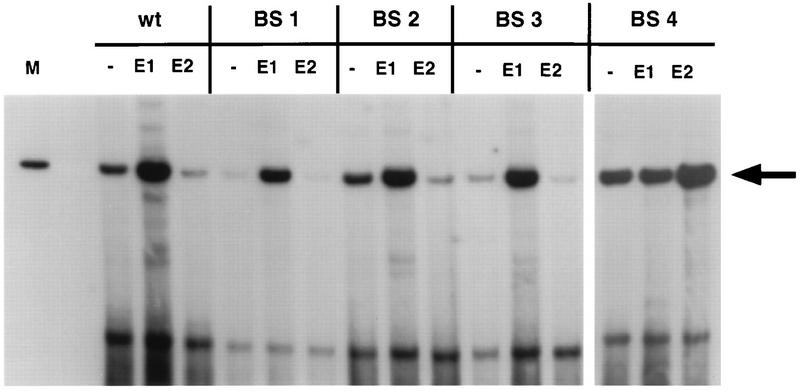
The transient replication assay measures replication as well as transcription properties of the viral genomes. It has been reported that BS4 is involved in the E2-mediated repression of the HPV16 P97 promoter as well as the early viral promoters of HPV18 and HPV11 (9, 10, 42, 50, 54, 55). It is possible that at least some replication factors are expressed from the P97 promoter. Therefore, we reasoned that the increased replication of this mutant could be due to a loss of E2-mediated downregulation of P97. To examine effects of exogenously altering the levels of replication proteins, transient replication assays were performed with genomes cotransfected with expression vectors for E1 or E2. We first performed titration experiments using different amounts of cotransfected E1 or E2 expression vectors with a constant amount of the wt genome. It was determined that inclusion of 0.5 μg of each vector was sufficient for the observed effects (data not shown). Cotransfection of E1 expression vectors resulted in increased transient replication of the HPV31-wt, -BS1, -BS2, and -BS3 genomes to levels approximately fourfold above that seen in the absence of expression vector (Fig. 3). This finding indicated that the amount of E1 was limiting during transient replication assays with the genomes alone. Interestingly, only a small increase in replication of the HPV31-BS4 genome was observed in the presence of the E1 expression vector. Cotransfection of the E2 expression vector reduced transient replication of the HPV31-wt, -BS1, -BS2, and -BS3 genomes to approximately 50% below that seen without added vector (Fig. 3). In contrast, replication of the HPV31-BS4 genome was increased twofold by E2.
FIG. 3.
Representative Southern analysis of transiently replicating HPV31 and E2BS mutant DNAs in the presence of expression vectors for E1 or E2. SCC-13 cells were cotransfected with 2 μg of religated HPV31 or HPV31-BS1, -BS2, -BS3, or -BS4 and 0.5 μg of pSG5 (−), pSG31:E1 (E1), or pSG31:E2 (E2). Low-molecular-weight DNA was isolated 120 h after transfection of SCC-13 cells and analyzed as described for Fig. 2. The marker lane (M) contains 100 pg of linearized HPV31 genome. The arrow indicates the position of the replicated DNA.
BS1, -3, and -4 are involved in establishment and stable maintenance of HPV31 genomes in keratinocytes.
To investigate the role of E2BS in the establishment and maintenance of HPV31 genomes, we made use of our recently developed methods for the generation of cell lines containing episomal HPV31 by transfection of cloned viral DNA (15). NHKs were transfected with HPV31-wt, -BS1, -BS2, -BS3, or -BS4 DNA together with pSV2neo, and following selection, cell lines were established. No significant differences in the number of G418-resistant colonies was evident, indicating that all DNAs were equally able to immortalize primary keratinocytes (data not shown). Total cellular DNA was isolated from cell lines at passage 3 and subjected to Southern analysis (Fig. 4). Restriction enzyme digests of the cellular DNA from the wt and BS2 cell lines with a noncutting restriction enzyme for the HPV31 genome (Fig. 4; lanes N) gave rise to several prominent species similar in mobility to supercoiled, open circle, and concatemer forms of viral DNA which we had previously characterized in the biopsy-derived CIN-612 cell line (1). This result indicated that the viral DNA in the wt and BS2 cell lines was present as episomes. There were no significant differences observed in the viral copy number between wt and BS2 cell lines, as measured by phosphorimager analysis of linearized replicated genomes (Fig. 4, lanes S). In contrast, cell lines generated after transfection of HPV31-BS1, -BS3, or -BS4 contained only high-molecular-weight hybridizing DNA, consistent with integrated forms of viral DNA. This was further confirmed by the presence of off-size bands in digests with an enzyme that cuts the HPV31 genome once. These off-size bands are indicative of joint fragments of integrated viral sequences and cellular DNA (Fig. 5; BS3, lane S). The isolated DNAs were also digested with restriction enzymes that specifically cut restriction sites engineered into the mutated sites, and the results confirmed that the cell lines contained the mutations in the E2BS (data not shown). DNA was also isolated from a HPV31-BS2 cell line after cryopreservation followed by an additional nine passages in culture and was found to contain only viral episomes (data not shown). We conclude that BS1, -3, and -4 are required for stable maintenance of HPV31 episomes in human keratinocytes, whereas BS2 is not essential.
FIG. 4.
Southern analysis of DNA from keratinocyte cell lines established after transfection of HPV31 or HPV31 genomes containing E2BS mutants. Ten micrograms of total cellular DNA was digested with BamHI (lanes N), which does not cut the HPV31 genome, or with EcoRV (lanes S), which cuts the HPV31 genome once. Genome copy number control lanes representing 50 and 5 copies per cell are shown on the left. Marker sizes are indicated in kilobases to the right. The migration of supercoiled (a), linearized (b), and open circle, concatemeric, or integrated (c) DNA forms is indicated to the right.
FIG. 5.

RNase protection analysis of RNA isolated from keratinocytes containing HPV31-wt (wt; LKP cell line [15]), HPV31-BS2 (BS2), or HPV31-BS3 (BS3) grown in monolayer cultures (M) or differentiated in raft cultures (R). Total cellular RNA (10 μg) was analyzed by RNase protection using the antisense RNA probe transcribed from linearized pRP742. Lane P contains undigested antisense probe. Lane S contains 32P-end-labeled 1-kb ladder (Gibco BRL), and the sizes are indicated in nucleotides to the right. Transcripts initiated at P97 or P742 are indicated by arrows to the left. The structure of the antisense RNA probe is shown below the autoradiograph. The major initiation sites for the late promoter P742 and the initiation site for the early promoter P97 are indicated by arrowheads, and parts of the E7 and E1 genes are shown below. The dashed line indicates that the P97 start site is not covered by the probe. The splice donor (SD) site at nt 877 is shown as an arrow.
BS2 is not required for differentiation-dependent viral transcription or DNA amplification.
In the previous experiments, no effect of the BS2 mutation was observed on the stable maintenance of episomes in monolayer culture. We next examined whether an intact BS2 is necessary for any of the differentiation-specific events of the viral life cycle. For these studies, HPV31-wt, -BS2, and -BS3 cell lines were grown in organotypic raft cultures for 16 days. Total cellular RNA was then isolated and analyzed by RNase protection using a probe that spans nt 678 to 919 and allows detection of both P97 and P742 transcripts (Fig. 5). P97 transcripts that were spliced at the nt 877 donor or unspliced were detected in monolayer cultures from HPV31-wt, -BS2, and -BS3 cell lines (Fig. 5, lanes M). These transcripts were found to be increased approximately two- to threefold in raft cultures from HPV31-wt and -BS2 cell lines but decreased in the HPV31-BS3 cell line. The reduction in P97 transcripts in the BS3 cell line was consistently seen, but as the viral genomes are integrated in this cell line the significance of this is unclear. Upon differentiation in rafts (Fig. 5, lanes R), we observed additional RNA species in the HPV31-wt and -BS2 cell lines that correspond to initiation at the late P742 promoter (24, 25). The heterogeneity in initiation sites of late messages has been described before and is consistent with TATA-box-less promoters (24, 25). No P742 transcripts are induced in the BS3 cell line, which is consistent with earlier observations that the episomal state of the viral DNA is required for P742 induction and genome amplification (15). Phosphorimager analysis indicated that transcripts which originated from P742 were induced to similar levels in both the wt and BS2 cell lines following differentiation. The slight increase in transcript levels in the BS2 mutant cell line as shown in Fig. 5 was not reproducibly observed. We conclude that the loss of BS2 function does not significantly alter induction of P742, but we cannot exclude the possibility that it induces a minor effect. We also investigated by RNase protection whether transcription of the viral structural genes L1 and L2 were changed in the BS2 cell line upon differentiation compared to the wt. The antisense RNA probe used covers the splice acceptor site at nt 5552 at the beginning of the L1 gene and allows detection of spliced L1 as well as unspliced L2/L1 messages that are induced upon differentiation (25). Total cellular RNA from HPV31-wt, -BS2, or -BS3 monolayer cells (Fig. 6, lanes M) or RNA from cells differentiated in raft cultures (Fig. 6, lanes R) was subjected to RNase protection analysis. No L1 transcription is seen in monolayer cultures or in BS3 raft cultures. Specific signals corresponding to unspliced and spliced L1 RNA can be detected in raft cultures of the wt and BS2 cell lines (Fig. 6). No significant difference in transcript levels could be observed between the wt and BS2 cell lines as judged by phosphorimaging analysis, indicating that the loss of BS2 does not influence transcription of the late gene region.
FIG. 6.
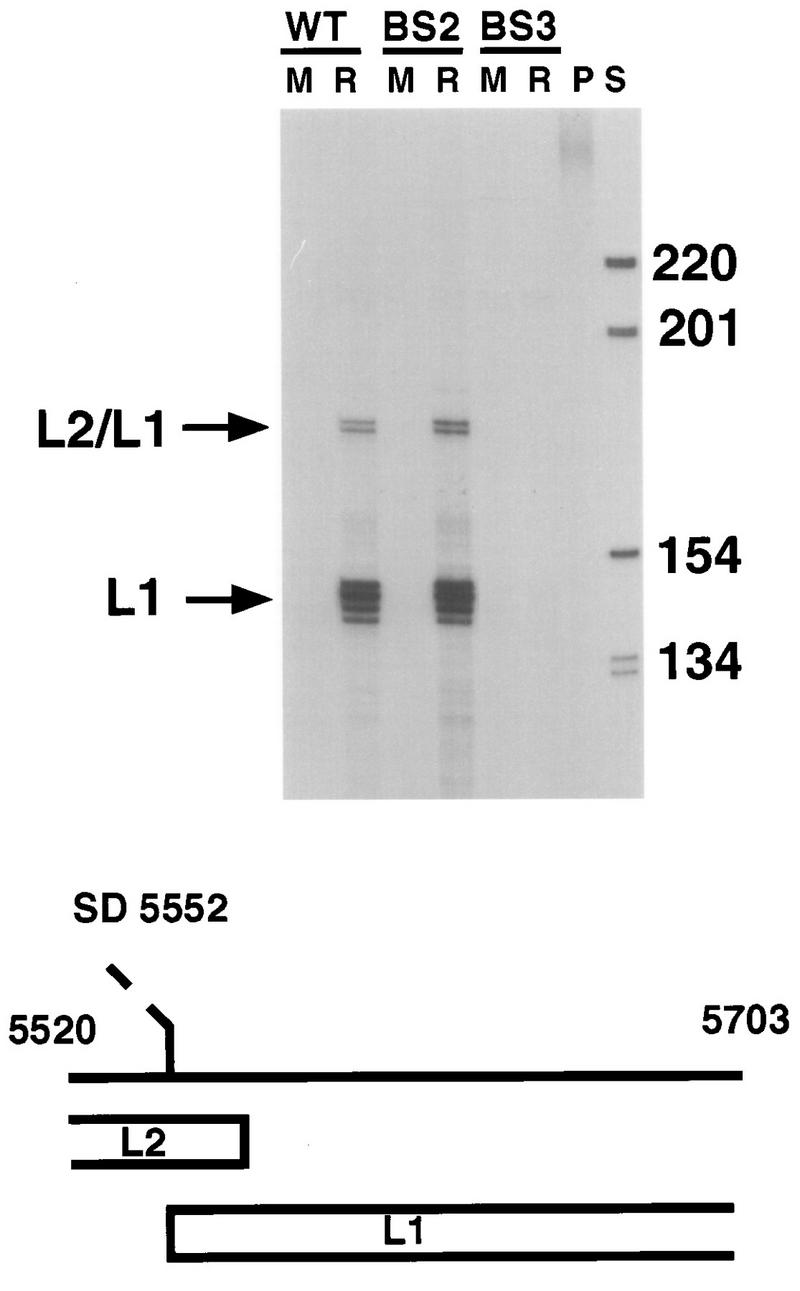
RNase protection analysis of total RNA (20 μg) isolated from keratinocytes containing HPV31-wt (WT; LKP cell line [15]), HPV31-BS2 (BS2), or HPV31-BS3 (BS3) grown in monolayer cultures (M) or differentiated in raft cultures (R). Lane P contains undigested antisense probe which was transcribed from pRPA31L1. Lane S contains 32P-end-labeled 1-kb ladder (Gibco BRL), and the sizes are indicated in nucleotides to the right. The structure of the antisense RNA probe is shown below. Parts of the L2 and L1 open reading frames are indicated. The splice acceptor site at nt 5552 is depicted by a dashed line. Spliced L1 and unspliced L2/L1 transcripts are indicated by arrows on the left of the autoradiogram.
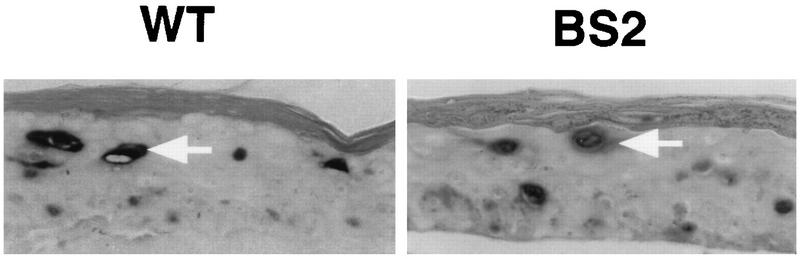
Based on the role of E2 as a replication factor in monolayer cultures, we next used DNA in situ hybridization to investigate whether the differentiation-dependent amplification of HPV31 DNA was altered in the HPV31-BS2 cell line. In raft cultures of both the wt and BS2 cell lines, we detected cells that contained amplified viral DNA (Fig. 7). As this assay is not highly quantitative, we cannot exclude the possibility that slight differences exist in the degree of amplification. However, based on the intensity of staining of individual cells and number of cells that have amplified DNA, there seemed to be no significant differences in amplification between HPV31-wt and -BS2 genomes.
FIG. 7.
DNA in situ hybridization analysis of HPV31-wt (WT) and HPV31-BS2 (BS2) cell lines differentiated in raft cultures. Tissue cross sections were hybridized to an HPV31/33/51-specific probe. Cells which have amplified viral DNA are darkly stained. A representative cell in each panel is indicated by a white arrow.
DISCUSSION
We have examined the role of each of the four conserved E2BS in the context of the complete HPV31 genome through a mutational analysis and analyzed the effects on transient and stable replication as well as viral transcription. All E2BS mutant genomes were found to replicate transiently but to different levels. Mutation of BS1 and BS3 decreased transient replication of HPV31, which could be due to a direct effect on replication or an indirect effect through modulating the expression levels of replication proteins. In previous studies, it has been reported that inactivation of BS3 or deletion of BS1 of HPV11 and -18 greatly reduced transient replication of origin constructs (5, 8, 40, 43).
Unexpectedly, we observed that genomes with a mutated BS4 replicated to levels three- to fourfold higher than wt levels. This is in contrast to previous reports that mutation of BS4 decreased replication efficiency of HPV11 and -18 origin constructs to the same extent as mutation of BS3 (8, 43). The reason for this discrepancy may be the expression source of viral replication proteins. Previous studies used expression vectors, while in our studies, expression of E1 and E2 was from the intact HPV31 genome. It is also possible that mutation of BS4 results in increased transcription from the P97 promoter, which may directly enhance replication as has been observed for several prokaryotic replicons (27). However, several observations argue against a direct coupling of transcription to papillomavirus replication: (i) in vitro replication of BPV1 is not affected by RNA polymerase II inhibitors (61), and (ii) mutation of the early TATA box of HPV11, -18, and -31 has only very minor effects on transient replication of origin plasmids (8, 23, 43). The alternative hypothesis is that HPV31-BS4 mutant genomes can replicate to higher levels because of a loss of E2-mediated P97 repression resulting in increased levels of E1 and/or E2. Repression by E2 of P97 transcription in HPV16 has been reported, and similar activities have also been demonstrated in HPV11 and -18 (9, 10, 42, 50, 54, 55). When an E1 expression vector was cotransfected with HPV31-wt or HPV31-BS mutants, all genomes except the HPV31-BS4 mutant replicated to levels which were four- to fivefold higher. We interpret this to mean that the E1 levels are rate limiting during transient replication of the HPV31-wt as well as the -BS1, -BS2, and -BS3 genomes. The HPV31-BS4 genome either is less responsive to E1 or expresses saturating levels of E1. Furthermore, replication of all genomes but HPV31-BS4 was repressed by cotransfection of an E2 expression vector. We suspect this may be due to an inability of E2 to repress E1 transcription from P97 in the HPV31-BS4 mutant.
We hypothesize that E1-encoding transcripts are expressed from P97 during the initial phases of HPV DNA replication and P97 transcription is negatively regulated by E2 via BS4. This would provide for a feedback loop in which E2 could regulate the extent of replication by modulating E1 transcription. In analyses of HPV31 transcripts expressed during the viral life cycle, the only E1 transcript which could be identified was one that encompassed all early open reading frames and initiated at P97 (28). Additional support for the idea that the concentrations of E1 and E2 control copy number has come from recent studies showing that overexpression of E1 and E2 in a cell line which stably maintains episomal copies of HPV31b results in an increase in the number of viral episomes (12, 13). Mechanisms in which the expression of the initiator protein is under negative control by a virus- or plasmid-encoded factor are common in prokaryotic systems, and similar processes may regulate copy number in HPVs (27, 36). Amplification of the genomes in the differentiating epithelium could then be achieved by expressing E1 from another promoter which may not be repressible by E2, such as the late P742 promoter. A further test of this model would require the demonstration that the levels of E1 or E2 are increased in BS4 mutant genomes.
In contrast to observations made in transient replication assays, neither the HPV31-BS1, -BS3, nor -BS4 genomes were able to establish themselves as episomes in transfected NHKs. The reason for this is unclear; however, it could be that other, yet unknown viral promoters which express factors necessary for maintenance are regulated by E2 through these binding sites. Another possibility is that the E2BS have additional functions aside from replication and transcription to ensure stable maintenance. One potential role may be partitioning of episomes. Evidence for an involvement of E2 in partitioning of BPV1 origin plasmids has been described by Piirsoo and coworkers, who demonstrated a requirement for at least seven E2BS for stable maintenance (38). Since only four E2BS are found in genital HPVs, differences may exist between HPVs and BPV1 in their requirements for stable maintenance. As three of the four mutant genomes tested in our assays deviated from the wt replication efficiency, it is possible that the copy number has to be close to 50 per cell for episomes to be maintained. This could also be due to requirements for sufficient levels of E6 and E7 proteins to immortalize NHKs. In another report, we have shown that HPV31 genomes with mutations in the splice acceptor at nt 3295 replicated efficiently in transient assays but were unable to establish episomes in NHKs (26). This provides further evidence that the requirements for stable maintenance are more stringent than for transient replication.
Genomes which contained a mutated BS2 behaved similar to wt in transient replication assays. We have also observed that in BS2 mutants the ability to maintain episomes as well as their copy number in immortalized keratinocytes was not changed from that of the wild type. Furthermore, differentiation-dependent induction of the P742 promoter and late gene transcription as well as amplification of the genome in HPV31-BS2 cell lines were not significantly different from that seen in wt lines. Therefore, our assays failed to uncover a role for BS2 in the viral life cycle. The conservation of the location of this binding site in the regulatory regions of HPVs among a large number of HPVs suggests a function during the viral life cycle. It is tempting to speculate that it plays a role in regulating other, yet uncharacterized promoters. Alternatively, it could provide a function after amplification and capsid protein expression, such as efficient packaging of the DNA into the virions (45). Alternatively, in vivo, the presence of this E2BS could provide a modest increase in the expression and replication of the virus which was not measurable in our assays.
ACKNOWLEDGMENTS
We acknowledge W. G. Hubert for constructing plasmid pHPV31-E2N-TTL, D. J. Klumpp for plasmid pRPA31L1, and M. Ruesch for pmodBR-HPV31. We thank R. Longnecker and J. T. Schiller for helpful comments on the manuscript.
This was supported by a grant from the NCI to L.A.L. and by a postdoctoral fellowship grant from the Deutsche Forschungsgemeinschaft to F.S.
REFERENCES
- 1.Bedell M A, Hudson J B, Golub T R, Turyk M E, Hosken M, Wilbanks G D, Laimins L A. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedrosian C L, Bastia D. The DNA-binding domain of HPV-16 E2 protein interaction with the viral enhancer: protein-induced DNA bending and role of the nonconserved core sequence in binding site affinity. Virology. 1990;174:557–575. doi: 10.1016/0042-6822(90)90109-5. [DOI] [PubMed] [Google Scholar]
- 3.Bernard B A, Bailly C, Lenoir M C, Darmon M, Thierry F, Yaniv M. The human papillomavirus type 18 (HPV18) E2 gene product is a repressor of the HPV18 regulatory region in human keratinocytes. J Virol. 1989;63:4317–4324. doi: 10.1128/jvi.63.10.4317-4324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blitz I L, Laimins L A. The 68-kilodalton E1 protein of bovine papillomavirus is a DNA binding phosphoprotein which associates with the E2 transcriptional activator in vitro. J Virol. 1991;65:649–656. doi: 10.1128/jvi.65.2.649-656.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang C M, Dong G, Broker T R, Chow L T. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J Virol. 1992;66:5224–5231. doi: 10.1128/jvi.66.9.5224-5231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang C M, Ustav M, Stenlund A, Ho T F, Broker T R, Chow L T. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Vecchio A, Romanczuk H, Howley P M, Baker C C. Transient replication of human papillomavirus DNAs. J Virol. 1992;66:5949–5958. doi: 10.1128/jvi.66.10.5949-5958.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demeret C, Le M M, Yaniv M, Thierry F. Control of HPV 18 DNA replication by cellular and viral transcription factors. Nucleic Acids Res. 1995;23:4777–4784. doi: 10.1093/nar/23.23.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demeret C, Yaniv M, Thierry F. The E2 transcriptional repressor can compensate for Sp1 activation of the human papillomavirus type 18 early promoter. J Virol. 1994;68:7075–7082. doi: 10.1128/jvi.68.11.7075-7082.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong G, Broker T R, Chow L T. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J Virol. 1994;68:1115–1127. doi: 10.1128/jvi.68.2.1115-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dostatni N, Lambert P F, Sousa R, Ham J, Howley P M, Yaniv M. The functional BPV-1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 1991;5:1657–1671. doi: 10.1101/gad.5.9.1657. [DOI] [PubMed] [Google Scholar]
- 12.Frattini M G, Hurst S D, Lim H B, Swaminathan S, Laimins L A. Abrogation of a mitotic checkpoint by E2 proteins from oncogenic human papillomaviruses correlates with increased turnover of the p53 tumor suppressor protein. EMBO J. 1997;16:318–331. doi: 10.1093/emboj/16.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frattini M G, Laimins L A. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc Natl Acad Sci USA. 1994;91:12398–12402. doi: 10.1073/pnas.91.26.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frattini M G, Laimins L A. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology. 1994;204:799–804. doi: 10.1006/viro.1994.1596. [DOI] [PubMed] [Google Scholar]
- 15.Frattini M G, Lim H B, Laimins L A. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haugen T H, Cripe T P, Ginder G D, Karin M, Turek L P. Trans-activation of an upstream early gene promoter of bovine papilloma virus-1 by a product of the viral E2 gene. EMBO J. 1987;6:145–152. doi: 10.1002/j.1460-2075.1987.tb04732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegde R S, Grossman S R, Laimins L A, Sigler P B. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 18.Hermonat P L, Spalholz B A, Howley P M. The bovine papillomavirus P2443 promoter is E2 trans-responsive: evidence for E2 autoregulation. EMBO J. 1988;7:2815–2822. doi: 10.1002/j.1460-2075.1988.tb03137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirochika H, Hirochika R, Broker T R, Chow L T. Functional mapping of the human papillomavirus type 11 transcriptional enhancer and its interaction with the trans-acting E2. Genes Dev. 1988;2:54–67. doi: 10.1101/gad.2.1.54. [DOI] [PubMed] [Google Scholar]
- 21.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 22.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, editors. Fundamental virology. New York, N.Y: Raven Press; 1996. pp. 947–978. [Google Scholar]
- 22a.Hubert, W. G. Unpublished observations.
- 23.Hubert, W. G., and L. A. Laimins. Unpublished observations.
- 24.Hummel M, Hudson J B, Laimins L A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hummel M, Lim H B, Laimins L A. Human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J Virol. 1995;69:3381–3388. doi: 10.1128/jvi.69.6.3381-3388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klumpp D J, Stubenrauch F, Laimins L A. Mutation of the splice acceptor at nucleotide 3295 of human papillomavirus type 31 disrupts stable but not transient viral replication. J Virol. 1997;71:8186–8194. doi: 10.1128/jvi.71.11.8186-8194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornberg A, Baker T. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman and Company; 1992. [Google Scholar]
- 28.Laimins L A. Human papillomaviruses target differentiating epithelia for virion production and malignant conversion. Semin Virol. 1996;7:305–313. [Google Scholar]
- 29.Laimins L A. The biology of human papillomaviruses: from warts to cancer. Infectious Agents Dis. 1993;2:74–86. [PubMed] [Google Scholar]
- 30.Li R, Knight J, Bream G, Stenlund A, Botchan M. Specific recognition nucleotides and their DNA context determine the affinity of E2 protein for 17 binding sites in the BPV-1 genome. Genes Dev. 1989;3:510–526. doi: 10.1101/gad.3.4.510. [DOI] [PubMed] [Google Scholar]
- 31.Lu J Z, Sun Y N, Rose R C, Bonnez W, McCance D J. Two E2 binding sites (E2BS) alone or one E2BS plus an A/T-rich region are minimal requirements for the replication of the human papillomavirus type 11 origin. J Virol. 1993;67:7131–7139. doi: 10.1128/jvi.67.12.7131-7139.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride A A, Romanczuk H, Howley P M. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 33.McCance D, Kopan R, Fuchs E, Laimins L A. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro. Proc Natl Acad Sci USA. 1988;85:7169–7173. doi: 10.1073/pnas.85.19.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyers C, Frattini M G, Hudson J B, Laimins L A. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 35.Mohr I J, Clark R, Sun S, Androphy E J, MacPherson P, Botchan M R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 36.Nordstrom K. Control of plasmid replication—how do DNA iterons set the replication frequency? Cell. 1990;63:1121–1124. doi: 10.1016/0092-8674(90)90405-4. [DOI] [PubMed] [Google Scholar]
- 37.Peden K W, Pipas J M, Pearson W S, Nathans D. Isolation of mutants of an animal virus in bacteria. Science. 1980;209:1392–1396. doi: 10.1126/science.6251547. [DOI] [PubMed] [Google Scholar]
- 38.Piirsoo M, Ustav E, Mandel T, Stenlund A, Ustav M. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 1996;15:1–11. [PMC free article] [PubMed] [Google Scholar]
- 39.Prakash S S, Horwitz B H, Zibello T, Settleman J, DiMaio D. Bovine papillomavirus E2 gene regulates expression of the viral E5 transforming gene. J Virol. 1988;62:3608–3613. doi: 10.1128/jvi.62.10.3608-3613.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remm M, Brain R, Jenkins J R. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic Acids Res. 1992;20:6015–6021. doi: 10.1093/nar/20.22.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rheinwald J G, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 42.Romanczuk H, Thierry F, Howley P M. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990;64:2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Ruesch, M. N. Unpublished results.
- 43.Russell J, Botchan M R. cis-acting components of human papillomavirus (HPV) DNA replication: linker substitution analysis of the HPV type 11 origin. J Virol. 1995;69:651–660. doi: 10.1128/jvi.69.2.651-660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders C M, Maitland N J. Kinetic and equilibrium binding studies of the human papillomavirus type-16 transcription regulatory protein E2 interacting with core enhancer elements. Nucleic Acids Res. 1994;22:4890–4897. doi: 10.1093/nar/22.23.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiller, J. T. Personal communication.
- 46.Sedman J, Stenlund A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo Y S, Muller F, Lusky M, Gibbs E, Kim H Y, Phillips B, Hurwitz J. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spalholz B A, Lambert P F, Yee C L, Howley P M. Bovine papillomavirus transcriptional regulation: localization of the E2-responsive elements of the long control region. J Virol. 1987;61:2128–2137. doi: 10.1128/jvi.61.7.2128-2137.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spalholz B A, McBride A A, Sarafi T, Quintero J. Binding of bovine papillomavirus E1 to the origin is not sufficient for DNA replication. Virology. 1993;193:201–212. doi: 10.1006/viro.1993.1116. [DOI] [PubMed] [Google Scholar]
- 50.Steger G, Corbach S. Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J Virol. 1997;71:50–58. doi: 10.1128/jvi.71.1.50-58.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stenlund A. Papillomavirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 679–698. [Google Scholar]
- 52.Sverdrup F, Khan S A. Two E2 binding sites alone are sufficient to function as the minimal origin of replication of human papillomavirus type 18 DNA. J Virol. 1995;69:1319–1323. doi: 10.1128/jvi.69.2.1319-1323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szymanski P, Stenlund A. Regulation of early gene expression from the bovine papillomavirus genome in transiently transfected C127 cells. J Virol. 1991;65:5710–5720. doi: 10.1128/jvi.65.11.5710-5720.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan S H, Leong L E, Walker P A, Bernard H U. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J Virol. 1994;68:6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thierry F, Howley P M. Functional analysis of E2-mediated repression of the HPV18 P105 promoter. New Biol. 1991;3:90–100. [PubMed] [Google Scholar]
- 56.Thierry F, Yaniv M. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 1987;6:3391–3397. doi: 10.1002/j.1460-2075.1987.tb02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ustav E, Ustav M, Szymanski P, Stenlund A. The bovine papillomavirus origin of replication requires a binding site for the E2 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:898–902. doi: 10.1073/pnas.90.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y J, Parker L M, Binder N E, Beckett M A, Sinard J H, Griffiths C T, Rheinwald J G. The mesothelial keratins: a new family of cytoskeletal proteins identified in cultured mesothelial cells and nonkeratinizing epithelia. Cell. 1982;31:693–703. doi: 10.1016/0092-8674(82)90324-5. [DOI] [PubMed] [Google Scholar]
- 61.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]