Abstract
It has been observed that AU-rich sequences form homologous recombination hot spots in brome mosaic virus (BMV), a tripartite positive-stranded RNA virus of plants (P. D. Nagy and J. J. Bujarski, J. Virol. 71:3799–3810, 1997). To study the effect of GC-rich sequences on the recombination hot spots, we inserted 30-nucleotide-long GC-rich sequences downstream of AU-rich homologous recombination hot spot regions in parental BMV RNAs (RNA2 and RNA3). Although these insertions doubled the length of sequence identity in RNA2 and RNA3, the incidence of homologous RNA2 and RNA3 recombination was reduced markedly. Four different, both highly structured and nonstructured downstream GC-rich sequences had a similar “homologous recombination silencing” effect on the nearby hot spots. The GC-rich sequence-mediated recombination silencing mapped to RNA2, as it was observed when the GC-rich sequence was inserted at downstream locations in both RNA2 and RNA3 or only in the RNA2 component. On the contrary, when the downstream GC-rich sequence was present only in the RNA3 component, it increased the incidence of homologous recombination. In addition, upstream insertions of similar GC-rich sequences increased the incidence of homologous recombination within downstream hot spot regions. Overall, this study reveals the complex nature of homologous recombination in BMV, where sequences flanking the common hot spot regions affect recombination frequency. A replicase-driven template-switching model is presented to explain recombination silencing by GC-rich sequences.
RNA recombination has been demonstrated for an increasing number of viruses (3, 6, 6a, 7, 15, 26, 27). It was found to occur not only between replication-competent viral RNAs but also between defective viral RNAs and between viral and host RNAs (15, 26, 27). RNA recombination may have several functions during the viral life cycle, including repairing defective RNA molecules, increasing sequence variability, and facilitating viral evolution and adaptation.
It has been proposed that RNA recombination occurs when viral replicase switches from one template to another, thus copying noncontiguous RNA sequences (6a, 9, 12, 13, 15, 19, 26). Both virus-encoded replicase protein(s) and sequences or secondary structures of RNA templates have been demonstrated to affect the selection of crossover sites. For instance, mutations within helicase-like protein 1a of brome mosaic virus (BMV) changed the distribution of nonhomologous recombination junctions relative to that seen with the wild-type (wt) enzyme (19). In addition, mutations that destabilized portions of the intermolecular duplex formed between the BMV RNAs resulted in a shift of crossovers toward the more stable portions of the heteroduplex (17). The role of sequence motifs and hairpin-loop structures in the selection of crossover sites is well described for turnip crinkle carmovirus (8–10). Stable hairpin-loop structures are not favored in a recombination system in tombusviruses, resulting in a shift of junction sites to nonstructured portions of the RNA (28, 29).
One of the best-characterized RNA recombination systems is that of BMV (6a, 7). BMV is a model positive-stranded RNA virus that belongs to the alphavirus supergroup. Its genome consists of three separate RNA components. While RNA1 and RNA2 code for RNA replication proteins 1a and 2a, respectively, RNA3 is dispensable for infection in barley protoplasts (1). Thus, most recombination studies have been done with the RNA3 component. In particular, it has been observed that mutations, deletions, or insertions (sequence duplications) introduced into the conserved 3′ noncoding region of BMV RNA3 are frequently repaired by recombination with the corresponding 3′ noncoding region of either RNA1 or RNA2 (16, 24).
Two major types of recombinants have been described for BMV, homologous and nonhomologous, with homologous recombination being the more frequent type (16, 24). The differences between the two recombination types are not only that nonhomologous recombination occurs between heterologous sequences while homologous recombination occurs between similar sequences but also that they have different sequence requirements. In particular, nonhomologous recombination requires short (30 nucleotides [nt] or longer) sequence complementarity between same-sense RNA substrates (17). It has been proposed by us that the formation of a local heteroduplex within the complementary region of the RNAs brings the donor and the acceptor RNAs into proximity and occasionally forces the replicase to switch templates (7, 17).
Homologous crossover events were found to occur within rather short (15- to 60-nt) similar sequences present in (common to) the recombining RNAs, giving rise to either precise or imprecise recombinants (18). According to a model proposed by us (18, 20), the nascent strand precisely anneals (or misanneals, in the case of imprecise events) to complementary sequences on the acceptor RNA strand before chain elongation is resumed by the replicase. In addition, we proposed that the formation of imprecise recombinants with nucleotide substitutions or extra (nontemplate) nucleotides could occur through replicase errors during crossover events (18, 20).
Importantly, however, not all homologous regions can support recombination in BMV (7, 17, 21). Characterization of BMV-derived and artificial recombination hot spots revealed that most of the precise and imprecise recombination events occurred within or close to short AU-rich sequences (20, 21). AU-rich sequences alone, when present on both recombining RNAs, were, however, only moderately active in homologous recombination. High recombination frequency was observed when, in addition to common AU-rich sequences, the recombining RNAs contained similar sequences of average or higher GC content (21). The relative positions of the common AU-rich and the less common AU-rich (i.e., GC-rich or average AU+GC content) sequences were also important factors, with the AU-rich sequences being located at the downstream location and the less AU-rich sequences being located at the upstream location in the most favored homologous recombination hot spots (21) (see also Fig. 1). In the proposed model, the BMV replicase may occasionally pause (stall) within or in the vicinity of the AU-rich sequence while copying the negative strand of RNA3. During the pause, the replicase may march backward on the primary template (12, 18, 21), thus allowing the 3′ end of the positive-stranded incomplete nascent RNA to dissociate from the primary template because of the weak A-U base pairing. Subsequently, the free 3′ end of the nascent strand may hybridize to the complementary target region present in negative-stranded acceptor RNA2. This model proposes that the role for the less AU-rich (GC-rich or average AU+GC content) upstream common regions would be to facilitate (stabilize) the hybridization of the nascent RNA with the complementary region in the acceptor RNA during the template switch (21). The final step in recombination is the resumption of strand elongation by the BMV replicase on the acceptor template (21).
FIG. 1.
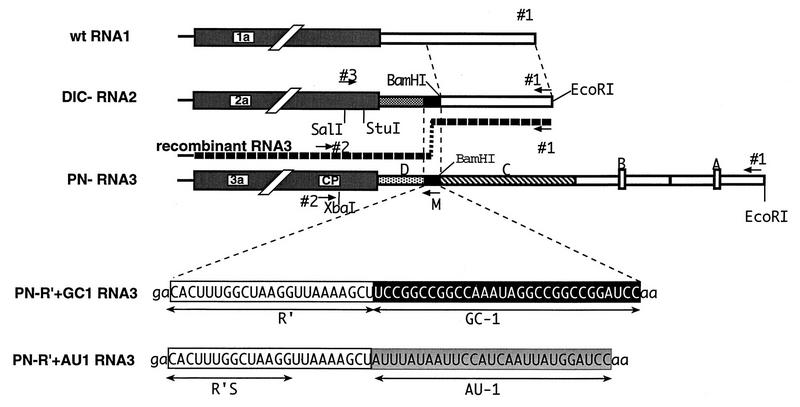
Schematic representation of the 3′ noncoding regions of wt BMV RNA1, DIC RNA2, and PN RNA3 constructs used for testing the effect of GC-rich flanking sequences on homologous recombination. The white box represents the 3′ noncoding region of wt RNA1, whose last 236 nt are also present in the DIC series of RNA2 constructs. The PN series of RNA3 constructs contains a ∼1,250-nt long chimeric 3′ noncoding region with four segments (regions A to D; see reference 21 for details). Regions A and B are the 3′ tRNA-like sequences with marker deletions (shown by small boxes), region C is a 3′ sequence from cowpea chlorotic mottle virus, and region D is the upstream, nonmodified portion of the 3′ noncoding region of wt RNA3. The extended 3′ noncoding region in parental RNA3 serves to facilitate recombinant RNA3 isolation. PN-R′+GC1 RNA3 contains a 23-nt-long sequence (marked as R′; positions 196 to 219 in wt RNA2; all positions are counted from the 3′ end in this work [2]) and a 30-nt artificial GC-rich region (designated GC-1). The locations of the restriction sites are indicated, while oligonucleotide primers used for PCR are shown by short horizontal numbered arrows. Arrow M depicts the position of mutagenesis oligodeoxynucleotides 93, 13, 148, and 28 (see Materials and Methods). In DIC-R′+GC1 RNA2, the 3′ noncoding sequence upstream of R′+GC1 is derived from wt RNA2 (positions 220 to 293 from the 3′ end), while the 3′-terminal region is derived from wt RNA1 (positions 1 to 236 from the 3′ end). Nucleotide sequences representing R′, R′S, GC-1, and AU-1 are shown by double-headed arrows.
In this work, we further investigated the role of RNA sequences in homologous recombination in BMV. In particular, the effect of short GC-rich flanking sequences on homologous recombination hot spots was examined. We demonstrated that insertions of different GC-rich sequences downstream of homologous recombination hot spots doubled the length of sequence identity but reduced the incidence of homologous recombination markedly. This “recombination silencing” effect of GC-rich sequences mapped to RNA2. We discuss our results in relation to mechanisms responsible for this novel recombination silencing phenomenon.
MATERIALS AND METHODS
Materials.
Plasmids pB1TP3, pB2TP5, and pB3TP7 (11) were used to synthesize in vitro-transcribed wt BMV RNA1, RNA2, and RNA3 components, respectively, and to engineer modified RNA2 and RNA3 constructs (see below). Plasmids PN-H65 (PN-R′) and DIC-0 (DIC-R′) were constructed as described previously (18, 20, 21). Moloney murine leukemia virus reverse transcriptase was from Gibco BRL (Gaithersburg, Md.), Taq DNA polymerase, restriction enzymes, and T7 RNA polymerase were from Promega Corp. (Madison, Wis.), and a Sequenase kit was from United States Biochemical Corp. (Cleveland, Ohio).
The following oligonucleotide primers were used in this study (the unique EcoRI and BamHI sites are underlined, and alternative bases are shown in parentheses): 1, 5′-CAGTGAATTCTGGTCTCTTTTAGAGATTTACAG-3′; 2, 5′-CTGAAGCAGTGC-CTGCTAAGGCGGTC-3′; 3, 5′-AGAAGGTCGACGATTACGCTACC-3′; 13, 5′-CAGTGGATCCGCCCGCCCTATTTGCCCGCCCG(T/A)-AGCTTTTAA(C/A)CTTAGCC-3′; 28, 5′-CAGTGGATCCAAGCCCCGGCCCCGG(A/C)CTTAGCCAAAGTG-3′; 93, 5′-CAGTGGATCCGGCCGGCC TAT T TGGCCGGCCGG( T/A)-AGC T T T TAA(C/A)C T TAGCC-3′; 148, 5′-CAGTGGATCCAAGCGTCGTACTACGACGC(T/C)TG(T/A)-AGCTTTTAA(C/A)CTTAGCC-3′; and 185, 5′-CAGTGGATCCGGCCGGCCAAATAGGCCGGCCGG(A/C)-CACTTTGGCTAAGGTTAAAAGC-3′ .
Engineering of plasmid constructs.
Plasmids of the DIC and N2 series are derivatives of pB2TP5, while plasmids of the PN series (described below) are derivatives of pB3TP7. A PCR-based approach (18) was used to generate the constructs DIC-R′+GC1, DIC-R′+GC2, DIC-R′+GC3, and DIC-R′S+GC5 by use of primer 3 and one of the following respective mutagenesis primers: 93, 13, 148, and 28 (shown schematically as primer M in Fig. 1). The PCR-amplified DNA fragments were digested with BamHI and StuI (Fig. 1) and used to replace the corresponding fragment in the DIC-0 RNA2 construct (20, 21). The N2-R′+GC1 construct was obtained by digesting DIC-R′+GC1 with BamHI and treating the digest with T4 DNA polymerase. The enzymes were heat inactivated at 75°C for 10 min, and the DNA was digested with EcoRI. After the restriction enzyme digestions, the large BamHI-EcoRI fragment, which included the plasmid sequences as well, was isolated from an agarose gel, followed by ligation with the ∼200-bp HindIII (after treatment with T4 DNA polymerase [25])-EcoRI fragment of pB2TP5.
To obtain plasmids PN-R′+GC1, PN-R′+GC2, PN-R′+GC3, and PN-R′S+GC5, a ∼200-bp 3′ cDNA fragment derived from PN-H65, a plasmid containing full-length cDNA of BMV RNA3 with a modified 3′ end that included R′ (18), was amplified by PCR with primer 2 and one of the following respective mutagenesis primers: 93, 13, 148, and 28. The amplified cDNA fragments were digested with BamHI and XbaI and then were used to replace the 3′ 166-bp BamHI-XbaI fragment in PN-H65.
Constructs DIC-R′+AU1+GC1 and DIC-R′+GC1+AU1 were obtained by digesting DIC-R′+AU1 and DIC-R′+GC1, respectively, with BamHI, followed by filling in of the ends with T4 DNA polymerase and subsequent heat inactivation (as described above) and digestion with EcoRI. The large fragments, which included the vector sequences, were isolated from agarose gels and ligated with the ∼260-bp HindIII (after treatment with T4 DNA polymerase)-EcoRI fragments of DIC-R′+GC1 and DIC-R′+AU1. The same approach was used to construct PN-R′+AU1+GC1 and PN-R′+GC1+AU1, but with PN-R′+AU1 and PN-R′+GC1, respectively, and the small fragment of either PN-R′+GC1 or PN-R′+AU1.
To obtain the PN-GC1+R′ RNA3 construct, the entire 3′-end fragment was amplified from PN-H65 by PCR with primers 1 and 185. The resulting PCR product was digested with BamHI (treated with T4 DNA polymerase) and EcoRI and then ligated between the EcoRV-EcoRI sites in PN-H149, which contained a unique EcoRV site corresponding to position 238 (from the 3′ end in pB3TP7) and the 3′-terminal EcoRI site.
Construct DIC-GC1+R′ was made as follows. The entire 3′-end fragment was amplified from DIC-0 by PCR with primers 1 and 185. The resulting PCR product was digested with BamHI (treated with T4 DNA polymerase) and EcoRI and then ligated between the SmaI-EcoRI sites in DIC-h3, which contained a unique SmaI site at position 219 (as counted from the 3′ end in pB2TP5) and the EcoRI site at the 3′ terminus. Construct DIC-h3 was obtained as follows. A cDNA fragment was PCR amplified from DIC-R′+AU1 with primers 3 and h3 (5′-CAGTGGATCCGACAGGGTCTCTACCTGCCTGACCAGGAG-3′), and the DNA was digested with StuI-BamHI restriction enzymes. The DNA was then ligated between the StuI-BamHI sites of DIC-R′+AU1 (21).
The entire PCR-amplified regions in all of the above-described constructs were sequenced to confirm the mutations introduced.
Full-length cDNA clones representing two different types of homologous recombinants (rec-R′+GC1 and rec-R′+GC3; see Fig. 5) were constructed by replacing the 3′-terminal BamHI-EcoRI inserts of PN-R′+GC1 and PN-R′+GC3, respectively, with the corresponding fragment of DIC-R′+GC1 (Fig. 1). The construction of rec-R′ has been described elsewhere (21).
FIG. 5.
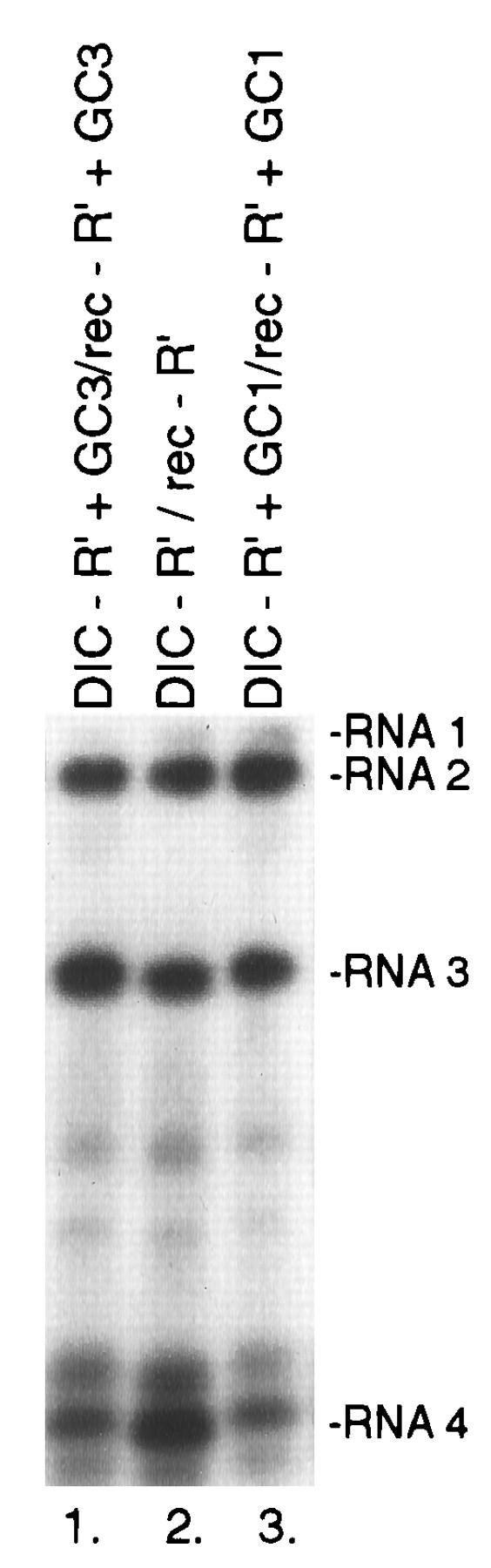
Analysis of levels of accumulation of various parental RNA2 mutants and reconstructed RNA3 recombinants in whole plants as determined by Northern blotting. Leaves of C. quinoa were inoculated with a mixture of in vitro-transcribed wt RNA1 and the RNA2 and RNA3 mutants (as shown above the lanes). C. quinoa plants were incubated for 10 days. Total RNA extracts were isolated from single local lesions, and equal amounts of RNAs were separated by electrophoresis in a 1% agarose gel. The RNA was transferred to a nylon membrane and probed with a 200-nt-long 32P-labeled RNA probe specific for the 3′ noncoding region of RNA1 to RNA4 as described in Materials and Methods.
Inoculation of plants, reverse transcription (RT)-PCR amplifications, cloning, and sequencing.
Leaves of Chenopodium quinoa were inoculated with a mixture of the transcribed BMV RNA components as described by Nagy and Bujarski (16, 18). Briefly, a mixture of ∼4 μg of each transcript was used to inoculate one fully expanded leaf. A total of six leaves were inoculated for each RNA3 mutant. Each experiment was repeated one or more times.
To clone a representative recombinant RNA3 from a given local lesion and to ensure that the isolated recombinants were independent, we cut out local lesions located far apart from each other on the inoculated leaves (only one to four lesions were chosen from each leaf; for details, see references 18 and 21). Total RNA was isolated from separate local lesions and used for RT-PCR amplification exactly as described previously (18). The 3′-end sequence of the progeny RNA3 was amplified with primers 1 and 2 (Fig. 1), and the sizes of the cDNA products were estimated by electrophoresis in 1.5% agarose gels (25). The cDNA fragments were digested with EcoRI-XbaI restriction enzymes and ligated between these sites in the pGEM3 zf(−) cloning vector (Promega). Sites of crossovers were localized by sequencing with Sequenase according to the manufacturer’s specifications. Only a single RT-PCR clone from a given local lesion sample was sequenced to avoid possible sibling clones (18, 21).
To analyze the accumulation of reconstructed RNA3 recombinants, total RNA was extracted 10 days postinoculation. One-fifth of the total RNA extract was separated by electrophoresis in a 1% agarose gel, followed by transfer to a Hybond N+ (Amersham) nylon membrane and hybridization to a 32P-radiolabeled BMV RNA probe as described by Kroner et al. (14).
The 3′ regions of parental RNA2 constructs were amplified as cDNA by RT-PCR with primers 1 and 3, and total RNA preparations were extracted from separate local lesions on C. quinoa 14 days after inoculation. Thereafter, the amplified cDNA was digested with SalI-EcoRI, followed by ligation into the corresponding sites of pGEM3 zf(−).
The possibility that the RNA3 recombinant represented RT-PCR artifacts was excluded by detection of similarly sized (shorter than parent sized) de novo homologous recombinants by Northern blotting of total RNA extracts from local lesions as described above. Also, RT-PCR control amplification of the RNAs present in the inoculum detected only parent-sized, not recombinant-sized, RNA3 recombinants (data not shown) (18, 20, 21).
RESULTS
Effect of downstream GC-rich sequences on homologous recombination hot spots.
The homologous recombination system that we used includes derivatives of BMV RNA2 and RNA3 constructs that have modified 3′ noncoding regions. The RNA3 construct designated PN-R′ RNA3 contains a 23-nt insert (R′) that was similar to the corresponding region in the RNA2 construct designated DIC-R′ RNA2 (Fig. 1). In DIC-R′ RNA2, the 3′-terminal 196 nt were replaced with the 3′-terminal 236 nt of wt RNA1 (21). This 3′ sequence arrangement separated R′ from the minus-strand initiation promoter and allowed for extensive modifications of R′ and the nearby sequences without debilitating the replication of RNA2. In addition, the extended 3′ noncoding region in PN-R′ RNA3 made this parental RNA less competitive (fit) than the shorter, de novo recombinant RNA3 constructs. This property facilitated the accumulation and isolation of recombinant RNA3 constructs generated in plants. The host system used for these studies was C. quinoa, on which parental BMV RNAs induce the formation of local lesions. These local lesions (regardless of whether they contain or lack de novo recombinant RNA3 constructs) are similar phenotypically, eliminating biased sampling. The common (i.e., present on both RNA2 and RNA3) R′ sequences were previously found to support homologous RNA2-RNA3 recombination in 39% of local lesions in C. quinoa plants (Fig. 2A) (21).
FIG. 2.
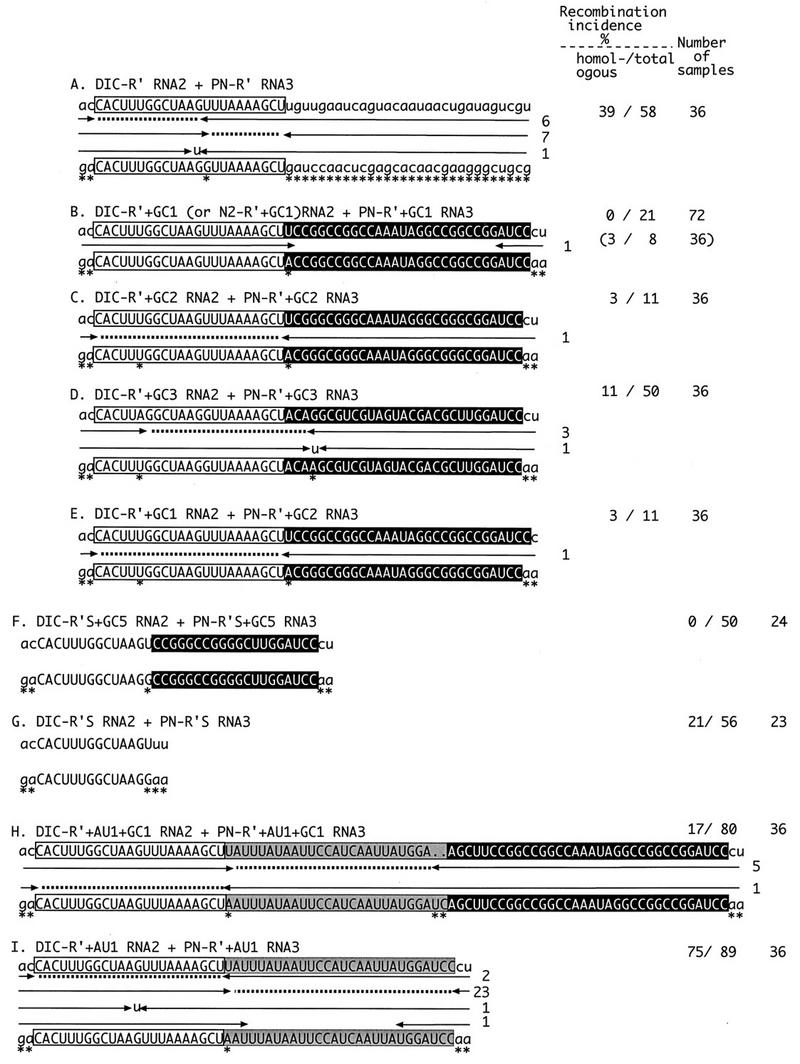
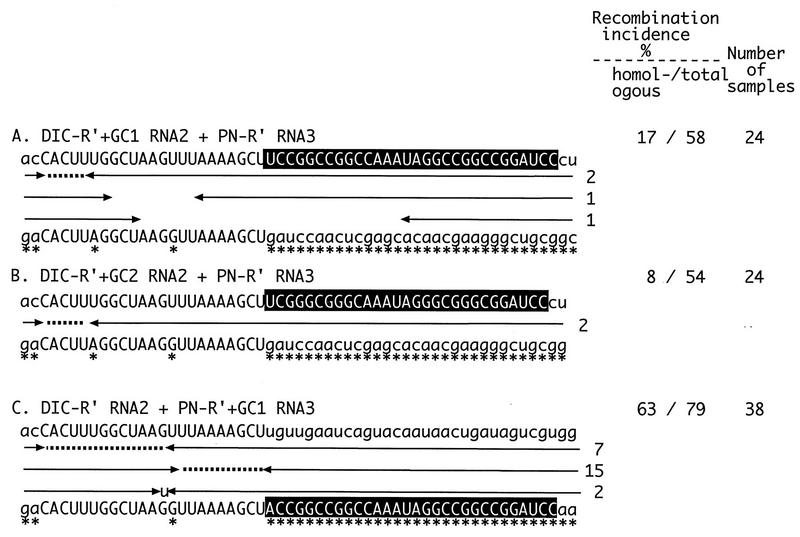
Diagram summarizing the recombination frequencies and distributions of crossover sites in the homologous RNA2-RNA3 recombinants isolated from infections with wt RNA1, the DIC series of RNA2 constructs, and the PN series of RNA3 constructs. Homologous RNA2 and RNA3 positive-sense sequences are shown on the top and bottom lines, respectively. Uppercase letters depict the homologous R′ segment (white box) and the GC-rich segment (black box). Marker mutations are indicated by asterisks under the RNA3 sequences. Each recombinant contains 3′-terminal sequences derived from RNA2 on the right side and 5′-terminal sequences derived from RNA3 on the left side (as shown schematically in Fig. 1). The incidence of each RNA3 recombinant is shown by numbers to the right of the arrows. Each entry represents an RNA2-RNA3 recombinant isolated from a separate local lesion. Each leftward-pointing arrow denotes the last nucleotide derived from RNA2, and each rightward-pointing arrow denotes the first nucleotide derived from RNA3. Dotted lines show ambiguous regions that could be derived from either RNA2 or RNA3 in the precise homologous recombinants. Gaps between opposing arrows show deleted nucleotides. Nontemplate nucleotides and nucleotide substitutions generated during the crossover events are shown by lowercase letters between the arrows. The nucleotide sequences in the imprecise recombinants with ambiguous crossovers were arbitrarily placed with the upstream junction. The percentages of homologous and total (the latter includes both homologous and background recombinants; see Results) recombination incidences were calculated by sequencing a representative number of cloned recombinant RNA3 molecules. The numbers of total RNA samples obtained from separate local lesions are shown on the right. The incidence of recombination shown in parentheses was obtained with N2-R′+GC1 RNA2. This construct contains R′, GC1, an upstream 3′ noncoding sequence (positions 220 to 293 from the 3′ end in wt RNA2), and a downstream 3′ noncoding sequence (positions 1 to 195 from the 3′ end in wt RNA2).
To test the effect of sequences flanking homologous recombination hot spots, we inserted a 30-nt artificial GC-rich sequence (designated GC1, with 73% G+C content) into DIC-R′ RNA2 and PN-R′ RNA3 downstream of 23-nt R′ (Fig. 1). The insertion of the GC1 sequence extended the length of sequence identity between RNA2 and RNA3 from 23 nt to 53 nt. The resulting cDNA constructs were used to generate in vitro RNA transcripts of DIC-R′+GC1 RNA2 and PN-R′+GC1 RNA3, followed by coinoculation with wt RNA1 onto leaves of C. quinoa (for simplicity, the use of the wt RNA1 component for all plant inoculations discussed below will not be mentioned). RT-PCR analysis of total RNA extracts obtained from 72 separate local lesions did not detect any homologous RNA2-RNA3 recombinants (Fig. 2B). Thus, the presence of downstream common GC1 sequences eliminated RNA2-RNA3 homologous recombination within R′ hot spot regions.
To examine whether other GC-rich sequences can inhibit the recombination activity of R′, two other artificial GC-rich sequences were tested. One of these sequences (designated GC3, 29 nt long) was less GC rich (59%) than GC1 (73%) but, like GC1, could predictably form a stable secondary structure (data not shown). The other GC-rich sequence (designated GC2, 29 nt long) contained as many GC nucleotides (72%) as GC1 but was likely to form a single-stranded region (data not shown). The incidence of homologous recombination was low for both GC2 and GC3 inserts (infections with DIC-R′+GC2 RNA2 and PN-R′+GC2 RNA3 or DIC-R′+GC3 RNA2 and PN-R′+GC3 RNA3, respectively; Fig. 2C and D). A heterologous combination of GC1 and GC2 sequences in RNA2 and RNA3 constructs (DIC-R′+GC1 RNA2 and PN-R′+GC2 RNA3; Fig. 2E) also reduced significantly the occurrence of homologous RNA2-RNA3 recombinants.
To test whether the GC-rich sequence-mediated recombination silencing was effective on sequences other than the above-described R′ recombinogenic sequence, we used R′S (14 nt; Fig. 1) and R′+AU1 (50 nt; Fig. 1) sequences. Common R′S and R′+AU1 sequences alone could support homologous recombination in 21% (infections with DIC-R′S RNA2 and PN-R′S RNA3; Fig. 2G) and 75% (infections with DIC-R′+AU1 RNA2 and PN-R′+AU1 RNA3; Fig. 2I) (see also reference 21) of local lesions. Insertions of GC5 (20 nt long, with 80% G+C content) downstream of R′S eliminated the appearance of homologous recombinants in DIC-R′S+GC5 RNA2 and PN-R′S+GC5 RNA3 infections (Fig. 2F). Insertions of GC1 at downstream locations decreased the incidence of homologous recombination within R′+AU1 to 17% (infections with DIC-R′+AU1+GC1 RNA2 and PN-R′+AU1+GC1 RNA3; Fig. 2H). These experiments confirmed the silencing effect of the downstream GC-rich sequences (see Discussion). Thus, four GC-rich sequences with different primary sequences and secondary structures greatly reduced the recombination incidence within different hot spot regions when present in both RNAs at downstream locations.
The possibility that GC-rich regions may switch the location of homologous crossovers to locations downstream of GC regions is not supported by the data in Fig. 2. Putative RNA2-RNA3 recombinants with downstream crossovers were expected to be fully replication competent and would be detectable due to the presence of appropriate marker mutations within the 3′-end sequences of both parental RNAs.
GC-rich sequence-mediated recombination silencing maps to RNA2.
To test if the presence of a GC-rich sequence at a downstream location in RNA2 alone is sufficient to silence homologous recombination, we used two different RNA2 constructs that, in addition to the common R′, carried either GC1 or GC2. The corresponding RNA3 constructs contained R′ alone. A markedly reduced incidence of homologous recombination was observed in these infections (infections with DIC-R′+GC1 RNA2 and PN-R′ RNA3 or DIC-R′+GC2 RNA2 and PN-R′ RNA3; Fig. 3A and B). These results confirmed that GC-rich sequences downstream of a homologous recombination hot spot reduced the incidence of homologous recombination when present in RNA2 alone.
FIG. 3.
Distribution of crossover sites in homologous recombinant RNA3 molecules and the incidence of recombination obtained with pairs of RNA2-RNA3 constructs that contained R′ while one of them lacked the GC-rich sequence. The total numbers of samples analyzed and other features are as described in the legend to Fig. 2.
To test if the presence of the GC1 sequence in RNA3 alone can inhibit recombination within the upstream R′ hot spot sequence, PN-R′+GC1 RNA3 was used for inoculation in combination with DIC-R′ RNA2. Surprisingly, we observed an increased incidence of homologous recombination (63% with DIC-R′ RNA2 and PN-R′+GC1 RNA3; Fig. 3C) compared with that in infections where both RNA2 and RNA3 constructs had R′ but lacked GC1 (39% with DIC-R′ RNA2 and PN-R′ RNA3; Fig. 2A) or had GC1 only in RNA2 (17% with DIC-R′+GC1 RNA2 and PN-R′ RNA3; Fig. 3A). Interestingly, a shift of junction sites toward the 3′ portion of R′ (i.e., toward GC1) was apparent with DIC-R′ RNA2 and PN-R′+GC1 RNA3 (Fig. 3C) compared with DIC-R′ RNA2 and PN-R′ RNA3 (Fig. 2A). These data suggested that a GC-rich sequence can increase the frequency of homologous recombination and can modify the profile of recombinant junctions when present in RNA3 alone at a downstream position. In addition, GC-rich sequences are detrimental to homologous recombination when present in both RNAs or in RNA2 alone (see Discussion).
Effect of upstream GC-rich sequences on homologous recombination.
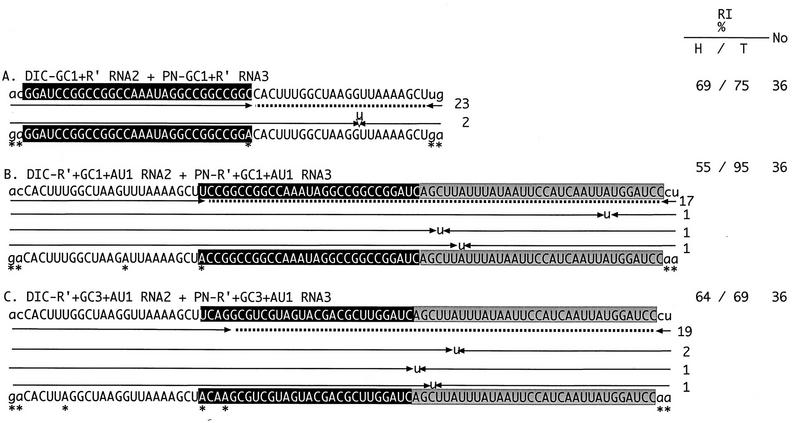
To test the effect of GC-rich sequences at upstream positions on homologous recombination, RNA2 and RNA3 constructs with GC1 and R′ sequences were used for inoculations. The incidence of homologous recombination was 69% for DIC-GC1+R′ RNA2 and PN-GC1+R′ RNA3 (Fig. 4A). Here, recombination occurred at a level much higher than that found with constructs having common R′ sequences alone (39%; Fig. 2A). Clustering of crossover sites within R′ (Fig. 4A) demonstrated that GC1 located upstream can stimulate homologous recombination at downstream positions (see Discussion).
FIG. 4.
Effect of upstream GC-rich sequences on the distribution of crossover sites in homologous recombinant RNA3 molecules and on the incidence of recombination (RI). The total numbers of samples analyzed and other features are as described in the legend to Fig. 2. The artificial AU1 sequences are shaded. H, homologous; T, total.
To further characterize the positional effect of GC-rich sequences on the selection of junction sites and on the incidence of homologous recombination, we used RNA2 and RNA3 constructs that carried GC1 (or GC3) between R′ and AU1 (RNA2 constructs DIC-R′+GC1+AU1 and DIC-R′+GC3+AU1 and RNA3 constructs PN-R′+GC1+AU1 and PN-R′+GC3+AU1; Fig. 4B and C). Infections with either DIC-R′+GC1+AU1 RNA2 and PN-R′+GC1+AU1 RNA3 or DIC-R′+GC3+AU1 RNA2 and PN-R′+GC3+AU1 RNA3 (Fig. 4B and C) showed a >50% incidence of RNA2-RNA3 recombination. All the homologous recombination junctions were located within the GC1+AU1 region. Thus, GC1 and GC3 inhibited homologous recombination within the upstream homologous hot spot (R′) (Fig. 2) but did not suppress it within the downstream sequence (AU1).
GC-rich sequences are maintained in parental RNA2 and RNA3 during infections.
To demonstrate that the artificial GC1 sequences were available for recombination in the above-described experiments, the RNA2 and RNA3 progeny were analyzed by sequencing of cDNA clones amplified by RT-PCR of total RNA extracts obtained from five separate local lesions induced by DIC-R′+GC1 RNA2 and PN-R′+GC1 RNA3 infections (Fig. 2B). Pairs of primers 1 and 3 or primers 1 and 2 (Fig. 1) were used to amplify, respectively, RNA2 and RNA3 3′ sequences. These experiments demonstrated that the parental sequences were stably maintained in RNA2 molecules and in nonrecombined RNA3 molecules (data not shown).
Growth characteristics of parental and recombinant RNAs.
It is possible that the reduced frequency or lack of isolation of recombinant RNA3 molecules in some of the above-described experiments was due to the reduced viability (fitness) of recombinant RNA3 molecules carrying GC-rich sequences at downstream positions. To test this possibility, we reconstructed full-length cDNA clones of the following recombinant progeny RNA3 molecules that were or were not isolated in the above-described experiments: (a) rec-R′+GC1, which represents a potential recombinant molecule that might have been generated by precise homologous recombination in DIC-R′+GC1 RNA2 and PN-R′+GC1 RNA3 infections (Fig. 2B); (b) rec-R′+GC3, which represents an infrequently isolated (it was detected in three local lesions) precise homologous recombinant RNA3 molecule in DIC-R′+GC3 RNA2 and PN-R′+GC3 RNA3 infections (Fig. 2D); and (c) rec-R′, which represents one of the most frequently isolated precise homologous recombinant molecules (the second recombinant from the top in Fig. 2A). All three types of recombinant RNA3 molecules were viable and accumulated to comparable levels in local lesions on C. quinoa when each was coinoculated with wt RNA1 and an RNA2 construct (DIC-R′+GC1, DIC-R′+GC3, or DIC-R′ RNA2; Fig. 5). This result argues that if these recombinants were generated at similar frequencies, they should have been detected at comparable frequencies in local lesions. The above-described experiments also demonstrated that the levels of accumulation of RNA2 mutants with three different combinations of RNA2 and RNA3 constructs were comparable (Fig. 5). This result suggests that the 3′ mutations in RNA2 did not alter the growth advantages (fitness) of RNA2 or, indirectly, that of recombinant RNA3 molecules.
In addition to the above-described targeted homologous recombinants, we have frequently isolated nontargeted (designated as background; see reference 21 for details) RNA3 recombinants. These background recombinants, together with the homologous RNA2-RNA3 recombinants, make up the total recombination incidence in Fig. 2 to 4. Since the background recombinants were generated by recombination between RNA3 and the 3′ noncoding region of RNA1 (which is present in both wt RNA1 and the DIC series of RNA2 molecules; Fig. 1) (data not shown) (21), we replaced DIC-R′+GC1 RNA2 with N2-R′+GC1 RNA2 (Fig. 2B). N2-R′+GC1 RNA2 contained the RNA2-derived 3′ noncoding region and lacked RNA1-derived sequences (data not shown). As expected, infections with N2-R′+GC1 RNA2 and PN-R′+GC1 RNA3 yielded reduced frequencies of background recombinants compared to DIC-R′+GC1 RNA2 and PN-R′+GC1 RNA3 infections, and the frequencies of targeted homologous RNA2-RNA3 recombinants were low in both cases (Fig. 2B) (more than a 20% difference in homologous recombination frequency between different combinations was considered significant during this work, based on a statistical analysis). We concluded that the background recombinants did not alter appreciably the frequency of homologous recombinant isolation in this system.
DISCUSSION
In this study, we demonstrated for the first time that GC-rich sequences can reduce the incidence of homologous recombination within nearby hot spot regions in BMV. The observed silencing effect of GC-rich sequences on recombination hot spots depends on several factors, including (i) the positions of the GC-rich sequences relative to the hot spot regions (i.e., the GC-rich sequences should be located downstream of the hot spot regions); (ii) the positions of the GC-rich sequences in the BMV RNA components (they must be present in both RNA2 and RNA3 or in RNA2 alone); and (iii) the percent G+C content in the GC-rich sequences (a G+C content of 59 to 80% within a 20- to 30-nt flanking sequence is required). Homologous recombination silencing, however, does not seem to depend on either the primary sequence of the GC-rich regions or the stability of their secondary (intramolecular) structures. These latter observations are in contrast with those for heteroduplex-mediated nonhomologous recombination in BMV and some recombination events in turnip crinkle virus and tombusviruses, where stable inter- or intramolecular structures in the RNA templates influenced the selection of crossover sites (8–10, 28, 29).
This study also revealed the complex nature of homologous recombination in BMV. For instance, GC-rich sequences not only can silence homologous recombination but also, on the contrary, can increase the frequency of homologous recombination when they are present upstream of the recombinogenic R′ sequence. Also, when present only in RNA3 at downstream positions, GC-rich sequences can increase the frequency of crossover events and alter the junction profile within the upstream hot spot sequence. The sites of crossovers were not located within the GC-rich sequences themselves, suggesting that GC-rich sequences may alter the pausing sites for the BMV replicase or disfavor the release of the aborted nascent strands from the donor template due to stable base pairing. An interaction between the downstream GC stretches of the parental or nascent RNAs is unlikely to occur, since heterologous GC-rich sequences in RNA2 and RNA3 (Fig. 2E) or a GC-rich sequence in RNA2 alone (Fig. 3B) reduced the frequency of recombination. Overall, these and previous data (21) showed that sequences around the homologous crossover sites can greatly influence both the incidence and the distribution of the crossovers.
It is unlikely that the observed homologous recombination silencing effect was due to the altered fitness of recombinants because reconstructed RNA3 recombinants containing GC-rich sequences accumulated to high levels in local lesions on C. quinoa (Fig. 5). Also, RNA2 mutants (as shown in Fig. 2 to 4) carrying various GC-rich sequences were viable. Further evidence supporting the idea that the frequency and sites of crossovers reflect the actual mechanism was obtained by use of RNA2 and RNA3 constructs with common R′+GC1+AU1 sequences. It was found that the homologous crossover sites were shifted into the artificial GC1+AU1 sequences from the upstream (virus-derived) R′ sequences (infections with DIC-R′+GC1+AU1 RNA2 and PN-R′+GC1+AU1 RNA3). This result supports our model of homologous recombination, in which sequence signals rather than biased fitness of the recombinants are responsible for the observed recombinant profiles. In addition, the fact that as homologous recombination declined (Fig. 2 to 4) total recombination levels in most cases did not change greatly is the consequence of an increased frequency of background RNA1-RNA3 recombinants in the DIC RNA2 and PN RNA3 systems. These background recombinants are generated at later times (14 to 21 days postinoculation), while homologous RNA2-RNA3 recombinants are generated at earlier times (7 to 14 days postinoculation) (see reference 21). Since single lesions usually contain only one type of recombinant RNA, it is likely that a “first come, first served” strategy of recombinant accumulation operates in the BMV-C. quinoa system. If a homologous recombinant is not generated in a particular lesion, there is an increased chance for the accumulation of a background recombinant.
To explain the silencing effect of downstream GC-rich sequences on homologous recombination, we propose that GC-rich sequences do not favor the formation of proper recombination intermediates. Such intermediates, as depicted in the introduction, likely are formed during positive-strand synthesis (21). Assuming the existence of single-stranded (free or protein-coated) negative RNA strands in eukaryotic virus-infected cells (5, 22), the observed GC-rich sequence-mediated homologous recombination silencing effect can be explained, for example, by the formation within the GC-rich regions of local intermolecular duplexes between the negative-stranded acceptor RNA2 molecules and the more abundant positive-stranded RNA2 molecules. This process might sequester (or “mask”) the AU-rich portions of the negative-stranded acceptor RNA; thus, those may not be available for interaction with the aborted, positive-stranded nascent RNA during homologous recombination. Overall, the chance of replicase-driven template-switching events occurring would be reduced.
Alternatively, since some of the data on eukaryotic virus replication support the existence of negative strands as part of replication intermediates (containing partially hybridized positive and negative strands; 4, 4a, 5, 23), we have proposed that AU-rich sequences can facilitate the formation of local non-base-paired (“bubble”) structures in acceptor replication intermediates (21) (Fig. 6). These bubbles can be the favorite “landing” sites for the nascent strands and/or the replicase. Formation of appropriate bubble structures within AU-rich sequences may be inhibited by GC-rich flanking sequences. This mechanism, however, can be only partially responsible for recombination silencing, since potentially both upstream and downstream GC-rich sequences should inhibit the formation of bubble structures in replication intermediates, yet only downstream GC-rich sequences were found to reduce the incidence of homologous recombination. It is possible that the BMV replicase has to enlarge the bubble structures in replication intermediates during and/or after the docking event on the acceptor RNA (Fig. 6). This step can be inefficient if the downstream portions of the replication intermediates are very stable due to their high G+C content. This stability can inhibit successful docking or reinitiation events by the replicase, resulting in a reduced incidence of recombination. Indeed, White and Morris (29) recently demonstrated that the preferred sites of crossovers were located behind (downstream) but not before a stable hairpin-loop in tombusviruses. This observation is similar to those of this study, suggesting that stable base-paired regions (formed by inter- or intramolecular interactions) can inhibit crossovers at upstream positions on acceptor RNAs.
FIG. 6.
Diagrammatic representation of a template-switching model explaining the silencing of homologous recombination hot spots by GC-rich sequences. (A) According to the model, template switching of the BMV replicase (represented by large shadowed double ellipses) occurs during positive-strand (represented by broken lines) synthesis when the replicase pauses at or near the AU-rich portion (represented by a curved line) present on the primary template RNA3 (21). Although partially double-stranded replication intermediates (RIs) are shown, the existence of single-stranded RNAs with negative polarity is also possible (not shown). (B) The released 3′ end of the nascent strand hybridizes to the acceptor strand; this hybridization is facilitated by the temporary formation of bubble structures (non-base-paired regions) within the AU-rich portion of the RI form of acceptor RNA2 (21). The resumption of chain elongation by the BMV replicase is indicated by a rightward-pointing arrow. When GC-rich sequences are present (indicated by a lock on the right), the formation of appropriate bubble structures within the AU-rich portion may be less favored (indicated by smaller loops), thus resulting in a reduced frequency of homologous recombination. It is also possible that the BMV replicase has to enlarge the bubble structures of the RIs during and/or after the docking event on the acceptor RNA. This step can be inefficient if the downstream portions of the RIs are very stable due to their high G+C content. This stability can inhibit successful docking or reinitiation events by the replicase, resulting in a reduced incidence of recombination (see also Discussion and reference 21).
As in our previous studies on homologous recombination in BMV (18, 20, 21), we did not observe a definite role for intramolecular secondary structures in homologous recombination silencing. For instance, both highly structured (GC1 and GC3) and nonstructured (GC2 and GC5) GC-rich sequences reduced the incidence of homologous recombination. According to our model (Fig. 6), the GC-rich regions in the RNAs interact with the complementary strands rather than forming intramolecular stem-loop structures. Of the three tested GC-rich sequences of comparable lengths, GC1 and GC2 could form the most stable intermolecular duplexes. Accordingly, downstream GC1 or GC2 inserts reduced homologous recombination in upstream R′ more effectively than GC3 inserts, supporting our model. Further experiments are needed to elucidate the proper structure of homologous recombination intermediates in BMV.
ACKNOWLEDGMENTS
We thank M. Figlerowicz, M. Graves, R. Olsthoorn, J. Pogany, and Andy White for comments and discussions.
This work was supported by grants from the National Institute for Allergy and Infectious Diseases (3RO1 AI26769), the National Science Foundation (MCB-9630794), the U.S. Department of Agriculture (96-39210-3842), and the Plant Molecular Biology Center at Northern Illinois University.
REFERENCES
- 1.Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist P, Dasgupta R, Kaesberg P. Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J Mol Biol. 1984;172:369–383. doi: 10.1016/s0022-2836(84)80012-1. [DOI] [PubMed] [Google Scholar]
- 3.Allison R F, Janda M, Ahlquist P. Sequence of cowpea chlorotic mottle virus RNAs 2 and 3 and evidence of a recombination event during bromovirus evolution. Virology. 1989;172:321–330. doi: 10.1016/0042-6822(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 4.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 4a.Bienz, K., D. Egger, and T. Pfister. 1994. Characteristics of the poliovirus replication complex. Arch. Virol. 9(Suppl.):147–157. [DOI] [PubMed]
- 5.Buck K W. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bujarski J J. RNA recombination and defective RNA formation. I. Introduction: experimental systems of genetic recombination and defective RNA formation in RNA viruses. Semin Virol. 1996;7:361–422. [Google Scholar]
- 6a.Bujarski J J, Nagy P D. Genetic RNA-RNA recombination in positive-stranded RNA viruses of plants. In: Paszkowski J, editor. Homologous recombination in plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–24. [Google Scholar]
- 7.Bujarski J J, Nagy P D, Flasinski S. Molecular studies of genetic RNA-RNA recombination in brome mosaic virus. Adv Virus Res. 1994;43:275–302. doi: 10.1016/S0065-3527(08)60051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter C D, Oh J-W, Zhang C, Simon A E. Involvement of a stem-loop structure in the location of junction sites in viral RNA recombination. J Mol Biol. 1995;245:608–622. doi: 10.1006/jmbi.1994.0050. [DOI] [PubMed] [Google Scholar]
- 9.Cascone P J, Carpenter C D, Li X H, Simon A E. Recombination between satellite RNAs of turnip crinkle virus. EMBO J. 1990;9:1709–1715. doi: 10.1002/j.1460-2075.1990.tb08294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cascone P J, Haydar T F, Simon A E. Sequences and structures required for recombination between virus-associated RNAs. Science. 1993;260:801–805. doi: 10.1126/science.8484119. [DOI] [PubMed] [Google Scholar]
- 11.Janda M, French R, Ahlquist P. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions of transcript infectivity. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 12.Jarvis T C, Kirkegaard K. The polymerase in its labyrinth: mechanisms and implications of RNA recombination. Trends Genet. 1991;7:186–191. doi: 10.1016/0168-9525(91)90434-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroner P, Richards D, Traynor P, Ahlquist P. Defined mutations in a small region of the brome mosaic virus 2a gene cause diverse temperature-sensitive RNA replication phenotypes. J Virol. 1989;63:5302–5309. doi: 10.1128/jvi.63.12.5302-5309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai M C M. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy P D, Bujarski J J. Genetic recombination in brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J Virol. 1992;66:6824–6828. doi: 10.1128/jvi.66.11.6824-6828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy P D, Bujarski J J. Targeting the site of RNA-RNA recombination in brome mosaic virus with antisense sequences. Proc Natl Acad Sci USA. 1993;90:6390–6394. doi: 10.1073/pnas.90.14.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy P D, Bujarski J J. Efficient system of homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J Virol. 1995;69:131–140. doi: 10.1128/jvi.69.1.131-140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy P D, Dzianott A, Ahlquist P, Bujarski J J. Mutations in the helicase-like domain of protein 1a alter the sites of RNA-RNA recombination in brome mosaic virus. J Virol. 1995;69:2547–2556. doi: 10.1128/jvi.69.4.2547-2556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy P D, Bujarski J J. Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J Virol. 1996;70:415–426. doi: 10.1128/jvi.70.1.415-426.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy P D, Bujarski J J. Engineering of homologous recombination hot spots with AU-rich sequences in brome mosaic virus. J Virol. 1997;71:3799–3810. doi: 10.1128/jvi.71.5.3799-3810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pata J D, Schultz S C, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 23.Pogue G P, Hall T C. The requirement for a 5′ stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J Virol. 1992;66:674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao A L N, Hall T C. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J Virol. 1993;67:969–979. doi: 10.1128/jvi.67.2.969-979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Simon A E, Bujarski J J. RNA-RNA recombination and evolution in virus infected plants. Annu Rev Phytopathol. 1994;32:337–362. [Google Scholar]
- 27.Strauss J H, Strauss E G. Evolution of RNA viruses. Annu Rev Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- 28.White K A, Morris T J. Recombination between defective tombusvirus RNAs generates functional hybrid genomes. Proc Natl Acad Sci USA. 1994;91:3642–3646. doi: 10.1073/pnas.91.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White K A, Morris T J. RNA determinants of junction site selection in RNA virus determinants and defective interfering RNAs. RNA. 1995;1:1029–1040. [PMC free article] [PubMed] [Google Scholar]