Abstract
The detailed mechanism which governs the choice between herpes simplex virus (HSV) latency and reactivation remains to be elucidated. It is probable that altered expression of cellular factors in sensory neurons leads to induction of HSV gene expression resulting in reactivation. As an approach to identify novel cellular genes which are activated or repressed by stimuli that reactivate HSV from latency and hence may play a role in viral reactivation, RNA from explanted trigeminal ganglia (TG) was analyzed by differential display reverse transcription-PCR (DDRT-PCR). Nearly 50 cDNAs whose mRNA level was modified by the stress of explantation were isolated and sequenced. We present a listing of a spectrum of altered RNAs, including both known and unknown sequences. Five of those differentially displayed transcripts were identified as interferon-related murine TIS7 mRNA. These results were confirmed in both infected and uninfected ganglia by quantitative RNase protection assay and immunostaining. Alpha and beta interferons and interferon regulatory factor-1 (IRF-1) were also induced by explantation. In addition, we have identified sequences that correspond to IRF-1 consensus binding sites in both HSV type 1 origins of replication. Our findings suggest that physiological pathways that include these cellular factors may be involved in modulating HSV reactivation.
Following primary infection, latent herpes simplex virus (HSV) persists in sensory ganglia of the peripheral nervous system. The virus can undergo sporadic reactivation to produce recurrent mucocutaneous lesions at peripheral sites innervated by the infected ganglia (reviewed in references 15, 46, and 53). Reactivation stimuli range from direct mechanical or pharmacological insults to the neuron and surrounding tissue to systemic changes in immune modulators and neurotransmitters (15, 16, 24). The earliest molecular events in neuronal cells that trigger reactivation of HSV remain unclear. It is thought that these events include altered expression of cellular factors such as induction of transcriptional activators and down-regulation of repressors. Identification of cellular factors which are induced during the reactivation process may lead to better understanding of the cellular environment during viral induction and may facilitate development of an effective treatment to prevent reactivation.
The present knowledge of the molecular pathogenesis of HSV latency and reactivation was generated from studies in laboratory animals including mice, guinea pigs, and rabbits (reviewed in reference 47). We and others have found current murine in vivo models to be inefficient in reactivation of the viral genome (13, 14, 21, 41, 48, 67). In contrast, the murine explant reactivation model is exceptionally useful for studying the molecular mechanisms of HSV reactivation, because infectious virus can be efficiently recovered upon explantation and culture of latently infected sensory ganglia (reviewed in reference 16). Using this model, we detect expression of viral early genes coincident with expression of immediate-early (IE) genes. Moreover, cellular IE factors Oct-1, Fos, Jun, and Myc are induced within the first 2 h following explantation of trigeminal ganglia (TG), prior to first detection of viral gene expression (54, 60).
Our goal in this study was to seek novel cellular factors which may have a role in the reactivation process. Moreover, we sought to identify factors whose expression is altered even in the absence of latent HSV type 1 (HSV-1) infection and induction, since such cellular factors would also be candidates for causative agents in viral reactivation. To identify such factors, we used differential display reverse transcription-PCR (DDRT-PCR), which allows the visualization and subsequent isolation of cDNAs corresponding to mRNAs displaying altered expression in different cell populations (34, 35). We compared the levels of gene expression in TG populations derived from various time points postexplantation (p.e.). Thus, any factors modulated during the first 4 h, the period in which induction of viral gene expression was previously detected (12, 54), may be important in the initial stimulation of latent viral genomes.
Recent studies have used DDRT-PCR to identify genes that are involved in neural stress and injury (26, 30, 42). For example, Kiryu et al. (30) demonstrated differential expression of the rat neuronal glutamate transporter in axotomized hypoglossal motor neurons. Since glutamate transporter expression was induced in response to neuronal injury, it may be involved in the regeneration process. Others used DDRT-PCR to isolate a novel neuropeptide, called melanin-concentrating hormone, in the hypothalamic response to starvation (42). DDRT-PCR has also been used to identify cellular genes modulated by simian virus 40 and Epstein-Barr virus (EBV) transformation (51, 68). However, it is a novel approach to study viral reactivation.
In this study, we isolated approximately 50 differentially displayed cDNAs representing transcripts whose levels were altered within the first 4 h following explantation of TG. Five cDNAs were identical to murine TIS7, whose sequence has been shown to be related to those of interferons (IFNs) (50, 58, 61). We also detected rapid induction of IFN-α and -β in neuronal cells of TG explants. In addition, other factors, such as the transcriptional activator IRF-1 (IFN regulatory factor-1) and the mitogen TNF-α (tumor necrosis factor alpha), were induced by explantation. Interestingly, we have identified sequences corresponding to IRF-1 consensus binding sites in both HSV origins of replication. We discuss the possibility that HSV reactivation involves the induction of a regulatory pathway shared with IFN-related genes and that specific inductive events of the viral genome occur in response to these regulatory factors.
MATERIALS AND METHODS
Infection of mice and explant reactivation.
Four- to six-week-old female BALB/c BYJ mice were obtained from Jackson Laboratory. Mice were anesthetized with intraperitoneal injection of ketamine (87 mg/kg)-xylazine (13 mg/kg) and then, after corneal scarification, inoculated in the eye with 104 PFU of HSV-1 17+ (7). At a minimum of 28 days postinfection, mice were sacrificed by cervical dislocation and TG were isolated. Groups of 6 to 10 explanted TG were incubated in Dulbecco’s modified Eagle medium supplemented with 5% fetal bovine serum at 37°C for 0, 1, 2, 4, or 24 h p.e. In a single experiment, TG were explanted in the absence of serum.
Extraction of RNA.
Ganglia used for RNA preparation were snap frozen in liquid nitrogen. RNA was isolated from TG and brain stems by using the TRIzol reagent, as described by manufacturer (Gibco BRL), followed by extensive digestion with RNase-free DNase I (Boehringer Mannheim Biochemicals) and ethanol precipitation. RNA concentrations were determined by spectrophotometer and agarose gel electrophoresis (37).
DDRT-PCR.
cDNA was prepared from 300 ng of RNA from latently infected TG at 0, 1, 2, and 4 h p.e., using a Differential Display kit (Display Systems Biotech, Inc., Los Angeles, Calif.) as described by the manufacturer. Primers used in this study are listed in Table 1. Briefly, RNA from each sample was incubated with one of nine downstream primers containing 11 T residues and two-nucleotide anchors (AA, AC, AG, CA, CC, CG, GA, GC, and GG) for 1 h at 40°C, followed by 5 min at 95°C to inactivate the Moloney murine leukemia virus enzyme. cDNA was stored at −70°C. Each cDNA was subjected to PCR amplification with DisplayTaq (Display Systems Biotech), using the original downstream primer, 1 of 24 10-mer 5′ primers, and [α-33P]dATP (65). PCR conditions were 35 cycles of 30-s denaturation at 94°C, 60-s primer annealing at 40°C, and 60-s extension at 72°C in a Perkin-Elmer Cetus Gene Amp PCR System thermocycler. A final extension reaction was then performed for 5 min at 72°C. Radiolabeled reaction products were subjected to high-resolution polyacrylamide-urea gel electrophoresis as described previously (35). Gels were dried on Whatman filters and analyzed by autoradiography. Differentially displayed PCR bands were cut out from the filter paper and dissolved in diethyl pyrocarbonate-treated water (Ambion, Austin, Tex.) for 30 min at room temperature followed by 10 min at 100°C.
TABLE 1.
Differential display primersa used in this study
| Name | Sequence |
|---|---|
| 5′ | |
| 1 | GATCATAGCC |
| 2 | CTGCTTGATG |
| 3 | GATCCAGTAC |
| 4 | GATCGCATTG |
| 5 | AAACTCCGTC |
| 6 | TGGTAAAGGG |
| 7 | GATCATGGTC |
| 9 | GTTTTCGCAG |
| 10 | TACCTAAGCG |
| 11 | GATCTGACAC |
| 12 | GATCTAACCG |
| 13 | TGGATTGGTC |
| 14 | GGAACCAATC |
| 15 | GATCAATCGC |
| 20 | GATCAAGTCC |
| 21 | GATCTCAGAC |
| 22 | GGTACTAAGG |
| 3′ | |
| 2 | TTTTTTTTTTTAC |
| 4 | TTTTTTTTTTTCA |
| 5 | TTTTTTTTTTTCC |
| 6 | TTTTTTTTTTTCG |
| 8 | TTTTTTTTTTTGC |
Obtained from Display Systems Biotech.
Reamplification PCR.
To ensure that each band analyzed contained a single cDNA species, each differentially displayed band was reamplified in four individual PCRs. Four T7-T11VVN 3′ primers were used, where VV was the original DDRT nucleotide anchor (Table 1), T7 was a 23-nucleotide portion of the T7 promoter (TAATACGACTCACTATAGGGCCC), and N was A, G, T, or C. The reamplification reactions included the original upstream primers, 2 μM deoxynucleoside triphosphate, and the Stoffel fragment of Taq polymerase (Perkin-Elmer Cetus). Reactions were performed under the original DDRT-PCR conditions. PCR products were separated by agarose electrophoresis, and the most prominent product among the four parallel reactions was isolated for automated sequencing and further confirmation.
Confirmation of differentially regulated expression.
Isolated reamplification bands were used as templates for synthesis of 32P-labeled riboprobes with a MAXIscript kit (Ambion) as described by manufacturer. RNase protection assays (RPA) were performed with a Hybspeed RPA kit (Ambion) and 0.5 to 1 μg of total RNA from a second, new set of latently infected and uninfected TG explants. Mouse β-actin riboprobes provided with the MAXIscript kit were used as controls. Probes and protected fragments were analyzed by denaturing polyacrylamide gel electrophoresis (PAGE) and phosphorimaging.
PCR amplification of cDNA.
The second confirmatory PCR using specific primer sets was performed as follows. cDNA was generated from 2 μg of total RNA by using Superscript preamplification kit priming with oligo(dT) and random hexamers (Gibco BRL). Reactions were performed in 25-μl volumes containing 4% cDNA, 200 μM each deoxynucleoside triphosphate (Pharmacia), 1 μM each primer, and 1.25 U of AmpliTaq Gold (Perkin-Elmer) in PCR buffer A (Fisher). Primer pairs used are described in Table 3. Primers specific for TIS7 were designed based on the published sequence (63). Cycling reactions were performed with a Perkin-Elmer Cetus Gene Amp PCR System thermocycler. After one cycle of 9-min denaturation at 94°C, cycles were as follows: (i) 1 min of denaturation at 94°C, (ii) annealing at 60°C for 1 min, and (iii) extension for 2 min at 72°C. The final cycle was terminated with a 7-min extension at 72°C. Amplification was carried out for 25 to 35 cycles. RT− reactions were included in each set of experiments as negative controls, and 10 ng of mouse DNA was used as a positive control. In every case, the size of PCR product bands corresponded to the predicted size.
TABLE 3.
PCR primer pairs used in this study
| Name | Sequence | Product (bp) | Reference |
|---|---|---|---|
| TIS7SA | CTCTTATCTCGGCATTTG | ||
| GGACAAGAGAAAGCAGCG | 342 | ||
| TIS7B | CGATGCCGAAGAACAAGA | ||
| CTGCCTGTCTTGTCTTCG | 300 | ||
| IFN-β | GAAAAGCAAGAGGAAAGATT | ||
| AAGTCTTCGAATGATGAGAA | 165 | 39 | |
| IFN-α | AATGACCTCCACCAGCAGCT | ||
| TCTCAGGTACACAGTGATCC | 201 | 39 | |
| IFN-α/βR | ACATGAGCCCCCCAGAAGTACG | ||
| ATGACCGGAGGAGGAGGGAGAA | 613 | 31 | |
| IRF-1 | CAGAGGAAAGAGAGAAGTCC | ||
| CACACGGTGACAGTGCTGG | 201 | 5 | |
| IRF-2 | CCTGAGTATGCGGTCCTGACTT | ||
| CCGGGTCTCCCGGTCTGGCCGA | 528 | 31 | |
| TNF-α | GAAAGCATGATCCGCGACGTGG | ||
| GTAGACCTGCCCGGACTCCGCAA | 678 | 31 | |
| β-Actin | ATAGCACAGCTTCCCTTTGAT | ||
| AACATGCATTGTTACCAACT | 452 | 54 | |
| Cyclophilin | ATTCGAGTTGTCCACAGTCAGCAATGG | ||
| ATGGTCAACCCCCACCGTGTTCTTCGAC | 469 | 6 |
Detection of PCR products.
Aliquots of 40% of the amplification products were fractionated on 2.5% NuSieve agarose (FMC). Gels were stained with ethidium bromide (Sigma), and the amounts of products were quantitated by fluorimetry. The relative amount of PCR product was determined in arbitrary numbers as the ratio between the PCR product band intensity to that of cellular housekeeping gene encoding cyclophilin or β-actin (12, 54). Statistical analysis was performed with Excel (Microsoft, Redmond, Wash.).
Immunohistochemical procedures.
Ganglia used for immunohistochemistry were immersed in 70% ethanol–150 mM NaCl for 24 h and then embedded in paraffin wax, and 6-μm serial sections were cut and processed as described elsewhere (45). Rabbit polyclonal antiserum to HSV-1 (Dako Corp., Carpinteria, Calif.) was used for detection of replicating virus as described elsewhere (1, 28). Rabbit polyclonal anti-mouse IFN-α/β (Lee Biomolecular Research, San Diego, Calif.) and rabbit polyclonal anti-mouse TNF-α (Genzyme Diagnostics, Cambridge, Mass.) were used to probe for cytokines. Rabbit polyclonal anti-TIS7 was a generous gift from B. Varnum, Amgen, Thousand Oaks, Calif. Antigen-expressing cells were detected by an indirect avidin-biotin immunoperoxidase method (Vectastain ABC kit; Vector Laboratories, Burlingham, Calif.), with 3,3′-diaminobenzidine as the chromagen (59).
RESULTS
We recently have shown that cellular IE factors, such as c-Jun, c-Myc, and Oct-1, are induced in neuronal cells at early times following explantation of murine TG, within the time frame of HSV-1 gene activation (54, 60). DDRT-PCR was used in this study as an approach to identify previously unknown cellular genes which are induced or repressed by explantation of TG.
Explantation of TG induces differential expression of multiple mRNAs.
RNA was prepared from latently infected TG at different time points (0, 1, 2, and 4 h) following explantation into culture media. Complementary DNA was amplified by using a set of arbitrary PCR primers. The PCR products were resolved by PAGE and visualized by autoradiography. Every pair of primers (216 primer combinations) identified a limited number of target sequences within the pool of cDNAs. Thus, a typical reaction generated 50 to 200 distinct radiolabeled PCR products between 50 and 600 bp in length. As expected from previous studies (34), the majority of PCR products were present at identical levels in samples derived from different time points (Fig. 1). However, over 50 differentially displayed PCR products were detected and isolated for further characterization.
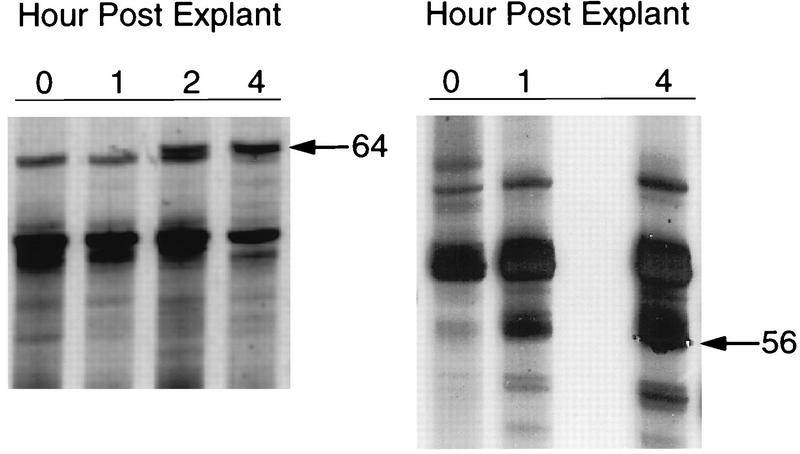
FIG. 1.
PCR differential display of cDNA derived from latently infected mouse TG following explantation. In the autoradiograph of radiolabeled DDRT-PCR products, arrows denote PCR products representing band 64 amplified with 3′ primer 2 and 5′ primer 7, band 56 amplified with 3′ primer 2 and 5′ primer 3.
Reamplified PCR products were subjected to sequencing followed by BLAST sequence analysis (2) and MasPar searches of the NCBI nonredundant nucleotide database. Of 48 bands sequenced, 26 were novel sequences without significant sequence homology in the database (results not shown), whereas others had multiple significant hits (P value was less than 1 e −10). The analysis of results, including GenBank accession numbers for the most significant hits, are summarized in Table 2. Five bands corresponded to structural genes encoding proteins such as laminin and tubulin, and two (bands 54 and 53) encoded repeating sequence elements. Fifteen database hits were similar to cellular enzymes or regulatory proteins of interest such as NADH ubiquinone oxyreductase (bands 218 and 20), the growth arrest gene GAS5 product (band 123), and mouse semaphorin (band 229). Bands 56, 64, 116, 125, and 201 were identical in sequence to the coding region of mouse TIS7 mRNA (61) (Table 2; Fig. 2) and were conserved with rat PC4 mRNA sequence (58). TIS7 and rat PC4 previously were shown to be highly related in protein sequence and are thought to be functional homologs involved in stress response (58, 61). These five overlapping products (Fig. 2B) were chosen for further characterization.
TABLE 2.
Sequence analysis of cDNAs differentially displayed in mouse TG explants
| Band no.a | 3′ primer | 5′ primer | Expression patternb | cDNA name | GenBank accession no. | P valuec |
|---|---|---|---|---|---|---|
| 64 | 2 | 3 | Ind | Mouse TIS7 | V00756 | 4.82 e −210 |
| 56 | 2 | 7 | Ind | Mouse TIS7 | J00424 | 8.84 e −75 |
| 116 | 6 | 2 | Ind | Mouse TIS7 | X17400 | 5.4 e −140 |
| 125 | 6 | 7 | Ind | Mouse TIS7 | X17400 | 2.7 e −37 |
| 201 | 8 | 2 | Ind | Mouse TIS7 | X17400 | 3 e −47 |
| 114 | 6 | 1 | Rep | Unknown mouse | W97484 | 1.6 e −66 |
| 123 | 6 | 5 | Ind | GAS5 mouse growth arrest gene | X67267 | 1 e −45 |
| 229 | 8 | 12 | Rep | Mouse semaphorin | X85990 | 1 e −189 |
| 65 | 2 | 4 | Rep | Mouse kallikrein tumor antigen | M18620 | 1 e −13 |
| 97 | 5 | 15 | Ind | Mouse T-cell antigen 4-1BB | U02567 | 3.14 e −52 |
| 27 | 4 | 9 | Ind | Rat ATPase | H39388 | 2.59 e −22 |
| 218 | 8 | 6 | Rep | Mouse lecithin cholesterolacyltransferase | X54095 | 2 e −15 |
| 20 | 4 | 20 | Rep | Human NADH ubiquinone oxyreductase subunit B14 | T58895 | 7.7 e −185 |
| 129 | 6 | 11 | Ind | Human nucleus-encoded mitochondrial NADH ubiquinone oxyreductase 24-kDa subunit | M25484 | 2.79 e −168 |
| 232 | 8 | 12 | Ind | Mouse laminin | T54408 | 1.17 e −116 |
| 41 | 2 | 2 | Rep | Mouse alpha-tubulin | H34265 | 1.18 e −73 |
| 138 | 6 | 15 | Ind | Mouse beta-tubulin | X04663 | 1 e −38 |
| 98 | 5 | 15 | Rep | Mouse beta-tubulin | X04663 | 2.09 e −176 |
| 21 | 4 | 21 | Rep | Human ribosomal protein S26 | X77770 | 0 |
| 54 | 2 | 6 | Ind | Mouse retrotransposon-like element | M21123 | 1.97 e −66 |
| 53 | 2 | 6 | Ind | Human retrovirus-related reverse transcriptase | K02590 | 1 e −153 |
Bands 56, 64, 116, 125, and 201 share sequences (see Fig. 2).
Ind, induced; Rep, repressed.
Represents Poisson distribution value. The cutoff number representing significant hits was 1 e −9.
FIG. 2.
Sequences of DDRT-PCR products. cDNAs isolated from differential display gels were reamplified by using primers that included the T7 promoter sequence, and PCR products were sequenced. Sequences were analyzed by BLAST searches. (A) BLAST output using band 56 as query sequence (P value = 7.7 × 10−49) demonstrates sequence identity with murine TIS7. (B) Alignment of each DDRT band with TIS7 mRNA. Four cDNA bands corresponded to mouse IFN-related gene TIS7 mRNA; band 56 (primers 2 and 7), band 64 (primers 2 and 3), band 116 (primers 6 and 2), and band 125 (primers 6 and 7).
Confirmation of differential display.
The intensity of each of the five PCR products (56 [Fig. 1], 64 [Fig. 1], 116, 125, and 201) was clearly increased in samples prepared 1 and 2 h p.e. We next determined whether RNA corresponding to the isolated bands was differentially expressed in either uninfected or latently infected TG explants. Reamplified PCR products were used as probes in quantitative RPA. RNA corresponding to band 56 was induced by 2 h following explantation of uninfected (Fig. 3A) and infected (results not shown) explants. Phosphorimager quantitation (Fig. 3B) indicated that the levels of RNA were induced nearly sevenfold by 4 h. Similar results were obtained with probes generated from band 64 (results not shown). Thus, RNA transcripts corresponding to differentially displayed bands 56 and 64 were significantly induced in uninfected TG by explantation.
FIG. 3.
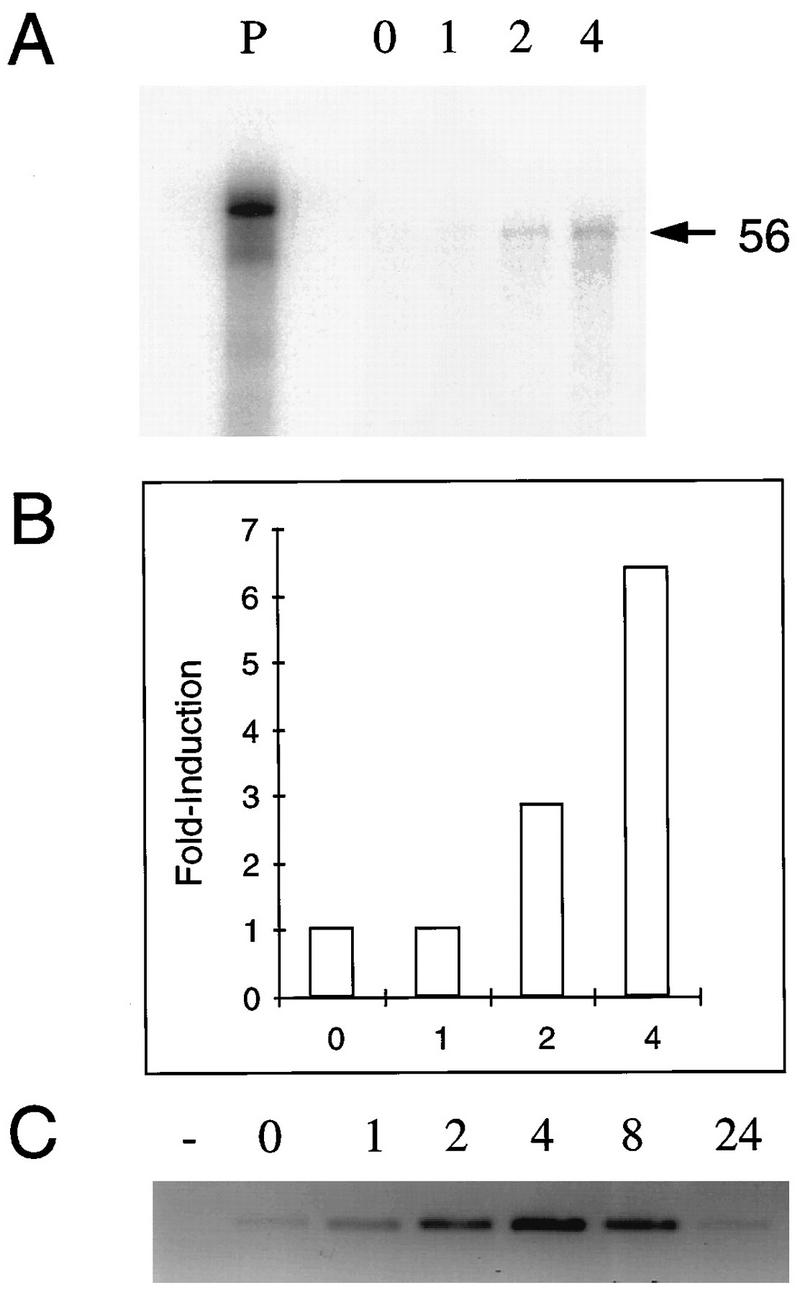
Confirmation of differential display. RNA was prepared from uninfected TG explants at 0, 1, 2, and 4 h p.e. Complementary DNA from differentially displayed band 56 was reamplified by PCR using 3′ primers that included the T7 promoter. PCR products were used as templates to prepare riboprobes labeled with [32P]UTP and added to each RNA sample. Following hybridization at 37°C and RNase digestion, samples were separated by PAGE. (A) The input probe (P) and protected fragments were visualized using phosphorimager screens. (B) The intensity of each protected fragment was quantitated with ImageQuant software. Fold induction was expressed as the ratio between each band to the zero time point. (C) Complementary DNA from latently infected TG explants at 0 to 24 h p.e. was subjected to PCR using primers specific for TIS7 (TIS7A set). Products were separated on 2.5% agarose gels stained with ethidium bromide and visualized by fluorimager analysis.
To further confirm the DDRT results, cDNA prepared from a different set of latently infected TG explants was subjected to PCR using primers specific for TIS7 (Table 3). Each PCR yielded a single product band whose size corresponded to that of TIS7 cDNA. Importantly, TIS7 detected with these primers was induced rapidly following explantation in both infected (Fig. 3C) and uninfected (not shown) explants, confirming the RPA results (Fig. 3A). Furthermore, by 24 h p.e., TIS7 expression returned to basal levels (Fig. 3C).
TIS7 expression is induced in neuronal cells.
HSV latency occurs primarily in neuronal cells which innervate the cornea (9, 43, 44). Infected neurons represent approximately 2% of the neuronal population in the TG (4, 38). To determine whether TIS7 expression colocalizes with reactivating virus in neuronal cells, TG sections from uninfected and latently infected explants were analyzed by immunostaining using affinity-purified polyclonal antisera. TIS7 protein expression was not detected at 0 h p.e. and was induced at 1, 2, and 4 h p.e. in both infected (data not shown) and uninfected (Fig. 4) explants. Furthermore, as judged by this technique, all neuronal cells expressed TIS7, demonstrated by brown staining of the cells. Since virus reactivates in approximately 1% of neuronal cells (60), we conclude that viral gene expression occurs in cells expressing TIS7. Thus, induced expression of TIS7 following explantation of ganglia was detected by four independent methods: DDRT-PCR, sequence-specific RT-PCR, RPA, and immunostaining.
FIG. 4.
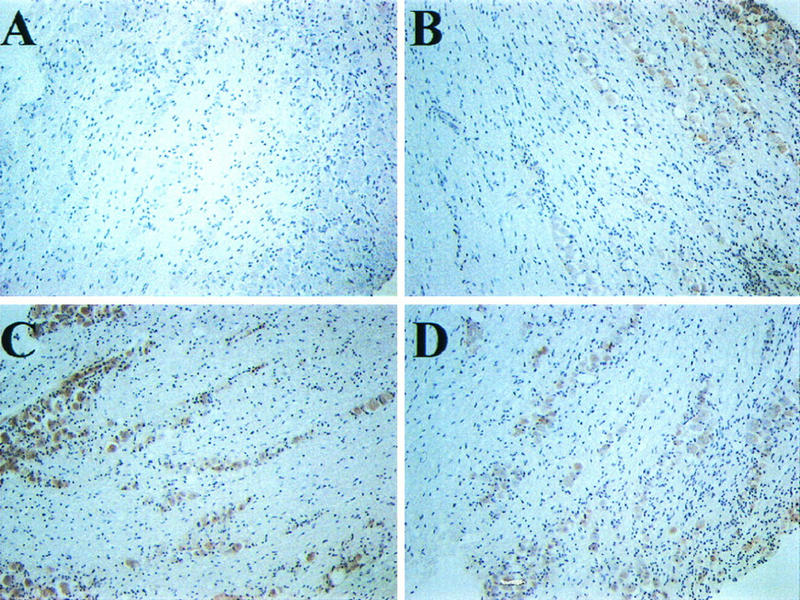
Immunostaining of TIS7 in TG following explantation. Latently infected BALB/c mice were sacrificed, and TG were excised and incubated in culture medium for 0 to 24 h. Paraffin-embedded sections of uninfected TG at 0 (A), 1 (B), 2 (C), and 4 (D) h p.e. were processed as described in Materials and Methods and reacted with rabbit polyclonal antiserum against TIS7. The experiment was repeated twice, and duplicate slides were screened.
IFN-β is induced by explantation of TG.
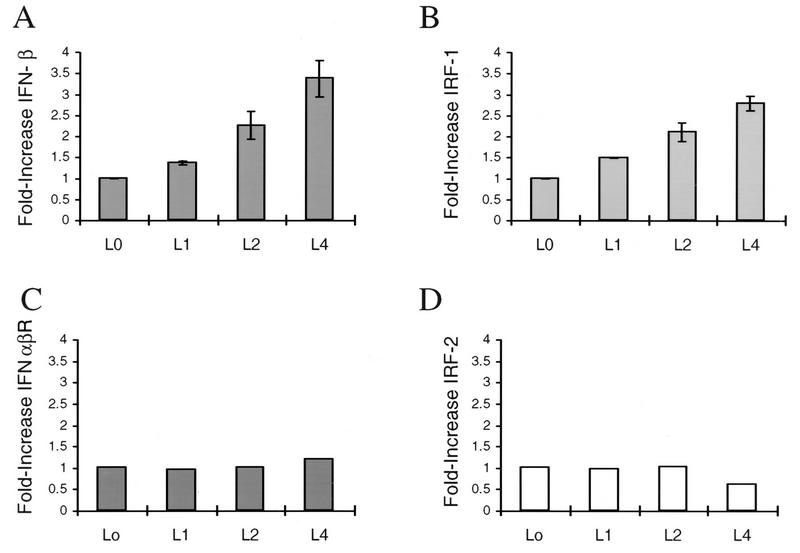
Several reports have indicated that TIS7 and PC4 are related in sequence to IFN-β (50, 58). To determine whether IFNs are also induced by explantation, we again used qualitative RT-PCR. cDNA derived from infected and uninfected TG explants was analyzed by PCR using primers specific for IFN-α or IFN-β (Table 3). Again, single specific PCR products were obtained from each reaction. IFN-β was induced 2- to 3.5-fold during the 4 h p.e. compared to the amount of the cellular housekeeping gene encoding cyclophilin (Fig. 5A). Similarly, IFN-α levels were 1.2- to 1.4-fold higher (data not shown). Both IFN-α and IFN-β were induced in uninfected samples as well (data not shown).
FIG. 5.
Detection of IFN-β, IRF-1, IFNα/βR, and IRF-2 transcripts in murine TG following explantation. RT-PCR was used to detect IFN-β (A), IRF-1 (B), IFNα/βR (C), and IRF-2 (D), and each was compared to the level of cyclophilin mRNA. Duplicate samples of TG explant RNA from 0, 1, 2, and 4 h p.e. were analyzed. Products were separated by agarose gel electrophoresis, followed by fluorimager scanning and analysis using ImageQuant software. The relative amount of cDNA is expressed in arbitrary units representing the ratio between the intensity of the PCR product band to the intensity of cyclophilin. The ratio at the zero time point is designated 1. L indicates latent.
IFN expression is induced in neuronal cells.
To determine whether IFN expression colocalizes with reactivating virus in neuronal cells, TG sections from latently infected and uninfected explants were analyzed by immunohistochemistry. IFN-α/β protein expression was not detected at 0 h p.e. and was induced at 4, 8, and 24 h p.e. in both infected and uninfected explants (Fig. 6). Furthermore, as observed for TIS7, all of the neuronal cells expressed IFN. Thus, we conclude that viral gene expression occurs in cells expressing IFN.
FIG. 6.
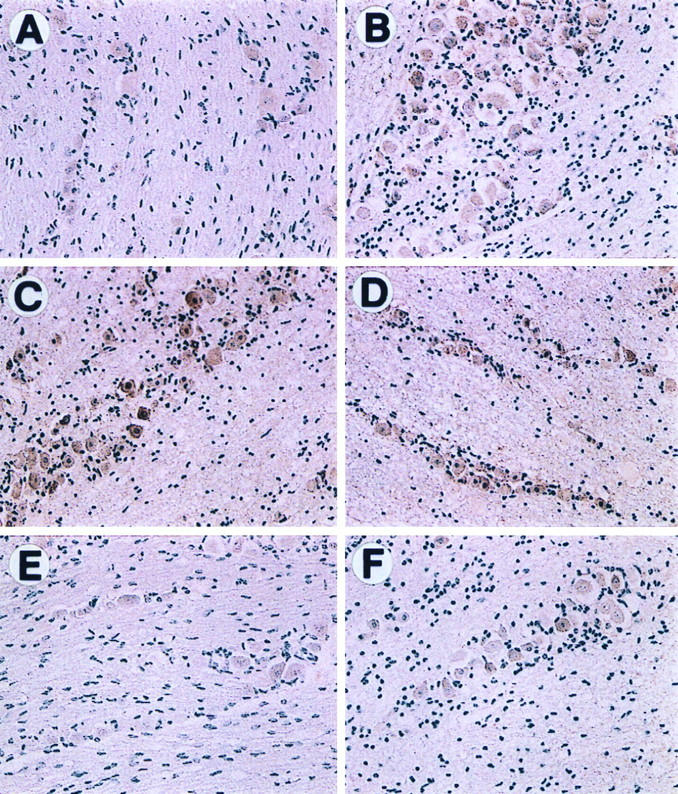
Immunostaining of IFN protein in TG following explantation. Latently infected or uninfected BALB/c mice were sacrificed, and TG were excised and incubated in culture medium for 0 to 24 h. Paraffin-embedded sections of latently infected TG at 0 (A), 4 (B), 8 (C), and 24 (D) h p.e. and of uninfected TG 0 (E) and 4 (F) h p.e. were processed as described in Materials and Methods and reacted with rabbit polyclonal antisera against IFN-α and -β. The experiment was repeated twice, and duplicate slides were screened.
Induction of IRF-1.
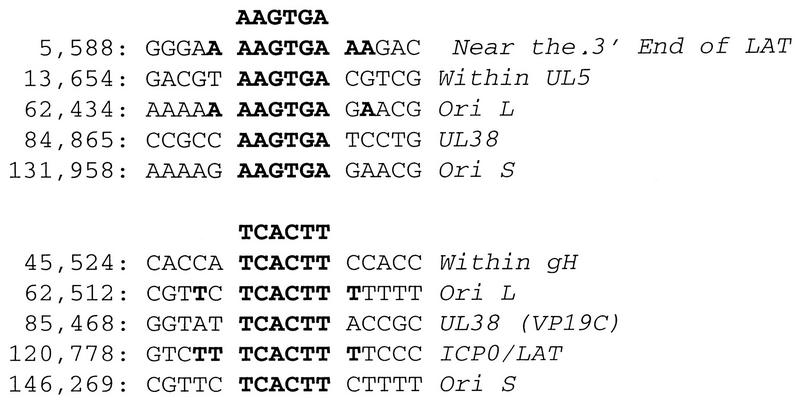
The IRFs bind to IFN consensus sequences found in many promoters of IFN gene family members (22, 56, 57). IRF-1 is an activator of IFN-β, whereas IRF-2 functions as a repressor (66). As shown in Fig. 5B, IRF-1 transcription was induced within the first hour p.e., and its profile of induction was strikingly similar to IFN-β (Fig. 5A). In contrast, no significant change was observed in the levels of the IFN-α/β receptor (IRFα/βR) or IRF-2 (Fig. 4D and 5C). Induction of IFN and IRF-1 also occurred in the absence of serum in the explantation media (not shown), indicating that serum factors are not the cause for increased gene expression. Induction of IFN and IRF-1 was detected reproducibly in both infected and uninfected preparations (not shown), as was also found for TIS7 (Fig. 3). Our results suggest that IFN induction after explantation of TG involves an IRF-1-dependent pathway. We used computational analysis to probe the HSV-1 genome for IRF-1 consensus binding sites (AAGTGA) (56). As shown in Fig. 7, 10 matches were identified in HSV-1 17+. Interestingly, two of the matches were in the ICP0/latency-associated transcript (LAT) region, and four mapped to palindromes in viral origins of replication OriL and OriS. These observations are discussed below.
FIG. 7.
Putative IRF-1 binding sites in the HSV-1 17+ genome, identified by sequence analysis using the Findpatterns function of the Genetics Computer Group (Madison, Wis.) software. The HSV-1 complete genome sequence (GenBank locus HE1CG accession no. X14112) was searched for the consensus IRF-1 binding site in IFN promoters the hexamer AAGTGA (56). Sequence matches are listed by sequence location, and gene name.
TNF-α is induced by TG explantation.
Soluble TNF-α enhances the reactivation frequency and replication of HSV-1 during explant reactivation (63). To determine whether endogenous TNF-α is induced during explantation, RT-PCR was performed with primers specific for TNF-α. TNF-α transcripts were induced rapidly following explantation (Fig. 8). However, we were unable to detect TNF-α, a secreted factor, in TG sections by immunostaining.
FIG. 8.
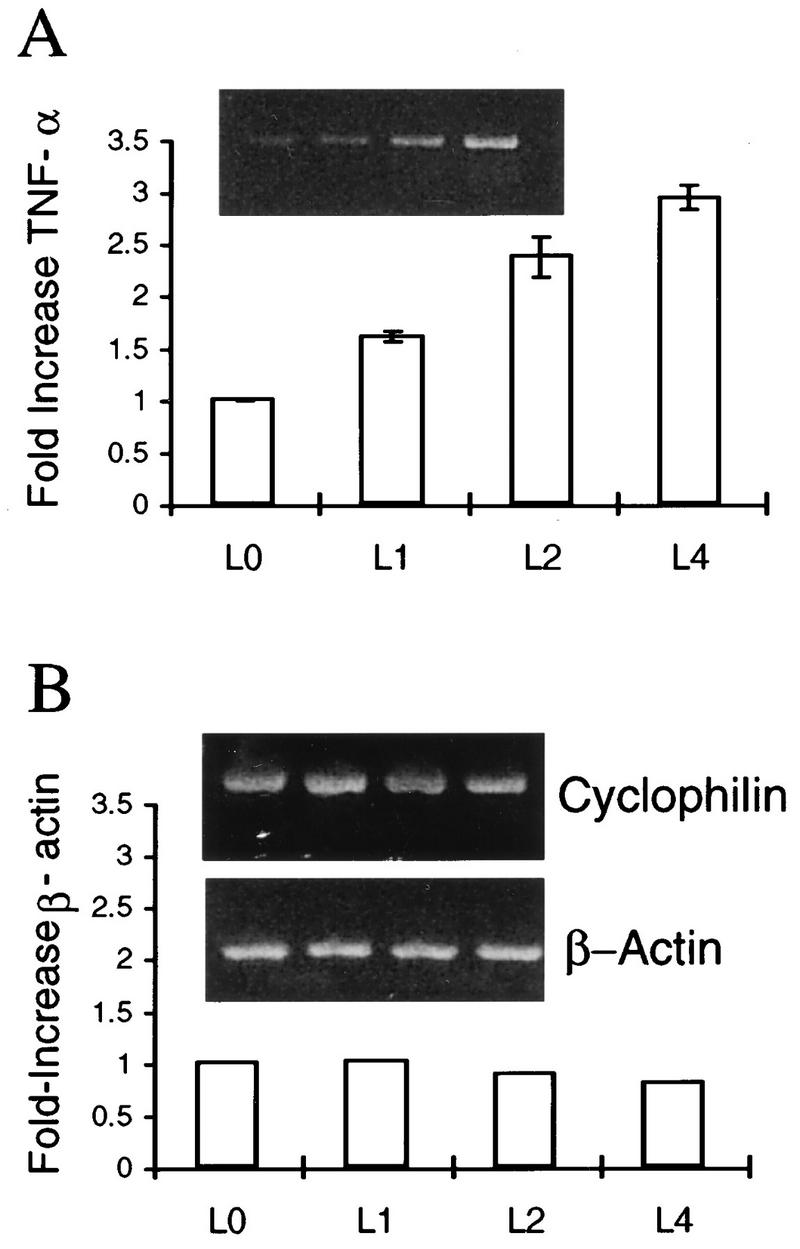
RT-PCR detection of TNF-α, cyclophilin, and β-actin transcripts in murine TG cultured for various times p.e. RNA from latently infected TG explants was prepared and analyzed by RT-PCR for TNF-α (A), and β-actin (B), and cyclophilin as described in Materials and Methods. Products were visualized by ethidium bromide staining as shown in the inserts. The graphs represent the ratio between the PCR product band and the cyclophilin band. The ratio at the time of explant (time zero) was determined as 1. Experiments were done in duplicate in four separate experiments. L indicates latent.
DISCUSSION
Cellular genes induced by the stress of explantation.
We have used DDRT-PCR to identify genes that are differentially expressed following the stress caused by explantation of murine TG. We previously established that these are conditions that induce viral gene activation and that certain HSV-1 genes are detected by 4 h p.e. (54). Mice were infected with HSV-1 by the corneal route; at 4 weeks postinfection, mice were sacrificed and the TG were explanted. RNA was prepared from TG at various times following explantation and then subjected to DDRT-PCR. The genes that we identified can be classified into three groups (Table 2): (i) 26 previously unknown sequences; (ii) known structural genes; and (iii) sequences corresponding to previously known genes associated with regulation of cellular processes (Table 2). While we recognize the possibility that none of these gene products directly influence the HSV reactivation process, the possibility that one or more are directly essential to viral activation is just as likely. Furthermore, the identification of these induced genes may reveal major stress-altered intracellular pathways, any component of which could be key to reactivation, including any of the previously unknown genes. Of the regulatory genes identified in the third group, the murine IFN-related TIS7 gene was represented multiple times. Because of this observation, our initial studies focused on TIS7, the related IFN genes, and known transcriptional regulators of these genes. Interestingly, these studies revealed a potential link between the IFN regulatory pathway and regulation of viral gene expression during reactivation.
TIS7 is induced by the stress of explantation.
Five differentially displayed bands were identified as overlapping regions of murine TIS7 (36, 61) (Fig. 2), and the results were confirmed by quantitative RPA, RT-PCR (Fig. 3), and immunostaining (Fig. 4). The TIS (TPA [tetradecanoyl phorbol acetate]-inducible sequences) family members are early-response genes (64) which are induced rapidly and transiently in Swiss 3T3 cells by the tumor promoter and mitogen TPA (36) or by serum (22). Most of the TIS genes also have been identified in rat PC12 cells following induction with nerve growth factor, TPA, epidermal growth factor, and depolarization (32). In addition, TIS induction has been detected in primary astrocyte cultures following mitogen induction (3). Thus, a family of TIS genes appear to constitute a common pathway or response to many cell stimulatory agents or physical stimuli.
The pattern of induction previously observed for TIS7 and PC4 genes in these systems revealed an increase in the levels of RNA or protein between 2 and 4 h poststimulation (62), similar to our observation in the mouse explant model (Fig. 2 to 5). Moreover, we have shown that another TIS transcript, TIS28 or c-fos, was induced rapidly following explantation (54, 60), and again, the kinetics matched those previously observed for c-fos following mitogen induction (3). These observations suggest that explantation and mitogen stimulation activate similar cellular early-response pathways which activate or induce TIS genes. One interesting possibility suggested by our results, although we have not yet obtained confirming evidence, is that these pathways also may be among the earliest events involved in the induction of latent herpesvirus.
IFN-β is induced by the stress of explantation.
TIS7 was originally identified as a gene induced in murine 3T3 cells following infection with Newcastle disease virus (50). Nucleotide sequence analysis revealed some similarity with human IFN-β and rat IFN-γ (58). We have shown that, like TIS7, both IFN-α and IFN-β are induced in neuronal cells within the first hour following explantation. These observations suggest that these IFN-related genes, although different in function, may share a common cellular pathway that may be involved in the early events of HSV reactivation.
IFN-β expression is modulated at the transcriptional level by multiple regulatory factors that bind upstream of the initiation site, such as the activators IRF-1 and NF-κB and the repressor IRF-2 (reviewed in reference 56). We have shown that IRF-1 was induced by the stress of explantation. In contrast, neither IRF-2 nor the IFN receptor was induced (Fig. 5). Since the induction of IRF-1 followed the same temporal pattern as that of IFN, IFN-α and -β are likely to be induced via an IRF-1-dependent pathway (27) in explanted TG cells. Induction of IFN expression has been previously observed following HSV infection (17, 18, 52). However, we detected induction of IFN at 1 to 2 h p.e., prior to viral gene induction, which we detected at 2 to 4 h p.e. (54). Furthermore, IFN induction was detected in neurons of both latently infected and uninfected TG. We also found that IFN-β and IRF-1 were induced in the TG in the absence of serum in the explantation medium. Taken together, these data indicate that IFN induction is a consequence of the stress of explantation and did not result from viral gene expression or from incubation in the presence of serum factors.
Possible relationship between reactivating HSV and IFNs.
IFN possesses antiviral properties and indeed is induced by HSV infection (17, 18, 52). Several mechanisms have been elucidated for specific effects of IFN on HSV. For example, IFN is an inhibitor of HSV IE gene activation by VP16 in vitro (33). We recently determined that both early and IE HSV genes are induced during the first hours of viral reactivation (54). Thus, the absence of viral IE gene expression prior to early gene expression during the first hours following explantation of TG may be a result of an inhibitory effect of IFN on a neuron-specific VP16 functional homolog.
Furthermore, IFN blocks both HSV morphogenesis and release of viral particles from infected cells (8). The presence of antiviral activity during the earliest times following tissue stimulation suggests an interesting relationship between IFN and reactivating HSV. Viral gene activation may induce one round of viral replication, allowing the virus to travel from TG neurons to the site of recrudescence in corneal epithelium. Immediate induction of IFN may prevent the spread of reactivating virus within the nervous system by inhibiting release of viral particles and activating host defense mechanisms such as natural killer cells (22). Moreover, in another virus system, neuroblastoma cells expressing high levels of IFN-β support persistent rabies virus infections (25). This finding suggests that the IFN response may be involved in ensuring the viability of infected host cells. IRF-1 has been implicated in the antiviral effects of IFNs on encephalomyocarditis virus by inhibiting viral replication. However, in IRF-1−/− fibroblasts derived from knockout mice, there was minimal reduction of IFN-γ-mediated inhibition of HSV-1 replication (29). This finding suggests that IRF-1 is not essential for IFN inhibition of HSV-1 replication in fibroblasts. Further latency studies in IRF-1−/− mice or ganglia may clarify the role of IRF-1 in HSV pathogenesis.
In a different scenario, IFN may inhibit viral reactivation in neurons, and in a few cells, other cellular factors override its effects by inducing high levels of viral gene expression. For example, Walev et al. (63) have shown that treatment of TG explants with soluble IFN inhibits reactivation, detected by reduction of infectious virus in the presence of IFN. In contrast, in the same study, TNF-α treatment increased the efficiency of reactivation. These data support the hypothesis that induction of IFN inhibits multiple rounds of viral replication in neuronal cells of the TG. Consistent with this scenario, we detected induction of TNF-α transcription in TG explants (Fig. 8) under conditions leading to viral reactivation. Thus, it is possible that the levels of TNF-α in few neurons (perhaps 1% of latent neurons) are higher than the levels of IFN, allowing the latent viral genome to reactivate in those neurons.
Are TIS7, IFN, and viral responses related?
TIS7 and IFNs are induced by viral infection (50, 56) and, as shown in this study, by the stress of explantation. While the precise function of TIS7 in the cell is not yet elucidated, it is clear that it plays a role in cellular growth and differentiation (19, 23). Furthermore, it is known that TIS7 has no antiviral activity (58), which is a property of IFNs (56). One unifying hypothesis is that these cellular components may share a common induction pathway with HSV. In particular, as suggested above, TIS7, IFN family members, and HSV share a common regulatory element. Thus, our initial detection of TIS7 and IFN increasing in explanted TG led us to test whether IRF-1 was also increasing. The observation that IRF-1 expression was increasing in explants in a similar time frame to IFN and TIS7, and prior to HSV gene activation, leads to our speculation that IRF-1 may play a direct role in reactivation of the HSV genome. As yet, we do not have direct experimental evidence to demonstrate that HSV-1 responds to IRF-1.
However, in support of this hypothesis, we have identified multiple sequences corresponding to potential IRF-1 binding sites in HSV-1 DNA (Fig. 7). As shown in Fig. 7, there are 10 perfect matches in HSV-1 17+ to the core IRF-1 consensus sequence (AAGTGA) (56). Of particular interest are several binding sites in the origins of replication OriS and OriL. Two are an inverted repeat within the palindrome in OriL, and the other two are in OriS; strikingly, all are conserved in HSV-1 and HSV-2 strains (not shown). These consensus sites are within regions previously identified as origin binding protein or UL9 binding site III. Also, of great interest are previous observations that, in addition to binding origin binding protein, binding site III interacts with yet unknown cellular proteins (10, 11). Taken together, these observations suggest that IRF-1 may bind to regulatory elements in the viral genome such as the origins of replication and ICP0/LAT and thereby upregulate viral replication or gene expression. Interestingly, functional IRF binding sites recently were identified in the herpesvirus EBV (40, 49). IRF-1 and IRF-2 bind directly to consensus sequence sites in the EBV type I latency promoter of EBNA-1 (Qp), and IRFs may play a primary role in transcriptional regulation of EBNA-1 in cell culture (40, 49). Experiments are in progress to determine whether IRF-1 or IRF-2 binds to IRF consensus sites in HSV-1 that we have identified.
The stress of hyperthermia treatment induces in vivo reactivation in mice (48). We recently determined that both TIS7 and IRF-1 are induced in murine TG by hyperthermia during the time frame of detection of viral gene activation (55). These results confirm our observations in the explantation model. Thus, it is possible that HSV has evolved reactivation mechanisms which take advantage of cellular activation pathways which are induced by stress. Further experiments are under way to test these hypotheses.
In summary, using DDRT-PCR as a starting point, we identified several genes involved in the cellular response to the stress of explantation. Although we have not established a causal link, the temporal correspondence of HSV-1 reactivation and induction of TIS7, IFN, and IRF-1 gene expression suggests the existence of common regulatory pathways. Also, our results yield insights into the cellular environment which is present during HSV reactivation. Therefore, PCR differential display represents an excellent method to screen for species of RNA which are transcriptionally regulated in TG following explantation. Genes identified using this method can be studied in other HSV reactivation systems, resulting in a database of specific genes which may be involved in the reactivation process.
ACKNOWLEDGMENTS
R.T.-S. and W.P. contributed equally to this work.
We thank S. Albelda (University of Pennsylvania) for providing PCR primers specific for mouse β-actin and John Y. Chan (SmithKline Beecham Pharmaceuticals) for bioinformatics support.
This research was supported by Public Health grant NS33768 to N.W.F. and S.L.B. and by funds from SmithKline Beecham Pharmaceuticals. R.T.-S. was supported in part by training grant CA09171 from the Public Health Service. T.M.L. was supported by the division of neurosurgery, Hospital of the University of Pennsylvania, and a clinical fellowship from the Measey Foundation.
REFERENCES
- 1.Adams R L, Springall D R, Levene M M, Bushell T E. The immunocytochemical detection of herpes simplex virus in cervical smears—a valuable technique for routine use. J Pathol. 1984;143:241–247. doi: 10.1002/path.1711430403. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gisg W, Miller W, Meyers E W, Lipman D J. BLAST: a local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Arenalder A T, Lim R W, Varnum B C, Cole R, Vellis J D, Herschman H R. TIS gene expression in cultured rat astrocytes: induction by mitogens and stellation agents. J Neurosci Res. 1989;23:247–256. doi: 10.1002/jnr.490230302. [DOI] [PubMed] [Google Scholar]
- 4.Arvidson B. Retrograde axonal transport of horseradish peroxidase from cornea to trigeminal ganglion. Acta Neuropathol. 1977;38:49–52. doi: 10.1007/BF00691276. [DOI] [PubMed] [Google Scholar]
- 5.Barber S A, Fultz M J, Salkowski C A, Vogel S N. Differential expression of interferon regulatory factor 1 (IRF-1), IRF-2, and interferon consensus sequence binding protein genes in lipopolysaccharide (LPS)-responsive and LPS-hyporesponsive macrophages. Infect Immun. 1995;63:601–608. doi: 10.1128/iai.63.2.601-608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergsma D J, Eder C, Gross M, Kersten H, Sylvester D, Appelbaum E, Cusiamano D, Levi G P, McLaughlin M M, Kasyan K, Prichett W P, Bossard M J, Brandt M, Levy M A. The cyclophilin multigene family of peptidyl-prolyl isomerases: characterization of three separate human isoforms. J Biol Chem. 1991;266:23204–23214. [PubMed] [Google Scholar]
- 7.Brown S M, Ritchie D A, Subak-Sharpe J. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to lineage groups. J Gen Virol. 1973;18:329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee S, Hunter E, Whitley R. Effect of cloned human interferons on protein synthesis and morphogenesis of herpes simplex virus. J Virol. 1985;56:419–425. doi: 10.1128/jvi.56.2.419-425.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook M L, Bastone V B, Stevens J B. Evidence that neurons harbor latent herpes simplex virus. Infect Immun. 1974;9:946–951. doi: 10.1128/iai.9.5.946-951.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dabrowski C E, Carmillo P J, Schaffer P A. Cellular protein interactions with herpes simplex virus type 1 oriS. Mol Cell Biol. 1994;14:2545–2555. doi: 10.1128/mcb.14.4.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dabrowski C E, Schaffer P A. Herpes simplex virus type 1 origin-specific binding protein: oriS-binding properties and effects of cellular proteins. J Virol. 1991;65:3140–3150. doi: 10.1128/jvi.65.6.3140-3150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devi-Rao G B, Bloom D C, Stevens J G, Wagner E K. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J Virol. 1994;68:1271–1282. doi: 10.1128/jvi.68.3.1271-1282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fawl R L, Gesser R M, Valyi-Nagi T, Fraser N W. Reactivation of herpes simplex virus from latently infected mice after administration of cadmium is mouse-strain-dependent. J Gen Virol. 1996;77:2781–2786. doi: 10.1099/0022-1317-77-11-2781. [DOI] [PubMed] [Google Scholar]
- 14.Fawl R L, Roizman B. Induction of reactivation of herpes simplex virus in murine sensory ganglia in vivo by cadmium. J Virol. 1993;67:7025–7031. doi: 10.1128/jvi.67.12.7025-7031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, N. W., J. G. Spivack, Z. Wroblewska, T. Block, S. L. Deshmane, T. Valyi-Nagy, R. Natarajan, and R. Gesser. 1991. A review of the molecular mechanism of HSV-1 latency. Curr. Eye Res. 10(Suppl.):1–14. [DOI] [PubMed]
- 16.Fraser N W, Valyi-Nagy T. Viral, neuronal and immune factors which may influence herpes simplex virus (HSV) latency and reactivation. Microb Pathog. 1993;15:83–91. doi: 10.1006/mpat.1993.1059. [DOI] [PubMed] [Google Scholar]
- 17.Gobl A E, Funa K, Alm G V. Different induction patterns of mRNA for IFN-alpha and -beta in human mononuclear leukocytes after in vitro stimulation with herpes simplex virus-infected fibroblasts and Sendai virus. J Immunol. 1988;140:3605–3609. [PubMed] [Google Scholar]
- 18.Green J A, Yeh T J, Overall J C., Jr Sequential production of IFN-alpha and immune-specific IFN-gamma by human mononuclear leukocytes exposed to herpes simplex virus. J Immunol. 1981;127:1192–1196. [PubMed] [Google Scholar]
- 19.Guardavaccaro D, Ciotti M T, Schafer B W, Montagnoli A, Tirone F. Inhibition of differentiation in myoblasts deprived of the interferon-related protein PC4. Cell Growth Differ. 1995;6:159–169. [PubMed] [Google Scholar]
- 20.Habu S, Akamatsu K, Tamaoki N, Okumura K. In vivo significance of NK cells on resistance against virus (HSV-1) infections in mice. J Immunol. 1984;133:2743–2747. [PubMed] [Google Scholar]
- 21.Harwick J, Romanowski E, Araullo-Cruz T, Gordon Y J. Timolol promotes reactivation of latent HSV-1 in the mouse iontophoresis model. Invest Ophthalmol Visual Sci. 1987;28:580–584. [PubMed] [Google Scholar]
- 22.Herschman H R. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 23.Herschman H R, Kujubu D A, Fletcher B S, Ma Q, Varnum B C, Gilbert R S, Reddy S T. The tis genes, primary response genes induced by growth factors and tumor promoters in 3T3 cells. Prog Nucleic Acid Res Mol Biol. 1994;47:113–148. doi: 10.1016/s0079-6603(08)60251-2. [DOI] [PubMed] [Google Scholar]
- 24.Hill T J. Herpes simplex virus latency. In: Roizman B, editor. The Herpesviruses. Vol. 4. New York, N.Y: Plenum Publishing Corp.; 1985. pp. 175–240. [Google Scholar]
- 25.Honda Y, Kawai A, Matsumoto S. Persistent infection of rabies virus (HEP-Flury strain) in human neuroblastoma cells capable of producing interferon. J Gen Virol. 1985;66:957–967. doi: 10.1099/0022-1317-66-5-957. [DOI] [PubMed] [Google Scholar]
- 26.Inokuchi K, Murayama A, Ozawa F. mRNA differential display reveals Krox-20 as a neural plasticity-regulated gene in the rat hippocampus. Biochem Biophys Res Commun. 1996;221:430–436. doi: 10.1006/bbrc.1996.0612. [DOI] [PubMed] [Google Scholar]
- 27.Kawakami T, Matsumoto M, Sato M, Harada H, Taniguchi T, Kitagawa M. Possible involvement of the transcription factor ISGF3 gamma in virus-induced expression of the IFN-beta gene. FEBS Lett. 1995;358:225–229. doi: 10.1016/0014-5793(94)01426-2. [DOI] [PubMed] [Google Scholar]
- 28.Kesari S, Randazzo B P, Valyi-Nagy T, Huang Q S, Brown S M, MacLean A R, Lee V M-Y, Trojanowski J Q, Fraser N W. Therapy of experimental human brain tumors using a neuroattenuated herpes simplex virus mutant. Lab Invest. 1995;73:636–648. [PubMed] [Google Scholar]
- 29.Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak T W, Taniguchi T. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 30.Kiryu S, Yao G L, Morita N, Kato H, Kiyama H. Nerve injury enhances rat neuronal glutamate transporter expression: identification by differential display PCR. J Neurosci. 1995;15:7872–7878. doi: 10.1523/JNEUROSCI.15-12-07872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kita M, Tanaka K, Shinmura K, Tanaka Y, Liu Y, Imanishi J. Expression des genes des cytokines et des genes associes a l’interferon chez l’embryon de la souris. C R Soc Biol. 1994;188:593–600. [PubMed] [Google Scholar]
- 32.Kujubu D A, Lim R W, Varnum B C, Herschman H R. Induction of transiently expressed genes in PC-12 pheochromocytoma cells. Oncogene. 1987;1:257–262. [PubMed] [Google Scholar]
- 33.LaMarco K, McKnight S. Purification of a set of cellular polypeptides that bind to the purine-rich cis-regulatory element of herpes simplex virus immediate early genes. Genes Dev. 1989;3:1372–1383. doi: 10.1101/gad.3.9.1372. [DOI] [PubMed] [Google Scholar]
- 34.Liang P, Bauer D, Averboukh L, Warthoe P, Rohrwild M, Muller H, Strauss M, Pardee A B. Analysis of altered gene expression by differential display. Methods Enzymol. 1995;254:304–321. doi: 10.1016/0076-6879(95)54022-9. [DOI] [PubMed] [Google Scholar]
- 35.Liang P, Pardee A B. Differential Display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 36.Lim R W, Varnum B C, Herschman H R. Cloning of tetradecanoyl phorbol ester-induced ‘primary response’ sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene. 1987;1:263–270. [PubMed] [Google Scholar]
- 37.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 38.Marfurt C F, Kingsley R E, Echtenkamp S E. Sensory and sympathetic innervation of the mammalian cornea. Investigative Ophthalmol Visual Sci. 1989;30:461–472. [PubMed] [Google Scholar]
- 39.Nickolaus P, Zawatzky R. Inhibition by interleukin-4 of constitutive beta interferon synthesis in mouse macrophages. J Virol. 1994;68:6763–6766. doi: 10.1128/jvi.68.10.6763-6766.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nonkwelo C, Ruf I K, Sample J. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J Virol. 1997;71:6887–6897. doi: 10.1128/jvi.71.9.6887-6897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Openshaw H, Asher S, Wohlenberg C, Sekizawa T, Notkins A L. Acute and latent infection in sensory ganglia with herpes simplex virus: immune control and virus reactivation. J Gen Virol. 1979;44:205–215. doi: 10.1099/0022-1317-44-1-205. [DOI] [PubMed] [Google Scholar]
- 42.Qu D, Ludwig D S, Gammeltoft S, Piper M, Pelleymounter M A, Cullen M J, Mathes W F, Przypek R, Kanarek R, Maratos-Flier E. A role for melanin-concentrating hormone in the central regulation of feeding behaviour. Nature. 1996;380:243–247. doi: 10.1038/380243a0. [DOI] [PubMed] [Google Scholar]
- 43.Ramakrishnan R, Levine M, Fink D J. PCR-based analysis of herpes simplex virus type 1 latency in the rat trigeminal ganglion established with a ribonucleotide reductase-deficient mutant. J Virol. 1994;68:7083–7091. doi: 10.1128/jvi.68.11.7083-7091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramakrishnan R, Poliani P L, Levine M, Glorioso J C, Fink D J. Detection of herpes simplex virus type 1 latency-associated transcript expression in trigeminal ganglia by in situ reverse transcriptase PCR. J Virol. 1996;70:6519–6523. doi: 10.1128/jvi.70.9.6519-6523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Randazzo B P, Kesari S, Gesser R M, Brown S M, Fraser N W. Treatment of experimental intracranial murine melanoma with a neuro-attenuated herpes simplex virus-1 mutant. Virology. 1995;211:94–101. doi: 10.1006/viro.1995.1382. [DOI] [PubMed] [Google Scholar]
- 46.Roizman B. Herpesviridae: a brief introduction. In: Fields B N, Knipe D M, editors. Fundamental virology. New York, N.Y: Raven Press, Ltd.; 1991. pp. 841–847. [Google Scholar]
- 47.Roizman B, Sears A E. An inquiry into the mechanisms of herpes simplex virus latency. Annu Rev Microbiol. 1987;41:543–571. doi: 10.1146/annurev.mi.41.100187.002551. [DOI] [PubMed] [Google Scholar]
- 48.Sawtell N M, Thompson R L. Rapid in vivo reactivation of herpes simplex virus in latently infected murine ganglionic neurons after transient hyperthermia. J Virol. 1992;66:2150–2156. doi: 10.1128/jvi.66.4.2150-2156.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaffer B C, Paulson E, Strominger J L, Speck S H. Constitutive activation of Epstein-Barr virus (EBV) nuclear antigen 1 gene transcription by IRF1 and IRF2 during restricted EBV latency. Mol Cell Biol. 1997;17:873–886. doi: 10.1128/mcb.17.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skup D, Windass J D, Sor F, George H, Williams B R, Fukuhara H, Maeyer-Guignard J D, Maeyer E D. Molecular cloning of partial cDNA copies of two distinct mouse IFN-beta mRNAs. Nucleic Acids Res. 1982;10:3069–3084. doi: 10.1093/nar/10.10.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sompayrac L, Jane S, Danna K J. Reduced levels of alpha1(XI) procollagen mRNA in SV40-transformed cells. Virology. 1996;218:412–416. doi: 10.1006/viro.1996.0212. [DOI] [PubMed] [Google Scholar]
- 52.Stanwick T L, Campbell D E, Nahmias A J. Cytotoxic properties of human monocyte-macrophages for human fibroblasts infected in herpes simplex virus: interferon production and augmentation. Cell Immunol. 1982;70:132–147. doi: 10.1016/0008-8749(82)90139-3. [DOI] [PubMed] [Google Scholar]
- 53.Stevens J G. Human herpesviruses: a consideration of the latent state. Microbiol Rev. 1989;53:318–332. doi: 10.1128/mr.53.3.318-332.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tal-Singer R, Lasner T M, Podrzucki W, Skokotas A, Leary J J, Berger S L, Fraser N W. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue culture. J Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tal-Singer, R., D. Sutton, and J. Leary. Unpublished data.
- 56.Tanaka N, Taniguchi T. Cytokine gene regulation: regulatory cis-elements and DNA binding factors involved in the interferon system. Adv Immunol. 1992;52:263–281. doi: 10.1016/s0065-2776(08)60877-9. [DOI] [PubMed] [Google Scholar]
- 57.Taniguchi T, Harada H, Lamphier M. Regulation of the interferon system and cell growth by the IRF transcription factors. J Cancer Res Clin Oncol. 1995;121:516–520. doi: 10.1007/BF01197763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tirone F, Shooter E. Early gene regulation by nerve growth factor in PC12 cells: induction of an interferon-related gene. Proc Natl Acad Sci USA. 1989;86:2088–2092. doi: 10.1073/pnas.86.6.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trojanowski J, Mantione R, Lee J, Seid D, You T, Inge L, Lee V. Neurons derived from a human teratocarcinoma cell line establishes molecular and structural polarity following transplantation into the rodent brain. Exp Neurol. 1993;122:283–294. doi: 10.1006/exnr.1993.1128. [DOI] [PubMed] [Google Scholar]
- 60.Valyi-Nagy T, Deshmane S L, Dillner A J, Fraser N W. Induction of cellular transcription factors in trigeminal ganglia of mice by corneal scarification, herpes simplex virus 1 type infection, and explantation of trigeminal ganglia. J Virol. 1991;65:4142–4152. doi: 10.1128/jvi.65.8.4142-4152.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varnum B C, Lim R W, Herschman H R. Characterization of TIS7, a gene induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene. 1989;4:1263–1265. [PubMed] [Google Scholar]
- 62.Varnum B C, Reddy S T, Koski R A, Herschman H R. Synthesis, degradation, and subcellular localization of proteins encoded by the primary response genes TIS7/PC4 and TIS21/PC3. J Cell Physiol. 1994;158:205–213. doi: 10.1002/jcp.1041580125. [DOI] [PubMed] [Google Scholar]
- 63.Walev I, Podlech J, Falke D. Enhancement by TNF-α of reactivation and replication of latent herpes simplex virus from trigeminal ganglia. Arch Virol. 1995;140:987–992. doi: 10.1007/BF01315409. [DOI] [PubMed] [Google Scholar]
- 64.Walz M A, Yamamoto H, Notkins A L. Immunological response restricts number of cells in sensory ganglia infected with herpes simplex virus. Nature. 1976;264:554–559. doi: 10.1038/264554a0. [DOI] [PubMed] [Google Scholar]
- 65.Warthoe P. Detection and identification of expressed genes by differential display. In: Dieffenbach C W, Dveksler G S, editors. PCR primers: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 421–438. [Google Scholar]
- 66.Watanabe N, Sakakibara J, Hovanessian A G, Taniguchi T, Fujita T. Activation of IFN-beta element by IRF-1 requires a posttranslational event in addition to IRF-1 synthesis. Nucleic Acids Res. 1991;19:4421–4428. doi: 10.1093/nar/19.16.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Willey D E, Trousdale M D, Nesburn A B. Reactivation of murine latent HSV infection by epinephrine iontophoresis. Invest Ophthalmol Visual Sci. 1984;25:945–950. [PubMed] [Google Scholar]
- 68.Yan J, Khanna K K, Lavin M F. Induction of inositol 1,4,5 triphosphate receptor genes by ionizing radiation. Int J Radiat Biol. 1996;69:539–540. doi: 10.1080/095530096145544. [DOI] [PubMed] [Google Scholar]