Abstract
Mutant forms of herpesvirus saimiri (HVS) subgroup C strain 488 with deletions in either STP-C488 or Tip were constructed. The transforming potentials of the HVS mutants were tested in cell culture and in common marmosets. Parental HVS subgroup C strain 488 immortalized common marmoset T lymphocytes in vitro to interleukin-2-independent growth, but neither of the deletion mutants produced such growth transformation. Wild-type HVS produced fatal lymphoma within 19 to 20 days of experimental infection of common marmosets, while HVS ΔSTP-C488 and HVS ΔTip were nononcogenic. Virus was repeatedly isolated from the peripheral blood of marmosets infected with mutant virus for more than 5 months. These results demonstrate that STP-C488 and Tip are not required for replication or persistence, but each is essential for transformation in cell culture and for lymphoma induction in common marmosets.
Herpesvirus saimiri (HVS), a gamma-2 herpesvirus or rhadinovirus, infects most squirrel monkeys without apparent disease (9, 13). In other nonhuman primates, however, HVS induces rapidly fatal T-cell lymphoproliferative diseases (14, 17). Sequence divergence among HVS isolates is most extensive at the left end of the unique L-DNA of the viral genome and is the basis for classification of HVS into subgroups A, B, and C (7, 9, 30). Variation in this region is correlated with differences in the capacity of the viruses to immortalize T lymphocytes in vitro and to produce lymphoma in nonhuman primates (4, 7, 8, 10, 24, 33). Viruses of both subgroups A and C immortalize common marmoset T lymphocytes to interleukin-2-independent proliferation (10, 34). Highly oncogenic subgroup C strains also immortalize human, rabbit, and rhesus monkey lymphocytes and can produce fulminant lymphoma in rhesus monkeys as well as in New World primates (1–4, 6, 31).
HVS subgroup A strain 11 mutants with deletions in the first open reading frame at the left end of the genome are capable of replication but fail to immortalize common marmoset T lymphocytes in vitro and do not induce lymphoma in vivo (7, 8, 10, 24, 33). This open reading frame encodes the saimiri transformation-associated protein (STP) (22). HVS subgroup C strain 488 (HVS C488) contains a divergent form of the STP gene along with an additional apparently unrelated open reading frame in the leftmost position (4, 16, 22). Similarities between STPs of HVS subgroup A strain 11 (STP-A11) and subgroup C strain 488 (STP-C488) include highly acidic amino termini, the presence of collagen-like repeats in the central parts of the proteins, and hydrophobic membrane-spanning regions at the carboxyl termini (18, 22). Both STP-C488 and STP-A11 are sufficient to transform rodent fibroblast cells in vitro, but STP-C488 is considerably more potent (16, 22). Transgenic mice expressing STP-C488 developed invasive epithelial cell tumors (32), while STP-A11 transgenic mice developed peripheral pleomorphic T cell lymphomas (25). Unlike STP-A11, which associates with Src kinase (26), STP-C488 associates with cellular Ras (19). Disruption of association between STP and Ras disrupts transforming activity of STP-C488 (19). To our knowledge, STP-C488 is the only virus-encoded protein that has been found to associate with cellular Ras in oncogenic transformation.
The product of the leftmost gene (orf1) of HVS C488 is expressed in transformed T cells and has been shown to associate with the tyrosine kinase Lck (5, 20), a member of the Src kinase family. This protein has consequently been designated Tip (for tyrosine kinase-interacting protein). Cell-free kinase assays have demonstrated that Tip is phosphorylated on tyrosine residues by Lck (20, 21). Lck-binding elements of Tip have been defined and include an SH3-binding sequence and a sequence homologous to the carboxyl terminus of Src-related kinases (20). Tip acts at an early stage of T-cell receptor signal transduction by downregulating Lck-mediated activation (21). Additionally, we have recently demonstrated that Tip also interacts with a novel cellular protein called Tap (for Tip-associated protein) (36). Coexpression of Tip and Tap in Jurkat T cells dramatically upregulates surface expression of adhesion molecules and activates NF-κB transcription factor activity (36). Therefore, both Lck and Tap are likely to be an important mediator of Tip functions, although it is not known whether these are separate or related functional activities.
In the present study, replication-competent deletion mutant viruses were constructed to assess the contributions of Tip and STP-C488 to oncogenic transformation in vitro and in vivo. We now show that Tip and STP-C488 are not required for viral replication or persistence but are essential for growth transformation of primary T cells in culture and for disease induction in vivo.
MATERIALS AND METHODS
Cell culture and virus propagation.
Owl monkey kidney cells (OMK 637) that were cultivated in minimal essential medium supplemented with penicillin, streptomycin, l-glutamine, and 10% (vol/vol) heat-inactivated fetal bovine serum (GIBCO BRL, Grand Island, N.Y.) were used for propagation of HVS C488. Low (<30)-passage OMK cells were used for transfections. Culture of common marmoset lymphocytes in immortalization assays with HVS mutants was performed in RPMI 1640 medium supplemented with penicillin, streptomycin, amphotericin B (Fungizone), l-glutamine, 20% (vol/vol) heat-inactivated fetal bovine serum, and 5 mg of β-mercaptoethanol per liter. COS-1 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) heat-inactivated fetal calf serum.
Virion DNA isolation.
HVS virion preparations were obtained from OMK cell lysates by removal of cell debris by low-speed centrifugation followed by pelleting of the virus at 18,000 rpm for 2 h in an SS-34 rotor. To purify intact virion DNA, the virus was disrupted at 60°C for 2 h in lysis buffer containing 10 mM Tris (pH 8.5), 1 mM EDTA, 1% (vol/vol) Sarkosyl, and 0.1 mg of proteinase K per ml. Extraction of the aqueous solution first with an equal volume of phenol and then twice with chloroform was sufficient to purify the virion DNA for use in transfections. Sterile cut pipette tips were used for manipulating virion DNA without shearing.
Reporter expression plasmid and SEAP assay.
As described previously (11), a reporter gene expression cassette containing the secreted engineered alkaline phosphatase (SEAP) gene under the control of the simian virus 40 (SV40) early promoter and enhancer (SV40-SEAP) was used as a selection marker for the identification of viral recombinants. SEAP production was detected by liquid scintillation counter measurement of chemiluminescence produced in assays of cell culture medium, using Phospha-Light reagents (Tropix Inc., Bedford, Mass.) according to the manufacturer’s recommendations.
Construction of STP and Tip deletion plasmids.
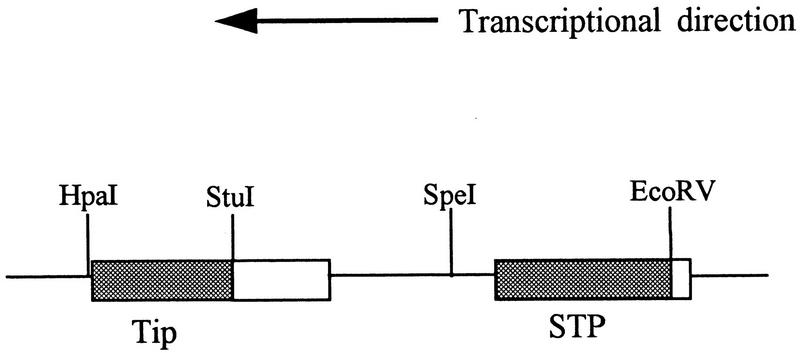
Deletions in plasmid pNEB193 containing Tip, STP-C488, and herpesvirus saimiri U RNAs (HSURs) within a 3.6-kb PstI/XbaI viral DNA fragment (C488PX) (4, 11) were made by restriction enzyme digestion followed by cloning of the reporter cassette into each deletion. Deletion of nucleotides 1318 to 1825 of HVS C488 by SpeI/EcoRV digestion removed 284 bp of STP-C488, which retained only the amino-terminal 8 amino acids of the total 102 amino acids of STP-C488 (Fig. 1). The possible stop codon after the deletion is 6 amino acids downstream of truncated STP-C488 gene. Similarly, StuI/HpaI digestion deleted HVS C488 nucleotides 438 to 879 for the carboxyl-terminal 139 amino acids of Tip (Fig. 1). This deletion removed the coding sequences for Lck-binding motifs and hydrophobic membrane-spanning region (20). The possible stop codon after the deletion is 17 amino acids downstream of truncated tip gene. A SEAP expression cassette was inserted into the deleted regions in plasmid DNA as previously described (33).
FIG. 1.
Schematic diagram of deletions in STP and tip genes. A 3.6-kb cloned HVS DNA fragment (C488PX) was used for deletion mutations. Shaded boxes indicate deletions in the genes; restriction enzyme sites are indicated at the top. Ninety-four of a total of 102 amino acids were deleted from STP-C488, while 139 of a total of 256 amino acids were deleted from Tip.
Transfections and isolation of HVS recombinants.
HVS C488 recombinants with specific gene deletions were generated by mixed transfection of virion and cloned DNA and identification of recombinants which express SEAP activity as described previously (8, 11, 33). Recombinants expressing SEAP were isolated in pure form by repeated passage of limiting dilutions of virus stock to OMK cell monolayers in 48-well tissue culture plates (Corning). SEAP production in individual wells showing cytopathic effect was assessed with the Phospha-Light chemiluminescence assay (Tropix) performed in opaque 96-well microtiter plates by using a MicroBeta scintillation counter (Wallac, Gaithersburg, Md.).
Since the SEAP expression cassette contains flanking AscI restriction enzyme sites which are not present in virion DNA, SEAP-positive virion DNA was digested with AscI, ligated overnight with T4 ligase, and transfected into OMK cells. Recombinant virus with the SEAP reporter deleted was isolated by limiting dilution and repeated selection of SEAP-negative virus. Presence of the deletion was assessed by two types of PCR: with primers homologous to deleted segments and with primers that flank the deletions. Expression of STP-C488 and Tip was determined by immunoblot and in vitro kinase assays with their specific antibodies.
To restore the deleted genes, virion DNA from HVS ΔSTP/SV40-SEAP and HVS ΔTip/SV40-SEAP was also similarly cotransfected into OMK cells with the unaltered 3.6-kb HVS DNA fragment containing intact STP-C488 and Tip. Recombinants with STP-C488 or Tip restored were then isolated by repeated limiting dilution and selection of SEAP-negative virus. PCR and DNA sequencing were performed to confirm the absence of the SEAP gene and presence of the STP-C488 or tip gene.
In vitro immortalization of common marmoset lymphocytes.
Assays of lymphocyte immortalization in vitro have been described previously (10). Peripheral blood mononuclear cells (PBMC) were isolated from 3-ml heparinized blood specimens from common marmosets (Callithrix jacchus) by centrifugation through lymphocyte separation medium (Organon Teknika Corp., Malvern, Pa.) followed by washing in RPMI 1640 culture medium. PBMC from each animal were individually washed, resuspended in RPMI 1640, and then distributed in 1-ml volumes containing approximately 106 cells into 12-well tissue culture plates. Cells were then infected at a multiplicity of infection ranging from 1 to 5 with 1 ml of purified HVS stocks. Cells were maintained with RPMI 1640 growth medium changed every 3 to 4 days. Immortalization or cell death was assessed microscopically.
Experimental infection of common marmosets.
In vivo oncogenicity of the HVS C488 recombinants was assessed by experimental infection of common marmosets. Marmosets were injected intramuscularly with 105 50% tissue culture infective doses of virus in a volume of 1 ml. Sera and blood cell pellets were collected and frozen at −70°C weekly during the first 4 weeks and every 2 weeks thereafter. Viral loads in PBMC specimens were assessed periodically by duplicate plating of 106 PBMC and serial threefold dilutions of PBMC on OMK cells in 24-well tissue culture plates. Selected culture supernatants from the PBMC viral load plates and selected sera were also tested for SEAP expression from recombinant HVS. Animals that became moribund were euthanized and received complete necropsies. Tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin.
Antibody responses against HVS virion proteins were assessed by enzyme-linked immunosorbent assay (ELISA) to purified lysed whole virus. Purified HVS was prepared from OMK cell lysates. Cell debris was removed by low-speed centrifugation and filtration through 0.45-μm-pore-size filters. Virus was then pelleted at 40,000 × g and resuspended in 1 ml of a solution containing 20 mM Tris, 100 mM NaCl, and 1 mM EDTA. Virus was then further purified by passage through a 10-ml Sepharose 4B column (10). Virion particles were collected in the void volume. Binding of antibodies in sera from infected marmosets to HVS was assayed on plates coated with 10 to 20 μg of purified HVS per 96-well plate. Antibodies to HVS in diluted sera were detected by using alkaline phosphatase-conjugated anti-human immunoglobulin G and measuring absorbance at 410 nm (10).
RESULTS
Isolation of STP-C488 and Tip deletion mutants.
HVS recombinants containing deletions in STP-C488 or Tip were generated by replacing selected portions of a 3.6-kb cloned HVS DNA fragment with a SEAP expression cassette driven by the SV40 early promoter. A 508-bp SpeI/EcoRV fragment including 91% of the STP-C488 gene was replaced with the SEAP expression cassette in the ΔSTP-C488 recombinant, while a 442-bp deletion containing more than half of the tip gene was replaced in the ΔTip virus (Fig. 1). Recombinants expressing SEAP were isolated by repeated limiting dilution passage in OMK cells.
To eliminate any possible inadvertent influence of the reporter in transformation assays, AscI restriction enzyme sites flanking the SEAP expression cassette were used to remove the reporter directly from the recombinant virion DNA. Transfection of ligated virion DNA into OMK cells produced SEAP-negative HVS ΔSTP and ΔTip recombinants with the specific gene deletions, which were again purified by repeated passage of limiting dilution of virus. Marker rescued viruses were also generated by recombination between virion DNA from the SEAP-expressing deletion mutants and the 3.6 kb of left-end L-DNA containing authentic STP-C488 and tip genes. Marker-rescued viruses were isolated by screening for SEAP-negative virus and confirming restoration of the deleted regions by PCR and DNA sequencing.
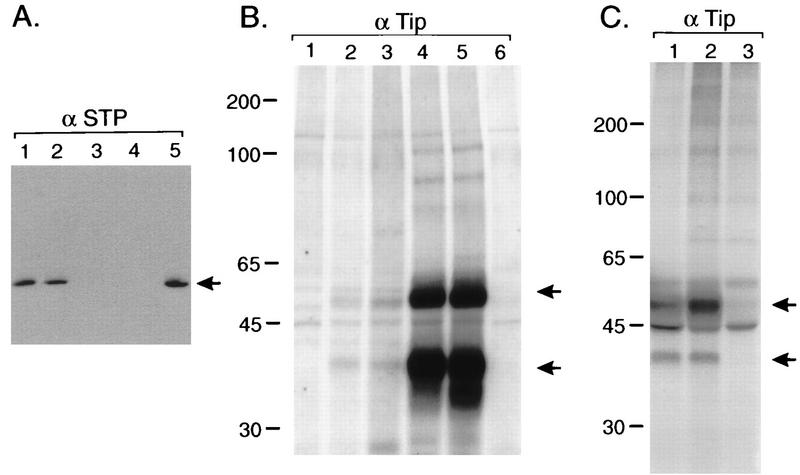
After construction of HVS deletion recombinants, STP-C488 expression was examined by immunoblot analysis. OMK cells were infected with each HVS deletion recombinant, and cell lysates were used for immunoblots with anti-STP-C488 antibody (16). The results revealed that STP was not expressed detectably in cells infected with the STP deletion mutants but was detected in HVS wild type (wt) and ΔTip/SV40-SEAP (Fig. 2A). When the same approach was used to examine Tip expression in deletion viruses, however, we were unable to detect the Tip expression with any of the viruses tested, including wt virus. This was likely caused by the low expression of Tip in HVS.
FIG. 2.
Expression of STP-C488 and Tip of HVS C488 recombinants. (A) Immunoblot with anti-STP (α STP) polyclonal antibody. Lysates of OMK cells infected with HVS C488 (wt) (lane 1), HVS ΔTip/SV40-SEAP (lane 2), HVS ΔSTP (lane 3), HVS ΔSTP/SV40-SEAP (lane 4), and HVS (STP restored) (lane 5) were used for immunoblot analysis. Arrow indicates STP-C488. (B) In vitro kinase reactions of anti-Tip (α Tip) immune complexes from transfected COS-1 cells. The C488PX, C488PXΔSTP, C488PXΔSTP/SV40-SEAP, and C488PXΔTip/SV40-SEAP constructs were cotransfected into COS-1 cells together with an Lck expression plasmid; 48 h after transfection, cell lysates were used for immunoprecipitation with an anti-Tip antibody, followed by in vitro kinase reaction. Lane 1, C488PXΔSTP/SV40-SEAP; lanes 2 and 3, C488PX; lanes 4 and 5, C488PXΔSTP; lane 6, C488PXΔTip/SV40-SEAP. Arrows indicate Lck (top) and Tip (bottom). (C) In vitro kinase reactions of anti-Tip immune complexes. Anti-Tip immune complexes from 2 × 107 primary common marmoset T cells immortalized with HVS C488 (wt) (lane 1) or HVS (Tip restored) (lane 2) and uninfected common marmoset PBMC (lane 3) were subjected to in vitro kinase reaction with [γ32P]ATP. Arrows indicate Lck (top) and Tip (bottom). In all panels, sizes are indicated in kilodaltons.
To demonstrate expression of Tip, we transfected COS-1 cells with the deletion plasmid constructs derived from a 3.6-kb cloned HVS DNA fragment (C488PX) containing wt STP and Tip (10). The C488PX, C488PXΔSTP, C488PXΔSTP/SV40-SEAP, and C488PXΔTip/SV40-SEAP constructs were cotransfected into COS-1 cells together with an Lck expression plasmid. At 48 h after transfection, cell lysates were used for immunoprecipitation with anti-Tip antibody, followed by in vitro kinase reaction. The results revealed that Tip was not expressed in C488PXΔSTP/SV40-SEAP and C488PXΔTip/SV40-SEAP constructs but was expressed in C488PX and C488PXΔSTP constructs (Fig. 2B). Thus, insertion of the SEAP reporter expression cassette at the STP locus appeared to block Tip expression. This result is not totally surprising since the tip gene is downstream of the STP gene in a bicistronic transcriptional unit (Fig. 1). Also, expression of the truncated form of Tip from the C488PXΔTip/SV40-SEAP construct was not detectable because of the lack of its association with Lck in this assay. Nonetheless, Tip was expressed well by the C488PXΔSTP construct. In fact, repeated experiments showed that Tip expression was dramatically higher with the C488PXΔSTP construct than with wt C488PX (Fig. 2B). In contrast, neither deletion of Tip nor insertion of the reporter cassette at the Tip locus affected STP expression (Fig. 2A).
In vitro T-cell immortalization.
Common marmoset T lymphocytes are immortalized efficiently to interleukin-2-independent growth by infection with HVS C488 (10, 34). This in vitro immortalization assay was therefore used to test the transforming activity of HVS recombinants. As shown in Table 1, deletion of STP-C488 or Tip resulted in loss of the ability to transform common marmoset T lymphocytes in vitro.
TABLE 1.
In vitro immortalization of common marmoset primary T cells
| Virus | No. immortalized/no. of attempts |
|---|---|
| None | 0/19 |
| HVS C488 | 23/24 |
| HVS ΔTip | 0/9 |
| HVS (Tip restored) | 15/15 |
| HVS ΔSTP-C488 | 0/14 |
| HVS (STP restored) | 12/12 |
To confirm that the loss of transforming activity of deletion virus was derived from the specific gene deletion and not from other unexpected alterations, the STP-C488 or tip gene was restored by homologous recombination of the respective deletion virus with a linearized cloned viral DNA fragment containing the corresponding wt gene. Restoration of STP-C488 or tip genes fully reconstituted the transforming ability of these viruses. Both HVS recombinants with a restored STP-C488 gene or a restored tip gene transformed primary T lymphocytes as efficiently as the wt HVS (Table 1). In addition, we examined the expression of restored STP-C488 by immunoblot analysis using OMK cells infected with the restored HVS. STP was detected at the same level in restored HVS as in the wt HVS C488 (Fig. 2A, lane 5). Also, Tip expression was examined in common marmoset T cells transformed with Tip-restored HVS by in vitro kinase reaction of anti-Tip immune complexes. The 40-kDa phosphorylated Tip along with phosphorylated 56-kDa cellular Lck were detected equally in HVS C488-transformed cells and Tip-restored HVS-transformed cells (Fig. 2C). Thus, the loss of transforming activity in the deletion mutant viruses was attributed solely to the specific STP-C488 or tip gene deletion and not to any inadvertently selected alterations.
In vivo lymphoma induction.
Experimental infections of common marmosets with virus deleted of either the STP-C488 or tip gene demonstrated that each of these genes was essential for induction of lymphoma in vivo. The two marmosets infected with wt HVS C488 died on days 19 and 20 after inoculation. However, animals infected with the STP-C488 and/or Tip-deleted recombinants (both with and without the SEAP reporter) remained healthy for the 12 months of observation after experimental infection (Table 2). Animals infected with wt HVS C488 developed fulminant multicentric lymphoma consistent with the disease as described previously (7). Organs involved included kidney, liver, eye, lung, adrenal gland, spleen, thymus, and lymph nodes, with the most severe infiltrates being observed in the kidneys. Isolation of virus from PBMC of all animals at 3 weeks and beyond verified that all animals were infected. Thus, the STP-C488 and tip genes are required not only for T-lymphocyte immortalization in vitro but also for lymphoma induction in vivo.
TABLE 2.
Experimental infection of common marmosets
| Virusa | No. of lymphomas inducted/infected marmosets | Survival (days) |
|---|---|---|
| HVS C488 (wt) | 2/2 | 19–20 |
| HVS ΔTip | 0/2 | >370 |
| HVS ΔTip/SV40-SEAP | 0/2 | >370 |
| HVS ΔSTP | 0/2 | >370 |
| HVS ΔSTP/SV40-SEAP | 0/2 | >370 |
In all cases, virus was recovered from both marmosets infected.
Persistent infection by attenuated HVS deletion viruses in vivo.
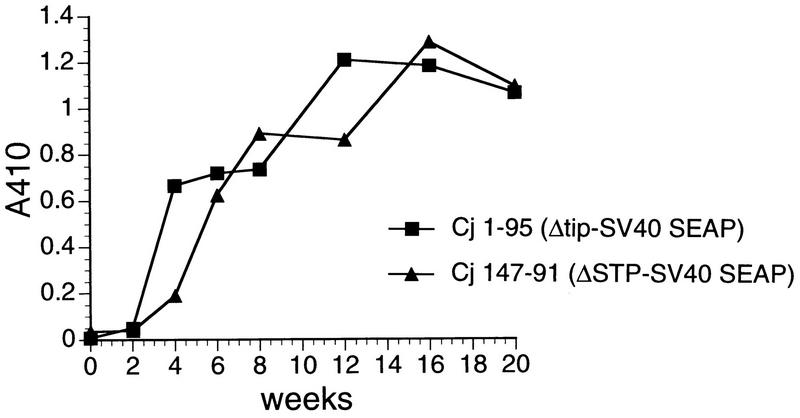
Antibody responses against HVS were measured by ELISA using plates coated with purified HVS virion proteins. The four marmosets infected with the HVS ΔSTP/SV40-SEAP or HVS ΔTip/SV40-SEAP recombinant showed strong antibody responses to HVS structural antigens. These antibodies persisted at high levels for the entire period of measurement, 20 weeks of infection (Fig. 3).
FIG. 3.
In vivo antibody responses against HVS virion proteins. Adsorption of antibodies to HVS in sera from infected marmosets was assay by ELISA using plates coated with 10 to 20 μg of purified HVS per plate. Alkaline phosphatase-conjugated anti-human immunoglobulin G was used to detect antibodies to HVS in diluted sera.
We have also developed procedures for evaluating virus load in vivo by measuring the numbers of PBMC required to isolate HVS. A similar method has been described previously for quantitating virus load of simian immunodeficiency virus (23). This assay measures the numbers of infectious cells in PBMC. Virus was readily recovered from PBMC of the animals infected with the deletion viruses for at least 5 months (Fig. 4). While both viruses were readily recovered on repeated occasions, the numbers of infectious cells were consistently higher with the ΔTip virus than the ΔSTP virus whether or not the SEAP gene was present (Fig. 4). Additionally, wt virus-infected marmosets showed a higher level of virus load at week 2 than deletion virus-infected marmosets (Fig. 4).
FIG. 4.
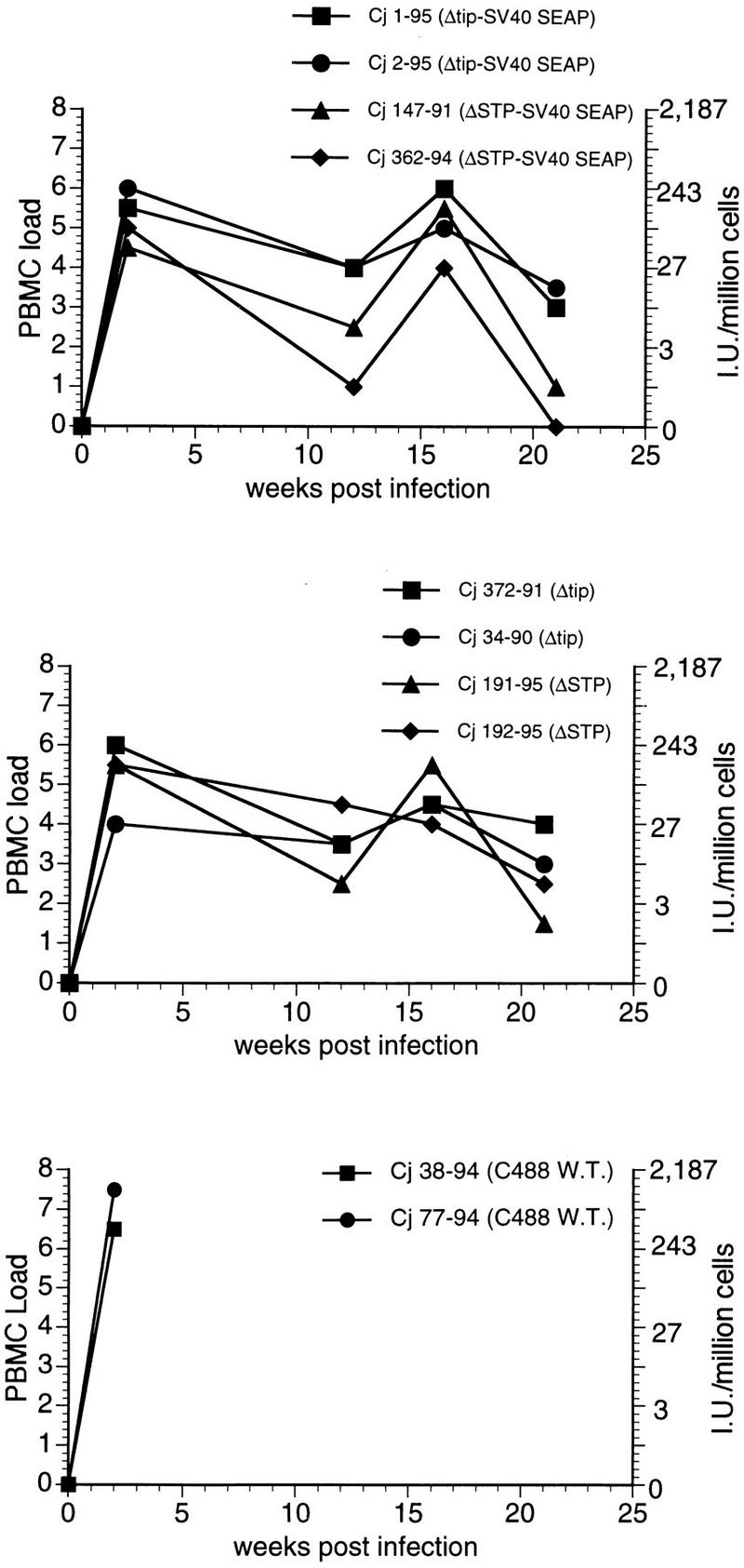
Cell-associated viral loads measured in common marmoset PBMC collected following experimental infection with HVS C488 recombinants. Virus loads on the y axis at the left indicate number of PBMC required to recover HVS, coded as follows: 0 = <106 (i.e., virus was not recovered); 1 = 106; 2 = 333,333; 3 = 111,111; 4 = 37,037; 5 = 12,345; 6 = 4,115; 7 = 1,371; and 8 = 457. On the y axis at the right, the virus loads are expressed as the number of cells per 106 PBMC that yielded HVS.
Assay of SEAP in culture supernatant samples from the viral load determinations at week 21 showed close correlation between detection of SEAP and detection of virus (data not shown). Production of virus capable of SEAP expression through week 21 further indicates persistence of the attenuated viruses in vivo and stability of the SV40-SEAP expression cassette during multiple viral replication cycles in vivo.
Plasma from animals infected with HVS ΔSTP/SV40-SEAP or HVS ΔTip/SV40-SEAP was used to measure the SEAP activity directly. SEAP production was sufficiently strong in the initial weeks after infection to be detected through week 4 (Table 3). Animals infected with the ΔTip/SV40-SEAP virus had consistently higher levels of SEAP activity in plasma during this period than animals infected with ΔSTP/SV40-SEAP (Table 3), consistent with the cell-associated viral load measured as described above.
TABLE 3.
SEAP detection in sera from common marmosets infected with HVS recombinants
| Wk | SEAP activity in sera of infected common marmosetsa
|
|||
|---|---|---|---|---|
| Cj1-95b | Cj2-95b | Cj147-91c | Cj362-94c | |
| 1 | 1,205 | 276 | 0 | 31 |
| 2 | 1,634 | 667 | 4 | 18 |
| 3 | 6,307 | 1,036 | 254 | 0 |
| 4 | 3,322 | 1,377 | 262 | 0 |
| 6 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 |
Expressed in chemiluminescent units; average of triplicate measurements.
Common marmoset infected with HVS ΔTip/SV40-SEAP.
Common marmoset infected with HVS ΔSTP/SV40-SEAP.
DISCUSSION
The results described here demonstrate that the STP-C488 and tip genes of HVS are required for T-lymphocyte transformation in vitro and lymphoma induction in vivo. Our ability to isolate recombinant viruses with deletions in each of the genes in vitro and to demonstrate persistence in vivo indicates that both genes are nonessential for replication and persistence. These data are consistent with previous studies demonstrating that sequences nonessential for replication but necessary for transforming capacity are present at the left end of HVS subgroup A genomes (7, 8, 10, 33). However, subgroup A and subgroup C HVS are distinguished by marked divergence in the composition of the leftmost open reading frames and by differences in ability to transform T cells from different species (4). Subgroup C viruses, unlike those of subgroup A, are capable of immortalizing human, rhesus monkey, and rabbit T lymphocytes (1, 3, 29).
Both STP-A11 and STP-C488 are sufficient to transform rodent fibroblasts in cell culture assays (16, 22) and are oncogenic in transgenic mice (25, 32). The extensive sequence divergence of these genes (4, 16, 22) and interaction of their products with distinct cellular targets (19, 26) emphasize the need to independently assess their contributions to oncogenesis through in vivo studies. STP-A11, which interacts with cellular Src, was previously shown to be essential for oncogenesis (33). While the corresponding gene from a different strain of HVS subgroup C has been shown to be necessary for short-term proliferation of T cells (28), STP-C488, which has been shown to associate with cellular Ras (19), has not been assessed previously. The lack of oncogenicity of ΔSTP-C488 deletion virus along with the transforming activity of STP-C488 indicates that STP-C488 is necessary for HVS C488 transformation of marmoset T lymphocytes.
Previously, a divergent form of Tip from HVS subgroup C strain 484 has been reported to be required for in vitro T-cell proliferation induced by HVS (27). However, this earlier report did not examine the transforming activity of HVS ΔTip immortalization of primary T cells and in lymphoma induction in New World primates. The data reported here show that deletion of Tip in HVS C488 renders the recombinant virus incapable of transformation in vitro and in vivo. The role of association between Tip and Lck has been controversial, with both activation (27, 35) and downregulation (15, 21) of Lck-mediated signal transduction being reported. Analysis of HVS with point mutations in the SH3-binding motif of Tip has recently demonstrated that interaction of Tip with Lck is not required for in vitro and in vivo transforming activity of HVS (12). A novel cellular partner of Tip, Tap, which interacts with Tip independently of the Tip-Lck association has recently been described (36). The interaction of Tip with Tap dramatically upregulates NF-κB transcriptional activity and induces the surface expression of lymphocyte adhesion molecules. This finding suggests that study of the interaction of Tip with Tap will be particularly important in defining roles of Tip in HVS oncogenesis.
In addition to insight into the roles of STP-C488 and Tip for viral oncogenesis, we report here new procedures to assess quantitatively the persistence in vivo of attenuated HVS deletion mutants. While the requirement of STP-C488 and Tip for transforming activity of HVS is now well documented, further detailed study will be required to define the mechanisms utilized by STP and Tip for achieving HVS transformation and for maintenance of high numbers of infected T cells in vivo in natural or nonnatural hosts.
ACKNOWLEDGMENTS
We thank J. Newton, T. Connors, and A. Hampson for manuscript preparation.
This work was supported by Public Health Service grants CA31363 and AI38131 and grant RR00168 from the Division of Research Resources.
REFERENCES
- 1.Alexander L, Du Z, Rosenzweig M, Jung J U, Desrosiers R C. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 nef alleles in lymphocyte activation. J Virol. 1997;71:6094–6099. doi: 10.1128/jvi.71.8.6094-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berend K R, Jung J U, Boyle T J, DiMaio J M, Mungal S A, Desrosiers R C, Lyerly H K. Phenotypic and functional consequences of herpesvirus saimiri infection of human CD8+ cytotoxic T lymphocytes. J Virol. 1993;67:6317–6321. doi: 10.1128/jvi.67.10.6317-6321.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biesinger B, Müller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biesinger B, Trimble J J, Desrosiers R C, Fleckenstein B. The divergence between two oncogenic herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology. 1990;176:505–514. doi: 10.1016/0042-6822(90)90020-r. [DOI] [PubMed] [Google Scholar]
- 5.Biesinger B, Tsygankov A Y, Fickenscher H, Emmrich R, Fleckenstein B, Bolen J B, Broker B M. The product of the herpesvirus saimiri ORF1 (Tip) interacts with T cell specific kinase p56lck in transformed cells. J Biol Chem. 1995;270:4729–4734. doi: 10.1074/jbc.270.9.4729. [DOI] [PubMed] [Google Scholar]
- 6.Bröker B M, Tsygankov A Y, Müller-Fleckenstein I, Guse A H, Chitaev N A, Biesinger B, Fleckenstein B, Emmrich F. Immortalization of human T cell clones by Herpesvirus saimiri. J Immunol. 1993;151:1184–1192. [PubMed] [Google Scholar]
- 7.Desrosiers R C, Bakker A, Kamine J, Falk L A, Hunt R D, King N W. A region of the Herpesvirus saimiri genome required for oncogenicity. Science. 1985;228:184–187. doi: 10.1126/science.2983431. [DOI] [PubMed] [Google Scholar]
- 8.Desrosiers R C, Burghoff R L, Bakker A, Kamine J. Construction of replication-competent herpesvirus saimiri deletion mutants. J Virol. 1984;49:343–348. doi: 10.1128/jvi.49.2.343-348.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desrosiers R C, Falk L A. Herpesvirus saimiri strain variability. J Virol. 1982;43:352–356. doi: 10.1128/jvi.43.1.352-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desrosiers R C, Silva D, Waldron L M, Letvin N L. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J Virol. 1986;57:701–705. doi: 10.1128/jvi.57.2.701-705.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duboise S M, Guo J, Desrosiers R C, Jung J U. Use of virion DNA as a cloning vector for the construction of mutant and recombinant herpesviruses. Proc Natl Acad Sci USA. 1996;93:11389–11394. doi: 10.1073/pnas.93.21.11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duboise, S. M., J. Guo, H. Lee, S. Czajak, M. Simon, M. Rosenzweig, R. C. Desrosiers, and J. U. Jung. 1997. Unpublished data.
- 13.Falk L, Wolfe L, Deinhardt F. Isolation of herpesvirus saimiri from blood of squirrel monkeys (saimiri sciureus) J Natl Cancer Inst. 1972;48:1499–1505. [PubMed] [Google Scholar]
- 14.Fleckenstein B, Desrosiers R C. Herpesvirus saimiri and herpesvirus ateles. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Publishing Corporation; 1982. pp. 253–332. [Google Scholar]
- 15.Guo J, Duboise S M, Lee H, Li M, Choi J-K, Rosenzweig M, Jung J U. Enhanced downregulation of Lck-mediated signal transduction by a Y114 mutation of herpesvirus saimiri Tip. J Virol. 1997;71:7092–7096. doi: 10.1128/jvi.71.9.7092-7096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung J U, Desrosiers R C. Identification and characterization of the herpesvirus saimiri oncoprotein, STP-C488. J Virol. 1991;65:6953–6960. doi: 10.1128/jvi.65.12.6953-6960.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung J U, Desrosiers R C. Herpesvirus saimiri and ateles. In: Webster R, Granoff A, editors. Encyclopedia of virology. Philadelphia, Pa: Saunders Scientific Publications, Inc.; 1994. pp. 614–622. [Google Scholar]
- 18.Jung J U, Desrosiers R C. Distinct functional domains of STP-C488 of Herpesvirus saimiri. Virology. 1994;204:751–758. doi: 10.1006/viro.1994.1590. [DOI] [PubMed] [Google Scholar]
- 19.Jung J U, Desrosiers R C. Association of the viral oncoprotein STP-C488 with cellular ras. Mol Cell Biol. 1995;15:6506–6512. doi: 10.1128/mcb.15.12.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung J U, Lang S M, Friedrich U, Jun T, Roberts T M, Desrosiers R C, Biesinger B. Identification of lck-binding elements in Tip of Herpesvirus saimiri. J Biol Chem. 1995;270:20660–20667. doi: 10.1074/jbc.270.35.20660. [DOI] [PubMed] [Google Scholar]
- 21.Jung J U, Lang S M, Jun T, Roberts T M, Veillette A, Desrosiers R C. Downregulation of Lck-mediated signal transduction by tip of herpesvirus saimiri. J Virol. 1995;69:7814–7822. doi: 10.1128/jvi.69.12.7814-7822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung J U, Trimble J J, King N W, Biesinger B, Fleckenstein B W, Desrosiers R C. Identification of transforming genes of subgroup A and C strains of herpesvirus saimiri. Proc Natl Acad Sci USA. 1991;88:7051–7055. doi: 10.1073/pnas.88.16.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kestler H W I, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1992;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 24.Koomey J M, Mulder C, Burghoff R L, Fleckenstein B, Desrosiers R C. Deletion of DNA sequences in a nononcogenic variant of herpesvirus saimiri. J Virol. 1984;50:662–665. doi: 10.1128/jvi.50.2.662-665.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kretschmer C, Murphy C, Biesinger B, Beckers J, Fickenscher H, Kirchner T, Fleckenstein B, Rüther U. A Herpes saimiri oncogene causing peripheral T-cell lymphoma in transgenic mice. Oncogene. 1996;12:1609–1616. [PubMed] [Google Scholar]
- 26.Lee H, Trimble J J, Yoon D-W, Regier D, Desrosiers R C, Jung J U. Genetic variation of herpesvirus saimiri subgroup A transforming protein and its association with cellular src. J Virol. 1997;71:3817–3825. doi: 10.1128/jvi.71.5.3817-3825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lund T, Medveczky M, Medveczky P. Herpesvirus saimiri Tip-484 membrane protein markedly increases p56lck activity in T cells. J Virol. 1997;71:378–382. doi: 10.1128/jvi.71.1.378-382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medveczky M M, Geck P, Sullivan J L, Srbousek D, Djeu J, Medveczky P G. IL-2 independent growth and cytotoxicity of herpesvirus saimiri-infected human CD8 cells and involvement of two open reading frame sequences of the virus. Virology. 1993;196:402–412. doi: 10.1006/viro.1993.1495. [DOI] [PubMed] [Google Scholar]
- 29.Medveczky M M, Szomolanyi E, Hesselton R, DeGrand D, Geck P, Medveczky P G. Herpesvirus saimiri strains from three DNA subgroups have different oncogenic potentials in New Zealand White rabbits. J Virol. 1989;63:3601–3611. doi: 10.1128/jvi.63.9.3601-3611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medveczky P, Szomolayi E, Desrosiers R C, Mulder C. Classification of herpesvirus saimiri into three groups based on extreme variation in a DNA region required for oncogenicity. J Virol. 1984;52:938–944. doi: 10.1128/jvi.52.3.938-944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mittrücker H-W, Müller-Fleckenstein I, Fleckenstein B, Fleishcher B. CD2-mediated autocrine growth of herpes virus saimiri-transformed human T lymphocytes. J Exp Med. 1995;176:900–913. doi: 10.1084/jem.176.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy C, Kretschmer C, Biesinger B, Beckers J, Jung J, Desrosiers R C, Müller-Hermelink H K, Fleckenstein B W, Rüther U. Epithelial tumors induced by a herpesvirus oncogene in transgenic mice. Oncogene. 1994;9:221–226. [PubMed] [Google Scholar]
- 33.Murthy S C S, Trimble J J, Desrosiers R C. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J Virol. 1989;63:3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szomolanyi E, Medveczky P, Mulder C. In vitro immortalization of marmoset cells with three subgroups of herpesvirus saimiri. J Virol. 1987;61:3485–3490. doi: 10.1128/jvi.61.11.3485-3490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiese N, Tsygankov A Y, Klauenberg U, Bolen J B, Fleischer B, Broker B M. Selective activation of T cell kinase p56lck by herpesvirus saimiri protein tip. J Biol Chem. 1997;271:847–852. doi: 10.1074/jbc.271.2.847. [DOI] [PubMed] [Google Scholar]
- 36.Yoon D-W, Lee H, Seol W, DeMaria M, Rosenzweig M, Jung J U. Tap; a novel cellular protein that interacts with tip of Herpesvirus saimiri and induces lymphocyte aggregation. Immunity. 1997;6:571–582. doi: 10.1016/s1074-7613(00)80345-3. [DOI] [PubMed] [Google Scholar]