Abstract
Retroviruses and their relatives, the LTR-retrotransposons, possess an integrase protein (IN) that is required for the insertion of reverse transcripts into the genome of host cells. Schizosaccharomyces pombe is the host of Tf1, an LTR-retrotransposon with integration activity that can be studied by using techniques of yeast genetics. In this study, we sought to identify amino acid substitutions in Tf1 that specifically affected the integration step of transposition. In addition to seeking amino acid substitutions in IN, we also explored the possibility that other Tf1 proteins contributed to integration. By comparing the results of genetic assays that monitored both transposition and reverse transcription, we were able to seek point mutations throughout Tf1 that blocked transposition but not the synthesis of reverse transcripts. These mutant versions of Tf1 were candidates of elements that possessed defects in the integration step of transposition. Five mutations in Tf1 that resulted in low levels of integration were found to be located in the IN protein: two substitutions in the N-terminal Zn domain, two in the catalytic core, and one in the C-terminal domain. These results suggested that each of the three IN domains was required for Tf1 transposition. The potential role of these five amino acid residues in the function of IN is discussed. Two of the mutations that reduced integration mapped to the RNase H (RH) domain of Tf1 reverse transcriptase. The Tf1 elements with the RH mutations produced high levels of reverse transcripts, as determined by recombination and DNA blot analysis. These results indicated that the RH of Tf1 possesses a function critical for transposition that is independent of the accumulation of reverse transcripts.
The reverse transcription of RNA encoded by retroviruses and retrotransposons generates DNA sequences that are inserted into the genomes of host cells. Although the retrotransposons that lack long terminal repeats (LTRs) simply prime cDNA synthesis, from breaks at insertion sites (34, 54), retroviruses and their relatives, the LTR-retrotransposons, possess an integrase protein (IN) that inserts the cDNA into the host genome after the bulk of reverse transcription is complete. The IN proteins of retroviruses and some LTR-retrotransposons must process the blunt ends of the reverse transcripts before integration by cleaving two nucleotides from the 3′ termini (18, 24, 44, 53). Some LTR-retrotransposons such as Ty1 produce reverse transcripts with blunt ends that do not require 3′ processing because the nucleotides that are joined to the target already exist as 3′-terminal residues (14, 39). Ultimately, the integration of the reverse transcript into the host genome is a concerted reaction that includes the cleavage of the insertion site and the joining of the 3′ termini of the cDNA to the 5′ ends of the cleaved target (3, 4, 18).
The IN proteins of human immunodeficiency virus (HIV), Moloney murine leukemia virus, Rous sarcoma virus, and Ty1 have been purified and were found in in vitro reactions with model substrates to be sufficient for strand transfer activity (6, 9, 23, 38). Although these results indicate that IN proteins possess the catalytic properties required for integration, the relative inefficiencies of the reactions, and in the case of HIV and Ty1, the low levels of two-ended insertions, suggested that other factors contribute to strand transfer in vivo (6, 38).
Further biochemical analysis of IN proteins has focused on the function of three essential domains. The IN proteins of retroviruses and LTR-retrotransposons contain the amino acid sequence motif HX3-7HX23-32CX2C (HHCC) near the amino terminus (11, 21, 25). The HHCC motif is reminiscent of the zinc finger and has been found to bind Zn in the case of HIV IN (7). A central region of the IN proteins is called the catalytic core domain and contains the motif DX39-58DX35E (17, 26, 45). This motif contains amino acid residues that are critical for catalytic activity (7, 12, 15, 26, 52). The C-terminal domain of IN proteins possess DNA binding activity that has equal affinity for both viral and nonspecific double-stranded DNA (35, 41, 46, 51). No motifs that are conserved among LTR-retroelements have been identified in the C-terminal domains of the IN proteins.
Schizosaccharomyces pombe is the host of Tf1, an LTR-retrotransposon that possesses integration activity in vivo. The transposition of Tf1 can be readily studied with techniques of yeast genetics (30, 32). Retrotransposons serve as useful retrovirus model systems because they are closely related to retroviruses and use many of the same proteins and mechanisms to replicate. Tf1 encodes Gag, protease (PR), reverse transcriptase (RT), and IN proteins that function similarly to the retroviral counterparts (1, 30, 32). The IN protein of Tf1 contains a Zn domain, a D,D35E motif, and a C-terminal domain that has not yet been tested for nonspecific DNA binding activity. The transposition activity of Tf1 can be measured in a strain of S. pombe that contains a plasmid copy of Tf1 fused to the inducible promoter from the nmt1 gene (30). Results from this in vivo assay demonstrate that Tf1 transposition requires the IN protein (28). We describe here modifications of the transposition assay that allowed us to identify Tf1 elements with point mutations that significantly reduced transposition without causing defects in reverse transcription. These mutant versions of Tf1 were candidates of elements that possessed defects in the integration step of transposition. Five mutations in Tf1 that caused low levels of integration activity in vivo were found to be substitutions in the IN protein. DNA blot analysis indicated that these mutations affected integration at a step after reverse transcription, since each of these strains produced wild-type levels of reverse transcripts. Two of the mutations were in the N-terminal domain, two were in the catalytic core, and one was in the C-terminal domain of IN. These results suggested that each of the three IN domains was required for Tf1 transposition. Because we randomly mutagenized the entire transposon, we were also able to identify two mutations in the RNase H (RH) domain of RT that resulted in low levels of integration. The Tf1 elements with the RH mutations produced high levels of reverse transcripts as determined by recombination assays and DNA blots. These results suggested that the mutations in RH blocked transposition during integration.
MATERIALS AND METHODS
Media.
The S. pombe minimal liquid and plate media were composed of Edinburgh minimal medium (EMM) (40). Selective plates contained EMM and dropout mix (2 g/liter), a powder with adenine and all amino acids (43). Each component of the dropout powder was present in equal gram quantities except for adenine, which was supplemented to 2.5 times the amount of the other nutrients. Ten micromolar vitamin B1 (thiamine) was added to EMM when indicated to repress the nmt1 promoter. The rich medium, YES, contained 5 g of yeast extract (Difco), 30 g of glucose, and 2 g of complete dropout mix per liter. YES–5-fluoroorotic acid (FOA; United States Biologicals, Swampscott, Mass.) plates were made by adding FOA (1 g/liter) to EMM supplemented with uracil (100 μg/ml). YES-FOA-G418 plates contained the components of YES plus 1 g of FOA and 500 mg (corrected for purity) of Geneticin (Gibco) per liter.
Strains and plasmids.
The yeast strains used in this study are listed in Table 1. All yeast strains were derived from YHL912. Each line in Table 1 represents a strain that differed from the others only with respect to the version of the Tf1 expression plasmid that it contained. Plasmid pHL449-1 contained the neoAI-marked version of Tf1 (Tf1-neoAI) that was used in the transposition and homologous recombination assays. The structure of pHL449-1 was identical to that of pHL414-2 (31) except that an artificial intron with a SpeI site (5′GTAGGTGCTATTTTACTAGTCTAAGCTAATCAATAG3′) was inserted into the NruI site of the neo gene such that this sequence was in the coding strand of the Tf1 transcript and the noncoding strand of the neo transcript. The intron inserted into neo was modeled after a closely related intron that showed splicing efficiency in S. pombe of 95% (19).
TABLE 1.
Yeast strains used
| Straina (reference) | Plasmid | Plasmid description |
|---|---|---|
| YHL1282 (28) | pHL449-1 | Wild-type Tf1-neoAI |
| YHL1554 (28) | pHL476-3 | Tf1-neoAI with IN frameshift |
| YHL1836 (1) | pHL490-80 | Tf1-neoAI with PR frameshift |
| YHL1887 | pHL449-1b | Tf1 expression plasmid that contained mutation 1 (A1311T) |
| YHL1888 | pHL449-1b | Tf1 expression plasmid that contained mutation 2 (P816S) |
| YHL1890 | pHL449-1b | Tf1 expression plasmid that contained mutation 3 (S1009L) |
| YHL1891 | pHL449-1b | Tf1 expression plasmid that contained mutation 4 (L929F) |
| YHL1895 | pHL449-1b | Tf1 expression plasmid that contained mutation 5 (S749L) |
| YHL1917 | pHL449-1b | Tf1 expression plasmid that contained mutation 9 (P903L) |
| YHL1826 | pHL449-1b | Tf1 expression plasmid that contained mutation 10 (E1142K) |
| YHL2053 | pHL495 | Tf1-neoAI plasmid with mutation 1 isolated from YHL1887 |
| YHL2057 | pHL496 | Tf1-neoAI plasmid with mutation 2 isolated from YHL1888 |
| YHL2061 | pHL498 | Tf1-neoAI plasmid with mutation 3 isolated from YHL1890 |
| YHL2065 | pHL499 | Tf1-neoAI plasmid with mutation 4 isolated from YHL1891 |
| YHL2069 | pHL502 | Tf1-neoAI plasmid with mutation 5 isolated from YHL1895 |
| YHL2085 | pHL519 | Tf1-neoAI plasmid with mutation 9 isolated from YHL1917 |
| YHL2089 | pHL575 | Tf1-neoAI plasmid with mutation 10 isolated from YHL1826 |
| YHL2165 | pHL680-1 | Tf1-neoAI plasmid with BsrGI-BamHI fragment from pHL495 |
| YHL2170 | pHL681-3 | Tf1-neoAI plasmid with BsrGI-BamHI fragment from pHL496 |
| YHL2174 | pHL682-3 | Tf1-neoAI plasmid with BsrGI-BamHI fragment from pHL498 |
| YHL2178 | pHL683-2 | Tf1-neoAI plasmid with BsrGI-BamHI fragment from pHL499 |
| YHL2182 | pHL684-2 | Tf1-neoAI plasmid with BsrGI-BamHI fragment from pHL502 |
| YHL2198 | pHL688-1 | Tf1-neoAI plasmid with BsrGI-BamHI fragment from pHL519 |
| YHL2202 | pHL689-2 | Tf1-neoAI plasmid with BsrGI-BamHI fragment from pHL575 |
| YHL2206 | pHL715-1 | Tf1-neoAI plasmid with AvrII-BsrGI fragment from pHL495 |
| YHL2230 | pHL731-1 | Tf1-neoAI plasmid with AvrII-BsrGI fragment from pHL496 |
| YHL2210 | pHL717-1 | Tf1-neoAI plasmid with AvrII-BsrGI fragment from pHL498 |
| YHL2214 | pHL718-1 | Tf1-neoAI plasmid with AvrII-BsrGI fragment from pHL499 |
| YHL2234 | pHL732-1 | Tf1-neoAI plasmid with AvrII-BsrGI fragment from pHL502 |
| YHL2222 | pHL723-1 | Tf1-neoAI plasmid with AvrII-BsrGI fragment from pHL519 |
| YHL2226 | pHL724-1 | Tf1-neoAI plasmid with AvrII-BsrGI fragment from pHL575 |
| YHL5282 | pHL1187-2 | PCR reconstruction of mutation 2 from plasmid pHL731-1 |
| YHL5274 | pHL1183-2 | PCR reconstruction of mutation 5 from plasmid pHL732-1 |
All plasmids were transformed into YHL912 (h− ura4-294 leu1-32) (2).
Contained hydroxylamine-generated mutation.
Hydroxylamine mutagenesis of Tf1-neoAI.
The plasmid that contained Tf1-neoAI, pHL449-1, was mutagenized in vitro by exposure to hydroxylamine with a protocol that was a modified version of a previously published description (48). A fresh solution of hydroxylamine was prepared just before use and stored on ice until needed. The hydroxylamine solution contained 0.35 g of hydroxylamine hydrochloride (Fluka) and 0.09 g of NaOH in 5 ml of water; 10 μg of purified pHL449-1 DNA (Qiagen kit; Qiagen Inc., Chatsworth, Calif.) was added to 0.5 ml of the hydroxylamine solution, and 0.1 ml was removed and left on ice as a 0-min time point. The remainder of the solution was incubated at 65°C, and 0.1-ml aliquots were placed on ice 15, 30, 60, and 90 min after the incubation was started. Each aliquot was then dialyzed on 0.025-μm pore-size filter discs (VSWP 025; Millipore, Bedford, Mass.) against 1 liter of TE (10 mM Tris base [pH 7.9], 1 mM EDTA) for 2 h. The treated DNA was transformed into Escherichia coli MH5 (trpC9830 pyrF::Tn5 galU galK hsdR strA lacΔX74; provided by M. Hall) by selecting for ampicillin-resistant (Ampr) colonies. The extent of mutagenesis could be evaluated for each time point because the URA3 gene in pHL449-1 complemented the pyrF::Tn5 deficiency in MH5. Approximately 2,000 Ampr colonies from each time point were replica printed to M9 plates that contained 0.2% Casamino Acids to identify the fraction of colonies that were auxotrophic for uracil. We chose to use the DNA from the 30-min sample because 0.38% of the colonies were auxotrophic for uracil and this low frequency of mutagenesis would result in few plasmids with double mutations. The URA3 gene is approximately 1.0 kb, and Tf1-neoAI is about 6.0 kb; therefore, we expected that approximately 2.3% of the Tf1 elements would show defects in transposition function. To amplify the population of DNA in the 30-min sample, we harvested 40,000 Ampr colonies and grew these cells for one doubling in liquid medium. Large quantities of plasmid DNA was isolated from these cells by using Qiagen Megacolumns. This DNA was then transformed into S. pombe cells by the lithium acetate procedure (40).
Isolation of plasmid DNA from S. pombe cells.
Plasmid DNA was isolated from S. pombe cells with a modified version of a previously published protocol (40). S. pombe cells were grown in 5 ml of EMM minus uracil medium until they reached an optical density at 600 nm (OD600) of 2.0. The cells were resuspended in 1.5 ml of 50 mM citrate-phosphate buffer (7.1 g of Na2HPO4 and 11.5 g of citric acid per liter [pH 5.6]) that contained 2 mg of Zymolase-20T (Seikagaku America, Rockville, Md.) per ml. After the cells were incubated at 37°C for 1 h, they were resuspended in 300 μl of TE, and 35 μl of 10% sodium dodecyl sulfate (SDS) was added. The mixture was incubated at 65°C for 5 min, and 100 μl of 5 M potassium acetate was mixed in thoroughly. After a 30-min incubation on ice, the cells were pelleted in a microcentrifuge that was spun at 17,000 × g and 4°C for 10 min. The supernatant was then mixed with 1.2 ml of binding buffer and 30 μl of matrix from a Prep-A-Gene kit (Bio-Rad Laboratories, Hercules, Calif.). The mixture was gently inverted for 10 min, and the matrix was pelleted and washed twice with 1 ml of wash buffer as described in the instructions provided with the kit. The DNA was eluted with 50 μl of TE, and 10 μl was transformed into bacteria to isolate plasmids encoding Ampr.
Subcloning of restriction fragments from mutant versions of the Tf1-neoAI plasmid.
Plasmids that were shown to carry mutations in Tf1 were Qiagen kit purified and digested with AvrII and BsrGI or with BsrGI and BamHI. The 2.3-kb AvrII-BsrGI products encoding the N terminus of IN and the 3.2-kb BsrGI-BamHI products encoding the majority of IN were gel purified from each defective plasmid so that they could be used to replace the same fragment from the wild-type Tf1-neoAI plasmid. The resulting plasmids were transformed into our wild-type expression strain, YHL912, and multiple transformants of each subclone were then analyzed by using our genetic assays to determine their transposition and recombination phenotypes. Subclones that exhibited high levels of recombination activity and showed defective transposition activities were sequenced throughout the length of the subcloned fragment. Sequencing was performed by using either a cycle protocol and a Applied Biosystems instrument or with Sequenase 2.0 (Amersham). The resulting sequence data were compared to the wild-type sequence of the Tf1 in pHL449-1 (32).
Reconstruction of single-base substitutions by fusion PCR.
PCR products termed fusion products that contained single-base substitutions within the middle of the fragment were templated by two half-fragments that had 30 bases of overlap positioned at the site of the mutation. Each of the half-fragments was also generated by PCR. The final fusion products contained the restriction sites AvrII and BsrGI at their termini so that they could be readily ligated into the vector fragment of pHL449-1 that resulted from an AvrII-BsrGI digest. The final plasmids were sequenced at the position of the mutation to verify the base substitution.
Transposition assay.
Strains containing a Tf1-neoAI plasmid were first grown as patches on agar plates that contained EMM with 10 μM thiamine to repress the nmt1 promoter and dropout powder minus uracil to select for the Tf1-neoAI plasmid. These patches were then replica printed to similar EMM agar plates that lacked thiamine to induce the nmt1 promoter. During this time, the artificial intron was spliced out of the Tf1 mRNA, and reverse transcription generated active copies of the neo gene. After 4 days of 32°C incubation, the plates were replica printed to EMM containing FOA to select against cells containing the Tf1-neoAI plasmid (2). These plates were then replica printed to YES medium containing G418 as well as FOA and incubated at 32°C for 2 days to determine the frequency at which Tf1-neo inserts into the genome (28, 30). Quantitative measurements of transposition frequencies were performed as follows. Strains were grown as patches of cells on EMM agar with minus uracil dropout powder for 4 days at 32°C. These cells were then resuspended to an OD600 of 1.0 and diluted approximately 100-fold. About 0.1 ml of the cells was then spread onto FOA plates, and the resultant colonies (about 7,000/plate) were printed to YES plates containing FOA and G418 (500 μg/ml) (31). The transposition frequency was the percentage of the FOAr colonies that were also G418r.
Homologous recombination assay.
The protocol is similar to the transposition assay in that strains that contained the neoAI-marked Tf1 plasmid were first grown as patches on agar plates that contained EMM (plus 10 μM thiamine and dropout powder) and then replica printed to similar EMM plates that lacked thiamine. After 4 days of 32°C incubation, the plates were replica printed directly to YES medium that contained G418 (500 μg/ml). Recombination between cDNA and cellular transposon sequences was scored on the G418 plates after 48 h of growth at 32°C. The quantitative version of this assay differs from the above protocol in that the patches from the plate lacking thiamine were resuspended to an OD600 of 1.0 and diluted serially before they were plated onto YES and YES-G418 plates. The recombination frequency was the percentage of the colonies on the YES plates that also grew on YES-G418 plates.
Protein preparations and immunoblots.
Crude protein extracts were made from 5-ml cultures grown without thiamine in EMM-minus-uracil dropout medium. The cells were washed first with water and then with extraction buffer consisting of 15 mM KCl, 10 mM HEPES-KOH (pH 7.8), and 5 mM EDTA. The cells were then resuspended in 0.4 ml of extraction buffer plus 5.0 mM dithiothreitol and 2.0 mM phenylmethylsulfonyl fluoride. Acid-washed glass beads 0.4 mm in diameter were added to the meniscus, and the sample was vortexed for 5 min; 0.1 ml of extraction buffer was mixed into the extract, and the liquid was removed. An equal volume of 2× sample buffer was added to the sample, and the mixture was boiled in preparation for SDS-gel electrophoresis.
The material for the immunoblots was loaded onto an SDS–10% polyacrylamide gel with equal amounts of total protein in each lane. Standard electrotransfer techniques were used (50) with Immobilon-P (Millipore) as the membrane. The detection method used was the ECL system as described by the manufacturer (Amersham) except that the secondary antibody, horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin, was used at a dilution of 10,000-fold. The primary polyclonal antisera used for each filter were from the production bleeds of rabbits 660 (anti-Gag) and 657 (anti-IN) (31).
cDNA preparations and blots.
About 109 cells (100 OD600 units) were resuspended in a 13- by 100-mm glass tube with 200 μl of EB (0.5 M NaCl, 0.2 M Tris-Cl [pH 7.5], 10 mM EDTA, 1% SDS), and 200 μl of PICA (1:1 mixture of EB equilibrated phenol and chloroform that contained 1/24 isoamyl alcohol) was added to enough acid-washed glass beads to fill past the miniscus. The mixture was vortexed vigorously for 30 min in a multivortexer (Baxter Scientific Products), and an additional 400 μl each of EB and PICA was added before a final 30-s vortex. The supernatant was reextracted with PICA two more times. The supernatant was ethanol precipitated, and the pellet was resuspended in 50 μl of TE; 5 μl of this mixture was restriction digested and subjected to agarose gel electrophoresis for DNA blot analysis. After transfer, the filters were hybridized with a 1.0-kb neo probe derived from a BamHI digest of pGH54 (22).
RESULTS
Recombination assay for measuring the level of Tf1 reverse transcripts in the nucleus.
Mutations in Tf1 that cause reduced transposition frequencies could affect any one of a number of processes that occur during particle formation, reverse transcription, or integration. Our interest in identifying sequences throughout the Tf1 transposon that contribute specifically to the integration process in vivo led us to apply a genetic assay for measuring the amount of Tf1 reverse transcripts present in the nucleus. The goal was to isolate a set of mutations in Tf1 that blocked transposition but nevertheless resulted in the accumulation of normal levels of cDNA in the nucleus. Mutations with these properties were candidates for substitutions that directly affected the integration process.
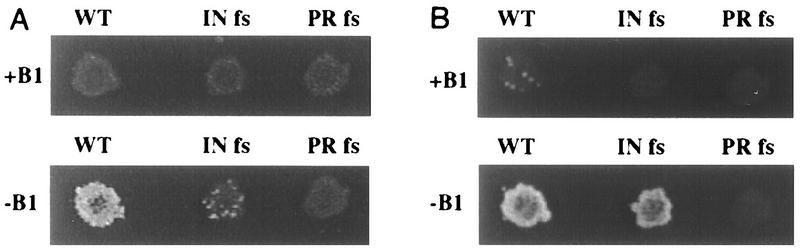
The assay used to measure the transposition activity of Tf1 elements has been previously described (28–30). The measure of transposition in S. pombe is based on the expression of Tf1 by an inducible nmt1 promoter from a multicopy vector (Fig. 1A). The plasmid copy of Tf1 also contained a bacterial neo gene that served as a marker for transposition. For reasons described below, this neo gene, neoAI, was disrupted by an artificial intron. Intact copies of neo resulted from the reverse transcription of Tf1-neoAI mRNA that had been spliced (Fig. 1B). Patches of cells that were induced for transcription of Tf1-neoAI were subsequently exposed to FOA to select for cells that no longer possessed the Tf1-neoAI plasmid. The presence of transposed copies of Tf1-neoAI was identified by replica printing the cells from the FOA medium to G418-containing medium. Figure 2A shows that patches of cells that contained wild-type Tf1-neoAI exhibit sufficient transposition to produce confluent growth on the G418 plate. A strain that contained a copy of Tf1-neoAI that lacked IN due to a frameshift exhibited transposition frequencies 18-fold below normal levels (Fig. 2A and Table 2).
FIG. 1.
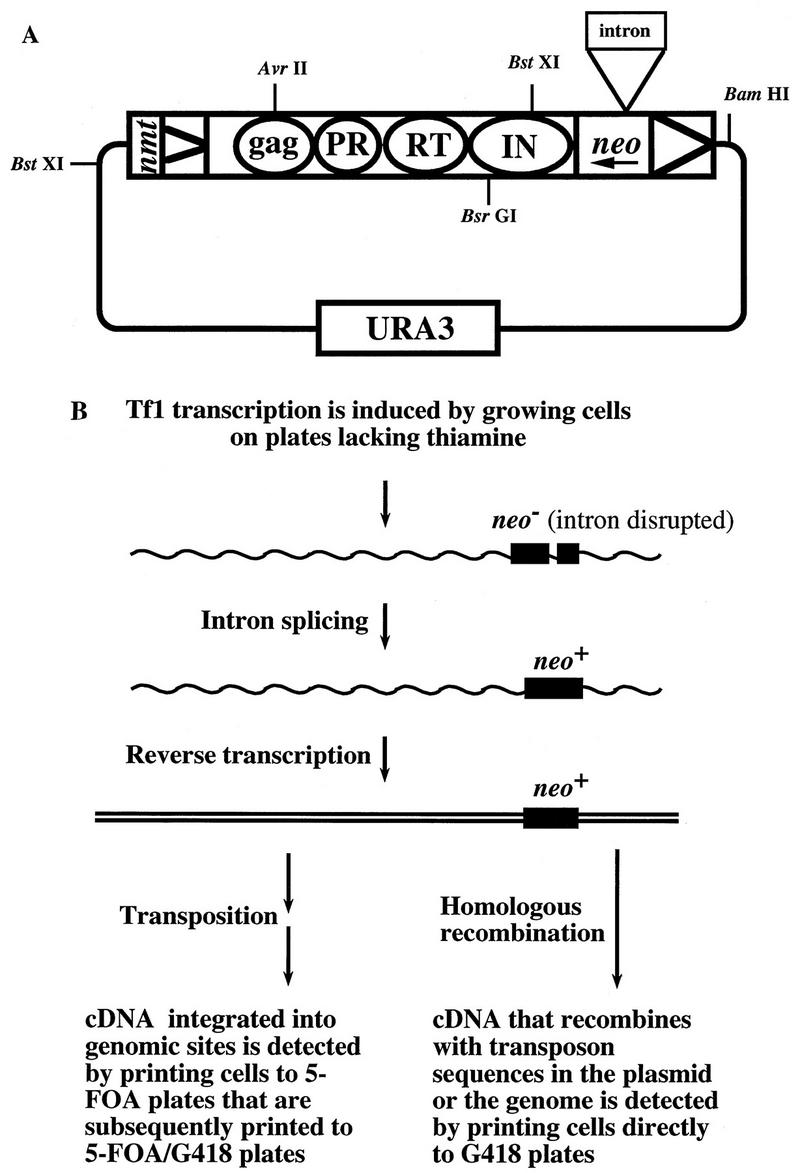
Genetic assays for measuring transposition and homologous recombination of Tf1. (A) Plasmid pHL449-1 was transformed into S. pombe cells and served as the source of Tf1 mRNA and protein for the transposition and recombination assays. The LTR sequences are represented by triangles, the Gag, PR, RT, and IN coding sequences are indicated by circles, and the neo, URA3, and intron positions are marked by rectangles. The arrow indicates that neo transcription proceeds in the opposite direction from Tf1 transcription. The position of the nmt1 promoter is indicated by nmt. The BstXI restriction sites were used for the DNA blot analysis of cDNA production, and the positions of restriction sites used to subclone mutations are shown. (B) Role of the artificial intron in the homologous recombination assay. The neo gene disrupted by the artificial intron is depicted by two adjacent rectangles. The wavy lines represent the Tf1 mRNA, and the double straight lines are the double-stranded cDNA. After splicing and reverse transcription, the neo gene was functional and was used to detect either the integration of the cDNA into genomic sequences or the homologous recombination of the cDNA with other sources of transposon sequences. The diagram also indicates that the differences in the protocols for these two assays were the agar media that the cells were grown on after Tf1 transcription was induced.
FIG. 2.
Transposition and recombination activities of wild-type Tf1 and of two versions with frameshift mutations. (A) Results of transposition assays of S. pombe strains that were either induced for Tf1 transcription (bottom) or repressed (top). The three strains tested contained versions of Tf1 that were either wild type (WT) or contained a frameshift (fs) mutation in IN or PR. The cells were first induced for transcription on medium that lacked vitamin B1 (B1). The induced cells were printed to plates that contained FOA to select against the presence of the Tf1-neoAI plasmid. The FOA-resistant cells were then replica printed onto plates that contained FOA and G418 to measure the frequency of transposition. (B) Results of recombination assays of S. pombe strains that were either induced for Tf1 transcription (bottom) or repressed (top). The cells were treated as described in the transposition assay except that after growth on medium lacking vitamin B1, the cells were replica printed directly onto medium containing G418.
TABLE 2.
Recombination and transposition frequencies of strains transformed with control plasmids
| Strain | Plasmid description | % Recombination | % Transposition |
|---|---|---|---|
| 1282 | Wild-type Tf1-neoAI | 2.9 | 1.8 |
| 1554 | Tf1-neoAI with IN frameshift | 1.9 | 0.1 |
| 1836 | Tf1-neoAI with PR frameshift | <0.001 | <0.01 |
The levels of reverse transcripts present in the nucleus were detected by measuring homologous recombination between Tf1 cDNA and copies of transposon sequence present either in an autonomously replicating plasmid or in the genome. The method for detecting recombination of Tf1 cDNA was adapted from a technique developed to study transposition in Saccharomyces cerevisiae of a Ty1 element that contained a neo gene that was interrupted by an artificial intron (10). It was shown that when Ty1 integration is blocked, Ty1 cDNA becomes an efficient substrate for homologous recombination (47). In our experiments, the cDNA was produced by the same Tf1 expression plasmid used to generate high levels of transposition events in vivo (Fig. 1A) (1, 28). The coding frame of the neo gene in pHL449-1 was interrupted by an artificial intron such that the intron was in the antisense strand of the neo transcript and could therefore not be spliced out to provide G418r. Because the neo gene with the intron (neoAI) was present in the opposite orientation relative to Tf1 transcription, the intron could be spliced out of the Tf1 transcript. This configuration of the artificial intron allowed us to detect homologous recombination events that resulted in the incorporation of reverse transcript sequences into plasmid or genomic sites since these products contained active copies of neo (Fig. 1B).
To measure recombination of the Tf1-neoAI cDNA, S. pombe cells were induced for Tf1 transcription on agar plates that lacked vitamin B1. After a 4-day induction period, the patches of cells were replica printed to rich medium supplemented with G418 to measure the levels of homologous recombination. Figure 2B shows that cells with the wild-type version of Tf1-neoAI produced confluent growth on G418 medium if the induction plates lacked vitamin B1. When the nmt1 promoter was repressed (presence of vitamin B1), no resistance to G418 was observed. To test what fraction of the cells became G418r as the result of integration, we measured the level of G418r cells produced by a version of Tf1-neoAI that lacked IN due to a frameshift mutation. Immunoblot analysis indicated that no IN can be detected in the cells with the IN frameshift (see Fig. 6B). This strain exhibited high levels of growth on G418 medium, indicating that much of the G418 resistance was not the result of transposition events. To quantify the fraction of cells that became G418r, we harvested patches from medium lacking vitamin B1 and plated dilutions of cells onto rich medium and G418 plates. Table 2 shows that the IN frameshift resulted in only a 35% drop in the number of G418r cells. In comparison, cells that contained Tf1-neoAI with a frameshift in PR that blocked expression of RT and IN produced no G418r cells. These results indicated that all of the events that resulted in G418r required reverse transcription, and a large fraction of the cDNA could be recombined to produce G418r even in the absence of integration.
FIG. 6.
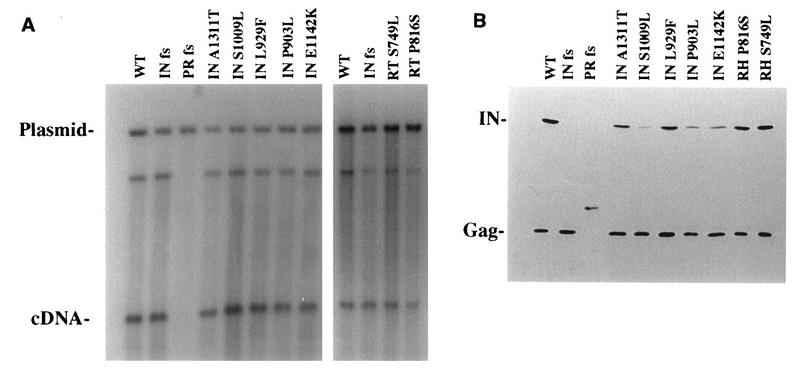
Effects of mutations in Tf1 on the accumulation of IN protein and cDNA were measured. (A) The results of DNA blot analysis were used to detect the effects of mutations in RH and IN on the accumulation of cDNA. Total DNA was extracted from S. pombe strains induced for Tf1 expression. The DNA was digested with BstXI and probed with neo-specific sequence. The 9.5-kb band was produced by vector sequence, and the 2.1-kb band was generated by Tf1 cDNA. The additional band is likely derived from single-LTR circles; however, this identification has not been definitively shown. The panel on the left contained DNA from cells grown in liquid culture, and the panel on the right contained DNA from cells grown on agar plates. WT, wild type. (B) The levels of Tf1 Gag and IN accumulated in strains with the Tf1 mutations were determined by immunoblot analysis. Equal amounts of total protein from each strains was subjected to immunoblot analysis, and the resulting membrane was probed simultaneously with anti-Gag and anti-IN antisera.
The molecular nature of the events that resulted in G418r produced by the Tf1-neoAI was further examined to determine if these events were generated by homologous recombination and to identify the type of sequences that participated in the recombination. Because the goal of these experiments was to develop an assay to measure cDNA recombination that was independent of integration, the parent strain used to generate this set of independent G418r progeny contained Tf1-neoAI with the IN frameshift. Sixteen independent patches of cells were induced for Tf1 expression and then printed directly to plates that contained G418. After colonies were purified from the G418r patches, isolates that lacked the Tf1-neoAI plasmid were generated. We then tested these strains for G418r to determine whether the active neo genes were associated with the Tf1 plasmids or with the S. pombe genomes. Of the original sixteen S. pombe strains, five lost resistance to G418 when their Tf1 plasmids were absent. This indicated that for 5 of the 16 strains, the drug resistance was due to alterations in the plasmid sequences. The rearrangements in these five plasmids were further characterized as representative examples of the processes that caused G418r. Plasmid DNA was extracted from the yeast strains and then transformed into bacteria. The bacteria transformants that possessed recombinant copies of neo were identified by patching cells onto LB plates containing kanamycin.
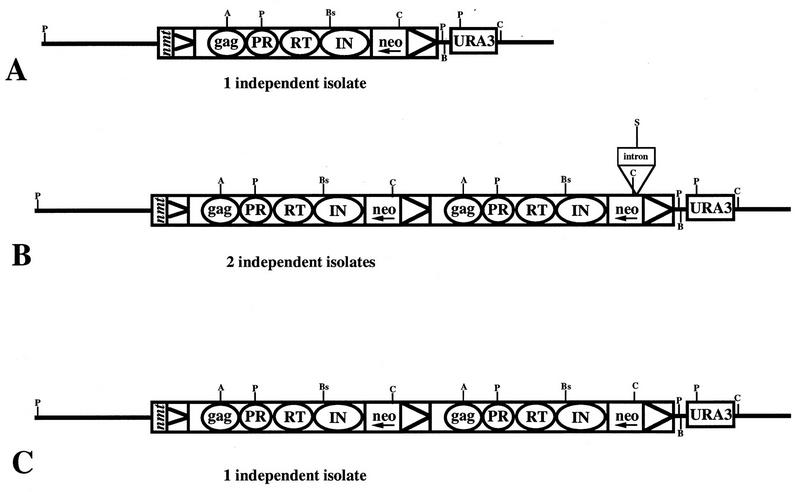
The structures of the plasmids isolated in bacteria were determined by extensive restriction site analysis. The three different structures observed and the relevant restriction sites are shown in Fig. 3. One of the independently isolated S. pombe strains contained a plasmid identical to the original except that the intron in the neoAI was absent (Fig. 3A). This was likely the result of homologous recombination between the Tf1 plasmid and a segment of cDNA with an intronless neo. The plasmids isolated from three other G418r strains contained tandem duplications of the Tf1 sequences. Two of the S. pombe strains contained plasmids that possessed tandem Tf1 copies with an intronless neo gene in the upstream copy of Tf1 and an intron containing neo in the downstream Tf1 (Fig. 3B). These plasmids may have resulted from full-length cDNA molecules that recombined with the 5′ LTR sequences found in the parent plasmid. Another S. pombe strain that contained a plasmid with tandem copies of Tf1 had intronless neo genes in each of the Tf1 elements (Fig. 3C). This plasmid may have resulted from the recombination of two cDNA molecules within the LTR sequences, thus producing a tandem cDNA with two intronless neo copies that subsequently recombined with the Tf1-neoAI plasmid. The last S. pombe strain carried a plasmid with a complex rearrangement that we have not been able to identify.
FIG. 3.
Rearranged versions of the Tf1-neoAI plasmid with the IN frameshift were selected during the recombination assay. The LTR sequences are represented by triangles, the Gag, PR, RT, and IN coding sequences are indicated by circles, and the neo, URA3, and intron positions are marked by rectangles. The arrow indicates that neo transcription proceeds in the opposite direction from Tf1 transcription. The position of the nmt1 promoter is indicated by nmt. Positions of the restriction sites that were used to reveal the structure of these plasmids are shown. Enzymes used: A, AvrII; P, PstI; Bs, BsrGI; C, ClaI; S, SpeI; B, BamHI. (A) One plasmid isolated from a G418r strain of S. pombe was identical to the parent except that the intron was absent. (B) Two other plasmids isolated from G418r strains of S. pombe were identical and contained tandem duplications of the transposon. The downstream copies of the neo genes contained introns, while the upstream copies lacked introns. (C) Another plasmid isolated from a G418r strain of S. pombe contained a tandem duplication of Tf1. Neither copy of neo in this plasmid contained intron sequence.
The rearrangements observed in all four plasmids were likely the result of homologous recombination with Tf1-neo+ cDNA. Because the recombination assay produced resistance to G418 that was entirely dependent on the expression of RT and only marginally dependent on expression of IN (Table 2), high levels of drug resistance indicated that reverse transcripts had been produced and were present in the nucleus. We were therefore able to use the homologous recombination assay to test a large set of defective Tf1 elements to determine which were nevertheless able to deliver significant amounts of cDNA to the nucleus.
Isolation of an extensive collection of point mutations in Tf1 that disrupt transposition.
Mutations throughout the Tf1-neoAI element in the transposition assay plasmid were created by treating the DNA with hydroxylamine in vitro. The extent of the mutagenesis reaction was monitored by using the complementation in E. coli of the pyrF gene by the URA3 gene of S. cerevisiae. After mutagenesis, the plasmid DNA was transformed into bacteria and the resulting colonies were tested for uracil prototrophy. The mutagenesis conditions selected produced bacterial transformants that exhibited uracil auxotrophy at a frequency of 0.4%. Because the URA3 gene is approximately 1.0 kb, we estimated that the fraction of plasmids that would carry a loss-of-function mutation in the 5.9 kb of Tf1-neoAI was about 2.4%. The mutagenized plasmid DNA was then transformed into S. pombe, and 4,000 colonies were assayed for transposition. The number of S. pombe transformants that exhibited obvious defects in transposition as measured by the patch assay was 147, or 3.7%. This frequency was similar to the predicted level of 2.4% that was based on the uracil prototrophy.
Each of the 147 strains defective for transposition was assayed for homologous recombination of the Tf1-neoAI cDNA. The results of patch tests indicated that 131 of the 147 strains tested showed significantly reduced recombination compared to wild-type Tf1-neoAI. Of the remaining 16 strains that showed high levels of homologous recombination, 6 possessed only minor defects in transposition. Therefore, 10 strains that had significant defects in transposition activity but nevertheless produced high levels of recombination were identified. The 10 strains were further characterized to evaluate whether the transposition defects were indeed associated with plasmid-encoded mutations.
The 10 candidate strains were extracted for plasmid DNA that was subsequently transformed into bacteria. The 10 plasmid preparations were then transformed back into S. pombe. Seven of the 10 retransformed strains exhibited the same transposition and recombination phenotypes as the original strains (data not shown). Since the phenotypes of three strains were not reproduced upon transformation of plasmid DNA into new strains, these plasmids were not characterized further. Thus, seven plasmids were identified as having mutations in Tf1 that disrupted transposition but not the participation of cDNA in homologous recombination.
Mapping the position of the mutations in Tf1.
To determine the positions of the mutations responsible for the low transposition activity, we inserted restriction fragments from the defective Tf1 elements into copies of Tf1 that were otherwise wild type. These subcloned versions of Tf1 were then analyzed to determine their transposition and recombination phenotypes.
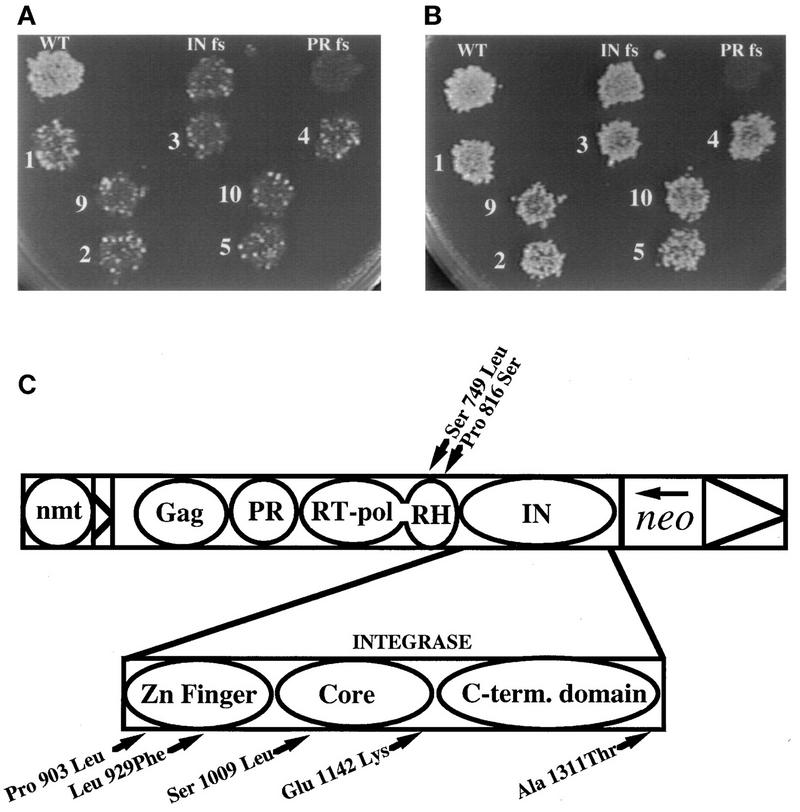
The majority of the Tf1 IN enzyme is encoded within a 3.2-kb BamHI-BsrGI fragment (Fig. 1A). The remaining 5′ end of IN is encoded within a 2.2-kb AvrII-BsrGI fragment which also contained sequences encoding RT, PR, and the 3′ half of Gag. The results summarized in Table 3 indicated that the transposition-defective phenotypes of mutants 2, 4, 5, and 9 were associated with the AvrII-BsrGI fragments whereas mutants 1, 3, and 10 had defects within the BamHI-BsrGI fragments. The defects in transposition exhibited by the subcloned versions of Tf1 are shown in Fig. 4A. The strains with the subcloned versions of Tf1 were also found to show the same high levels of homologous recombination that the original strains possessed (Fig. 4B).
TABLE 3.
Positions of mutations in Tf1 that affected transposition
| Mutant | Fragment with mutation | Substitution | Protein domain |
|---|---|---|---|
| 1 | BsrGI-BamHI of pHL680-1 | Ala1311→Thr | DNA binding domain of IN |
| 2 | AvrII-BsrGI of pHL731-1 | Pro816→Ser | RH domain of RT |
| 3 | BsrGI-BamHI of pHL682-3 | Ser1009→Leu | Catalytic core of IN |
| 4 | AvrII-BsrGI of pHL718-1 | Leu929→Phe | Zn finger domain of IN |
| 5 | AvrII-BsrGI of pHL732-1 | Ser749→Leu | RH domain of RT |
| 9 | AvrII-BsrGI of pHL723-1 | Pro903→Leu | Zn finger domain of IN |
| 10 | BsrGI-BamHI of pHL689-2 | Glu1142→Lys | Catalytic core of IN |
FIG. 4.
Transposition and recombination activities of seven mutated versions of Tf1. (A) Seven versions of Tf1 with mutations and three control strains were measured for transposition activity. The plate contained FOA plus G418 so that the level of growth represented the transposition activity of each strain. Each patch of cells is labeled with its mutation number or the nature of the control plasmid that it contained. fs, frameshift. (B) The same strains were also tested for homologous recombination activity. The plate shown contained G418, and the level of growth represented the level of cDNA recombination produced by each strain.
Each of the restriction fragments that contained transposition mutations was sequenced to identify all base changes. Five of the seven mutants (mutants 1, 3, 4, 9, and 10 in Table 3) were found to consist of single base changes within the IN domain. This finding largely substantiated our reasoning that elements which were unable to transpose but exhibited wild-type cDNA recombination would possess defects in integration. These mutations were located throughout the entire IN protein including the Zn domain, the conserved catalytic core, and the C-terminal domain (Fig. 4C).
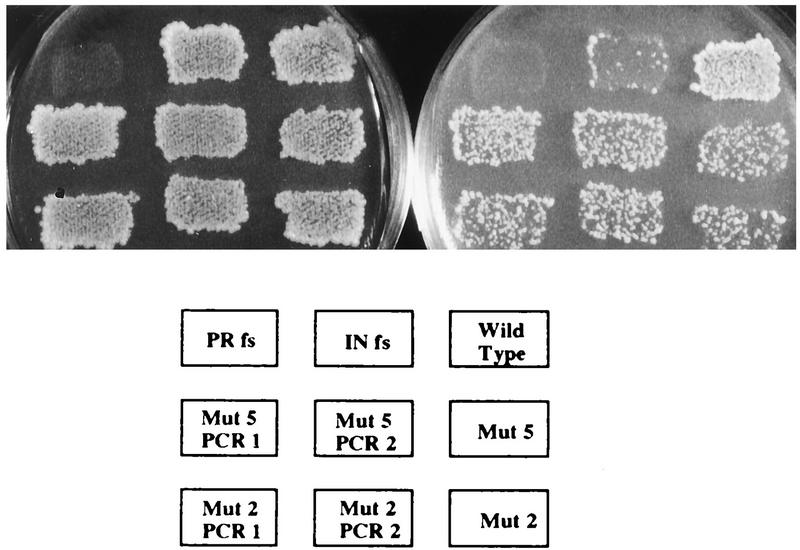
Complete sequencing of the AvrII-BsrGI fragment from mutant 5 and partial sequencing from the AvrII-BsrGI fragment of mutant 2 identified single-base changes within the RH domain of the Tf1 RT. This was a surprising result since data from the recombination assay suggested that reverse transcription occurred at wild-type levels in these strains. To determine if each of these mutations in RH was truely the single mutation responsible for the transposition defects, we recreated each single change in the context of wild-type Tf1 by subcloning into the assay plasmid two independent PCR fragments that contained each mutation. Once the sequence of the mutated sites was verified, the new plasmids were transformed into S. pombe and the transformants were assayed for transposition and recombination. The assays in Fig. 5 verified that the S. pombe strains expressing the reconstruction of mutation 2 (YHL5282) had transposition and recombination phenotypes indistinguishable from those of the original mutant (YHL2230). Likewise, the strain expressing the reconstructed mutant 5 (YHL5274) showed transposition and recombination phenotypes indistinguishable from those of the original mutant strain (YHL2234). We quantitated the transposition frequencies of each of these strains to determine if the activities of the reconstructed Tf1 elements matched those of the original isolates (Table 4). The strains transformed with the PCR-reconstructed mutants exhibited transposition defects at frequencies that were approximately 10-fold below wild-type frequencies. These values were very similar to the transposition frequencies of the cells with the original amino acid substitutions.
FIG. 5.
Transposition and recombination activities of the Tf1 elements with the original substitutions in RH, mutations (Mut) 2 and 5, and activities of equivalent elements reconstructed by PCR. The strains on each plate are identified by the template shown below the photograph. The plate on the left contained G418 and shows the results of the recombination assay; the plate on the right contained FOA plus G418 and shows the results of the transposition assay. fs, frameshift.
TABLE 4.
Quantitative transposition frequencies of mutants 2 and 5 and PCR reconstructions
| Strain | Plasmid description | G418r colonies/FOAr colonies | % Transposition |
|---|---|---|---|
| 1282 | Wild-type Tf1-neoAI | 78/3,796 | 2.05 |
| 1554 | Tf1-neoAI with IN frameshift | 3/5,086 | 0.06 |
| 1836 | Tf1-neoAI with PR frameshift | 0/4,290 | <0.02 |
| 2234 | Mutant 5 (S749L) | 6/5,252 | 0.11 |
| 5274 | Mutant 5; PCR reconstruction 1 | 6/3,376 | 0.18 |
| 5278 | Mutant 5; PCR reconstruction 2 | 5/3,331 | 0.15 |
| 2230 | Mutant 2 (P816S) | 7/5,745 | 0.12 |
| 5282 | Mutant 2; PCR reconstruction 1 | 7/4,707 | 0.15 |
| 5286 | Mutant 2; PCR reconstruction 2 | 5/2,322 | 0.22 |
DNA blot analysis of cDNA produced by mutant copies of Tf1.
To determine directly the levels of cDNA produced in each of these mutant strains, we used DNA blot analysis. Total DNA was extracted from S. pombe strains expressing the mutant copies of Tf1 as well as from strains expressing wild-type, IN frameshifted, and PR frameshifted versions of Tf1. These DNAs were restriction digested with BstXI and subjected to DNA blot analysis with a neo-specific probe. The Tf1 expression plasmid contained two BstXI sites, one within vector sequence and the other near the 3′ end of the Tf1 element (Fig. 1A). Of the two products resulting from BstXI digestion of the plasmid, only the 9.5-kb band is recognized by the neo probe. Since the completed reverse transcript contained the single BstXI site upstream of neo, a 2.1-kb band specific to the reverse transcription product was also detected by the neo probe. The DNA blot in Fig. 6A showed that all strains tested exhibit similar band intensities of the 9.5-kb plasmid. This indicated that approximately equivalent amounts of total DNA were loaded in each lane. Lane 2 contained DNA products extracted from a strain that expressed the IN frameshifted version of Tf1. This strain generated wild-type levels of cDNA. DNA extracted from the PR frameshifted Tf1 that expressed no RT or IN indicated that this strain produced no detectable cDNA (lane 3). All five of the mutants that possessed substitutions in IN produced approximately wild-type levels of reverse transcripts (Fig. 6A). This result confirmed our expectation that strains competent for recombination would contain high levels of cDNA.
The DNA blot analysis of reverse transcripts produced by the Tf1 elements with mutations in RH revealed that the Ser749→Leu (S749L) substitution (mutant 5) did not cause a reduction in cDNA levels. However, the Pro816→Ser (P816S) substitution (mutant 2) resulted in a 1.9-fold reduction in the amount of reverse transcripts detected by phosphoimager analysis (Fig. 6A). The wild-type levels of cDNA generated by the Tf1 with the S749L mutation in RH was consistent with the homologous recombination data but was surprising because RH is not thought to be required for transposition once reverse transcription is complete. In light of this result, we thought it important to ensure that the growth conditions of the cells extracted for DNA match the conditions used for the transposition assays. We therefore prepared DNA from the strains with the mutations in RH by harvesting patches of cells that were fully induced for transposition on agar plates. This DNA was used in the blot analysis shown in Fig. 6A. In this way, we were able to show that wild-type levels of cDNA were produced by the very same patch of cells that exhibited low transposition due to the S749L mutation.
Immunoblot analysis of IN accumulation.
The finding that the transposition-defective mutants produced wild-type levels of reverse transcription products suggested that the principal defect in these mutants was in their ability to complete the integration step of the transposition pathway. To investigate whether the integration defects were a manifestation of decreased stability of the IN or Gag protein, we performed immunoblot analysis with antibodies raised against the Tf1 IN and Gag. Total cell proteins were extracted from log-phase cultures of strains expressing the wild-type, the IN frameshifted, the RT frameshifted, and the seven integration-defective versions of Tf1. Equal amounts of total protein were loaded in each lane of an SDS-polyacrylamide gel that was transferred to an Immobilon-P membrane and probed with both IN- and Gag-specific antibodies. From previous results, it is known that the Tf1 Gag and IN proteins are expressed at equal levels from within a single primary translation product and that during the log-to-stationary-phase transition of the culture, the IN protein is specifically targeted for degradation (1). Figure 6B indicated that all seven of the mutant versions of Tf1 produced detectable levels of IN protein in log phase and adjusted their Gag-to-IN ratios during the log-to-stationary-phase transition (data not shown). Decreased IN signal was observed for vegetative cells with the Ser1009→Leu (S1009L), Pro903→Leu (P903L), and Glu1142→Lys (E1142K) substitutions. In multiple experiments, these three strains showed decreased although variable levels of stable IN protein relative to the IN levels of the strain expressing wild-type Tf1.
DISCUSSION
The presence of the Zn domain and the D,D35E motif (26) in Tf1 IN as well as the highly conserved TG…CA nucleotides at the termini of the Tf1 LTRs indicated that the mechanism of Tf1 integration is closely related to the mechanisms of the other LTR-elements. To evaluate this conclusion and to extend our understanding of LTR-retroelement integration, we sought to develop an in vivo method for identifying mutations that specifically blocked integration. This approach was designed to allow us to determine which amino acids within any of the transposon proteins were critical for integration. Our test for defects in integration was based on genetic measurements of Tf1 cDNA levels. By measuring homologous recombination between Tf1 cDNA and plasmid copies of Tf1-neoAI, we were able to identify mutations in Tf1 that had little effect on the accumulation of cDNA but nevertheless resulted in low levels of transposition. The version of Tf1 that contained a frameshift in the beginning of the IN sequence served as an initial example of this type of mutation. Although the IN frameshift resulted in an 18-fold drop in transposition activity, only a 35% decrease in the level of G418r was detected with the recombination assay. The relatively high level of recombination observed correlated well with the results of DNA blots that showed the strain with the IN frameshift produced wild-type levels of Tf1 cDNA. The 35% reduction caused by the IN frameshift indicated that when IN was present, about a third of the events detected by the recombination assay were due to transposition.
To understand how recombination of Tf1 cDNA caused G418r, we examined the structure of five Tf1-neoAI plasmids that encoded G418r as the result of rearrangements that occurred during the recombination assay. Although the parent plasmid contained a neo gene disrupted by an artificial intron, each recombinant plasmid possessed active copies of neo that lacked the intron. These plasmids either acquired intron-free versions of neo either in the context of a complete Tf1 duplication or were identical to the parent except that the intron was absent from neo. In either case, the structures were consistent with parent plasmids that homologously recombined with Tf1-neo cDNA that lacked the artificial intron. The relatively high frequency (2.9%) of the recombination events that generate G418r may be due to the linear structure of the Tf1 cDNA.
The recombinant events that we isolated were similar to those obtained in a study of cDNA recombination of Ty1 sequences in S. cerevisiae (47). Although we characterized only the recombination events that resulted in plasmid-encoded G418r, tandem duplications of Ty1 were found to form both in plasmid sequences and in the S. cerevisiae genome. Whereas the recombination produced in S. cerevisiae was found to be dependent on a gene required for homologous recombination, RAD52, we were unable to demonstrate a similar dependence on cellular recombination factors because no gene in S. pombe that has the high mitotic activity of RAD52 has been identified. Nevertheless, the high frequencies of G418r produced by versions of Tf1 that lack IN allowed us to use this assay as a measure of cDNA levels.
Mutations in IN reduce transposition.
The homologous recombination assay was used to screen 147 strains that exhibited low transposition activity. The seven isolates that had severe defects in transposition but produced high frequencies of G418r in the recombination assay were thought to possess low integration activity. This reasoning was supported by the finding that the defects in five of these strains were caused by single amino acid substitutions in IN.
Three of the five mutations in IN were positioned at or near highly conserved residues that are known to be required for IN activity in other retroelements. The mutation P903L is located 14 residues upstream of the first conserved histidine in the HHCC Zn domain of the amino terminus of Tf1 IN. Immunoblotting of this mutant indicated that a destabilized IN protein may be the cause of the integration defect exhibited by strains expressing this mutant version of Tf1. The mutation Leu929→Phe (L929F) is just five residues downstream from the second conserved histidine in the Zn domain. Results from immunoblotting showed that the defective IN from this strain was stable and therefore lacked activity due to a loss of function. Since the two conserved histidines in the Zn domain of HIV IN are required for DNA strand transfer activity (15), it was possible that the L929F mutation disrupted the essential function of the histidine residues in the Zn domain of Tf1. This interpretation is complicated by results showing that in the context of live HIV, the histidines in the Zn domain are required for reverse transcription (36). Since the L929F mutation had no observable effect on reverse transcription, this substitution may have only disrupted the function of the Zn domain in the integration reaction.
The mutation in IN, S1009L, was located at a position highly conserved among retroviruses that is 21 residues downstream of the first D in the D,D35E motif of the central core domain (15). The immunoblot results indicated that the S1009L mutation greatly destabilized the Tf1 IN protein. This finding is consistent with studies of HIV IN with mutations at the analogous residue, serine 81. The substitution of serine 81 caused HIV IN to become insoluble when expressed either from yeast or bacteria (27, 52). The position of serine 81 in the crystal structure of HIV IN is at the top of a turn between beta strands 2 and 3. This location appears to bisect the center of the IN protein in a way that may tether the two domains of alpha helices together (13).
The two other single substitutions in Tf1 IN were located in regions that possess no similarity to other IN proteins. The E1142K mutation was positioned approximately at the end of the core domain, 59 residues downstream of the glutamic acid in the core motif D,D35E. The results from immunoblot analysis suggested that the E1142K mutation greatly reduced the stability of the IN protein. The significant destabilization caused by this substitution may have resulted from the change of a negatively charged residue to a positive amino acid. Another mutation in Tf1 IN, Ala1311→Thr (A1311T), was located 20 residues from the C terminus and had little effect on the stability of the protein. The 248 residues in Tf1 IN downstream of the D,D35E motif have little homology to other IN proteins and is much larger than the C-terminal domains of retroviruses. The significant reduction in transposition caused by the A1311T mutation represents the first indication that the C-terminal domain in Tf1 IN is required for function.
Mutations in RT reduce transposition.
The finding that two of the seven mutations were located in RT was very surprising as all of the mutations were isolated on the basis of recombination results that indicated cDNA levels were normal. That both of these mutations were found specifically in the RH domain of RT was no less surprising since the RH activity of retroviruses is required during several stages of reverse transcription (8). The primary role of retroviral RH in reverse transcription is the degradation of the RNA template as it is copied into DNA. This releases newly synthesized minus-strand strong-stop DNA so that it can reanneal to the 3′ LTR in the mRNA and prime synthesis of the full-length minus strand. The primer of plus-strand DNA synthesis is generated by a specialized activity of RH that recognizes and cleaves the 3′ end of the polypurine tract (PPT). In addition, mutations in two catalytic residues of RH in Tf1 indicated that RH activity is required for the initiation of reverse transcription (29).
One of the substitutions in Tf1 RH that we isolated, P816S (mutant 2), resulted in a version of the element that exhibited low levels of transposition despite the high frequencies of G418r colonies generated in the homologous recombination assay. The results of immunoblots showed that the P816S mutation did not affect the levels of IN protein expressed by Tf1 and DNA blots indicated that the levels of cDNA that accumulated were reduced 1.9-fold. Taken together with the result that a twofold reduction in double-stranded reverse transcripts can result in a fivefold drop in transposition (33), this finding indicates that it is possible that the P816S mutation caused a significant loss of transposition because of a relatively minor defect in RH function. If the defect in RH activity specifically affected the cleavage of the PPT, the lack of plus-strand cDNA would be expected to lower transposition without greatly reducing the frequency of homologous recombination. The recombination events of Ty1 cDNA have been shown to occur efficiently even if only minus-strand reverse transcripts are produced (47). Low levels of plus-strand cDNA would also result in a reduced signal on a DNA blot as was produced by the strain with the P816S mutation. Interestingly, the position of the P816S mutation is eight residues upstream from a highly conserved aspartic acid that in HIV (Asp549) has been shown to contribute to the ability of RH to recognize the PPT (42).
The other substitution in RH, S749L (mutant 5), was located three residues upstream of the highly conserved E752 that corresponds to E478 in HIV RT. Our DNA blots demonstrated that Tf1 with the S749L substitution produced quantities of double-stranded cDNA that were indistinguishable from that of wild-type Tf1. The immunoblots showed that Tf1 with the S749L mutation accumulated wild-type levels of IN protein. These results indicated that the low level of Tf1 integration caused by the S749L mutation was not due to the lack of IN protein or its DNA substrate. Another function of retroviral RH that has been described is the degradation of the tRNA and the PPT once they have served as the primers of DNA synthesis. It is possible that the S749L substitution did reduce the specific degradation of the minus-strand and or plus-strand primers. It is also possible that the presence of the minus-strand or the plus-strand primers at the 5′ ends of the otherwise completed double-stranded reverse transcripts inhibited the function of IN. An additional possibility is that other aspects of replication such as virus-like particle (VLP) assembly or localization of the VLP to the nucleus has been affected by this mutation in RT. We have shown that VLPs produced by this mutant strain are indistinguishable from VLPs produced by strains expressing wild-type Tf1 as determined by sucrose gradient analysis (1a). If there is a subtle defect in particle assembly in this mutant, it may result in a reduced import of a protein component of the preintegration complex (PIC).
An alternative is the possibility that the mutation S749L inhibited transposition because the substitution disrupted an unidentified function of RH that may contribute directly to the integration reaction. One compelling hypothesis is that the strong structural similarity observed between the IN core domain and the RH domain of HIV (13) could lead to a heterodimerization of these proteins that could be important for robust integration activity. Consistent with this possibility is the report that the large nucleoprotein complex termed the PIC purified from HIV-infected cells contains RT (5, 37), and the HIV PIC produces double-ended insertions in vitro with significantly greater efficiency than purified IN (16). Interestingly, interactions between RT and IN of other retroviruses have been reported. The immunoprecipitation of murine leukemia virus particles that are boiled or treated with 2-mercaptoethanol indicates that disulfide bonds exists between RT and IN (20). Another example of an interaction between a retroviral IN and RT is the heterodimeric RT of avian sarcoma-leukosis virus. One of the polypeptides contains both RT and IN domains (49). The importance of the IN domain in the avian sarcoma-leukemia virus RT is not yet understood. Whether the S749L mutation in Tf1 disrupted an RH function specifically required for integration or affected a previously described function of RH, further analysis will be required to reveal how a mutation in RH can block transposition without affecting the levels of cDNA.
REFERENCES
- 1.Atwood A, Lin J, Levin H. The retrotransposon Tf1 assembles virus-like particles with excess Gag relative to integrase because of a regulated degradation process. Mol Cell Biol. 1996;16:338–346. doi: 10.1128/mcb.16.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Atwood, A. Unpublished data.
- 2.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 3.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champoux J. Roles of ribonuclease H in reverse transcription. In: Skulka A, Goff S, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 103–117. [Google Scholar]
- 9.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 10.Curcio M J, Garfinkel D J. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doolittle R F, Feng D F, Johnson M S, McClure M A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 12.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 13.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 14.Eichinger D J, Boeke J D. A specific terminal structure is required for Ty1 transposition. Genes Dev. 1990;4:324–330. doi: 10.1101/gad.4.3.324. [DOI] [PubMed] [Google Scholar]
- 15.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 17.Fayet O, Raymond P, Polard P, Prère M F, Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990;4:1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 19.Gatermann K B, Hoffmann A, Rosenberg G H, Kaufer N F. Introduction of functional artificial introns into the naturally intronless ura4 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1989;9:1526–1535. doi: 10.1128/mcb.9.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu S C, Court D L, Zweig M, Levin J G. Murine leukemia virus pol gene products: analysis with antisera generated against reverse transcriptase and endonuclease fusion proteins expressed in Escherichia coli. J Virol. 1986;60:267–274. doi: 10.1128/jvi.60.1.267-274.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson M S, McClure M A, Feng D F, Gray J, Doolittle R F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joyce C M, Grindley N. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J Bacteriol. 1984;158:636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 24.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 25.Khan E, Mack J P, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. . (Erratum, 25:1358.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leavitt A D, Shiue L, Varmus H E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 28.Levin H L. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol Cell Biol. 1995;15:3310–3317. doi: 10.1128/mcb.15.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin H L. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol Cell Biol. 1996;16:5645–5654. doi: 10.1128/mcb.16.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin H L, Boeke J D. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J. 1992;11:1145–1153. doi: 10.1002/j.1460-2075.1992.tb05155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin H L, Weaver D C, Boeke J D. Novel gene expression mechanism in a fission yeast retroelement: Tf1 proteins are derived from a single primary translation product. EMBO J. 1993;12:4885–4895. doi: 10.1002/j.1460-2075.1993.tb06178.x. . (Erratum, 13:1494, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin H L, Weaver D C, Boeke J D. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol Cell Biol. 1990;10:6791–6798. doi: 10.1128/mcb.10.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin J, Levin H. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes Dev. 1997;11:270–285. doi: 10.1101/gad.11.2.270. [DOI] [PubMed] [Google Scholar]
- 34.Luan D D, Korman M H, Jakubczak J L, Eickbush T H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 35.Lutzke R A, Vink C, Plasterk R H. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda T, Planelles V, Krogstad P, Chen I S. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore S P, Garfinkel D J. Expression and partial purification of enzymatically active recombinant Ty1 integrase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:1843–1847. doi: 10.1073/pnas.91.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore S P, Powers M, Garfinkel D J. Substrate specificity of Ty1 integrase. J Virol. 1995;69:4683–4692. doi: 10.1128/jvi.69.8.4683-4692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 41.Mumm S R, Grandgenett D P. Defining nucleic acid-binding properties of avian retrovirus integrase by deletion analysis. J Virol. 1991;65:1160–1167. doi: 10.1128/jvi.65.3.1160-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rausch J W, Le Grice S F. Substituting a conserved residue of the ribonuclease H domain alters substrate hydrolysis by retroviral reverse transcriptase. J Biol Chem. 1997;272:8602–8610. doi: 10.1074/jbc.272.13.8602. [DOI] [PubMed] [Google Scholar]
- 43.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 44.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 45.Rowland S J, Dyke K G. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 46.Schauer M, Billich A. The N-terminal region of HIV-1 integrase is required for integration activity, but not for DNA-binding. Biochem Biophys Res Commun. 1992;185:874–880. doi: 10.1016/0006-291x(92)91708-x. [DOI] [PubMed] [Google Scholar]
- 47.Sharon G, Burkett T J, Garfinkel D J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski R S, Boeke J D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1990;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 49.Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1993. [PubMed] [Google Scholar]
- 50.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Gent D C, Elgersma Y, Bolk M W, Vink C, Plasterk R H. DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2. Nucleic Acids Res. 1991;19:3821–3827. doi: 10.1093/nar/19.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Gent D C, Groeneger A A, Plasterk R H. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci USA. 1992;89:9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varmus H, Brown P, editors. Retroviruses. Washington, D.C: American Society for Microbiology; 1989. [Google Scholar]
- 54.Zimmerly S, Guo H, Perlman P S, Lambowitz A M. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82:545–554. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]