Abstract
The complete DNA sequence of bovine adenovirus type 3 is reported here. The size of the genome is 34,446 bp in length with a G+C content of 54%. All the genes of the early and late regions are present in the expected locations of the genome. However, the late-region genes are organized into seven families, instead of five as they are in human adenovirus type 2. The deduced amino acid sequences of open reading frames (ORFs) in the late regions and early region 2 (E2) and for IVa2 show higher degrees of homology, whereas the predicted amino acid sequences of ORFs in the E1, E3, and E4 regions and the pIX, fiber, and 33,000-molecular-weight nonstructural proteins show little or no homology with the corresponding proteins of other adenoviruses. In addition, the penton base protein lacks the integrin binding motif, RGD, but has an LDV motif instead of an MDV motif. Interestingly, as in other animal adenoviruses, the virus-associated RNA genes appear to be absent from their usual location. Sequence analysis of cDNA clones representing the early- and late-region genes identified splice acceptor and splice donor sites, polyadenylation signals and polyadenylation sites, and tripartite leader sequences.
Bovine adenoviruses (BAVs) belong to the Mastadenovirus genus (which includes adenoviruses of mammals) of the family Adenoviridae and are involved in respiratory and enteric infections of calves (42). The 10 presently accepted serotypes are divided into two subgroups based on a number of characteristics, which include the ability to multiply in different cell cultures of bovine origin and the presence or absence of a complement-fixing antigen common to mastadenoviruses (5). The representatives of subgroup 1 (BAV type 1 [BAV-1], BAV-2, -3, and -9) grow relatively well in established bovine cell lines and contain a common complement-fixing antigen. However, the members of subgroup II (BAV-4, -5, -6, -7, -8, and -10) do not cross-react with any other mammalian adenovirus in the complement fixation test and can be propagated exclusively in low-passage cultures of calf testicular or thyroid cells.
BAV-3 was first isolated by Darbyshire and coworkers in Britain from the conjunctiva of an apparently healthy cow (19). Although the virus has been subsequently isolated from cattle suffering from respiratory and enteric infections, experimental infection of calves has resulted in virus replication with only mild or no clinical symptoms (40). BAV-3 has previously been shown to be capable of transforming rodent cells in vitro (64) and inducing tumors in newborn hamsters (18). Like other adenoviruses, BAV-3 is a nonenveloped icosahedral particle of 75 nm in diameter (46) containing a linear double-stranded DNA molecule, which has been physically mapped with different restriction enzymes (23, 36). Limited nucleotide sequencing has identified a few BAV-3 genes, which encode homologs of proteins found in other adenoviruses (10, 24, 30, 41, 58, 75).
Adenoviruses have gained considerable importance as vectors for gene therapy and vaccination (9, 32). However, the use of human adenoviruses (HAVs) as vectors for gene therapy has been hampered because of the presence of preexisting neutralizing antibodies against HAVs, which may interfere with entry and replication of recombinant virus, and because of the possibility of recombination and/or complementation between recombinant virus and the preexisting wild-type HAV. Therefore, animal adenoviruses, which are highly species specific, are being considered as vectors for gene therapy and recombinant vaccines (74). A prerequisite for the development of BAV-3 as a vector is molecular characterization of its genome. As a step toward that goal, this paper describes the complete nucleotide sequence and transcription map of BAV-3.
MATERIALS AND METHODS
Virus and viral DNA.
The WBR-1 strain of BAV-3 was propagated in Madin-Darby bovine kidney (MDBK) cells. MDBK cells were grown in Eagle’s minimum essential medium supplemented with 5% fetal bovine serum. Eagle’s minimum essential medium with 2% fetal bovine serum was used for the maintenance of infected cells. The purification of virus and extraction of DNA from virus were carried out as previously described (41).
Plasmids and genomic DNA sequencing.
Selected restriction enzyme fragments of BAV-3 DNA (23) were cloned into pGEM-3Zf(+) and pGEM-7Zf(+) plasmids (Promega). Subclones of these BAV-3 DNA fragments were cloned into pGEM-3Zf(+) and (−). Nucleotide sequences were determined on both strands of the genome by the dideoxy chain-termination method (55) with Sequenase enzyme (U.S. Biochemicals) and by the dye-terminator method with an Applied Biosystems (Foster, Calif.) DNA sequencer. To determine the overlapping sequence of different BAV-3 DNA clones and to confirm the sequence at various parts of the genome, viral DNA was directly sequenced with internal primers. The identity of each nucleotide was verified at least four times. A homology search of the GenBank database was made by using BLAST for the deduced amino acid sequence for each open reading frame (ORF). Sequence alignments were carried out by using the PALIGN and CLUSTAL programs of the PC-GENE sequence analysis software package (Oxford Molecular) and Clone Manager (version 4).
cDNA library.
The cDNA library was generated from polyadenylated [poly(A)] RNA extracted from BAV-3-infected MDBK cells at 8 to 10 h and 18 to 24 h postinfection. Double-stranded cDNAs were made with reagents from Stratagene and cloned into Lambda ZAP vector. Plaques which hybridized to specific restriction enzyme fragments of BAV-3 DNA were plaque purified twice. Plasmids containing cDNA were excised from Lambda ZAP vector according to the manufacturer’s protocol. The resulting plasmid clones were characterized by restriction endonuclease analysis and sequencing of both ends of cDNA with T3- and T7-specific primers. Selected cDNA clones were completely sequenced with internal primers.
Nucleotide sequence accession number.
The complete nucleotide sequence reported here has been submitted to GenBank and assigned accession no. AF030154.
RESULTS AND DISCUSSION
Portions of the BAV-3 genome sequence have been previously reported and are listed in Table 1. As the complete nucleotide sequence is now available, the numbers given in Table 1 are the true distances from the left end of the genome, except for the DNA sequence assigned GenBank accession no. X74265, which contains an extra 35-bp repeat (24).
TABLE 1.
Summary of published BAV-3 sequences
| GenBank accession no. | Reference | Sequenced region |
|---|---|---|
| AF030154 | This report | 1–34446 |
| M17113 | 57 | 1–195 |
| X74265 | 24 | 1–4091 |
| D21353 | 75 | 1–4059a |
| U57334 | 58 | 5235–5891 |
| K01264 | 30 | 17736–20584 |
| X53990 | 10 | 20408–21197 |
| D16839 | 41 | 26032–31130 |
This sequence contains one more base (nt 3704) than does the sequence reported here.
General properties and organization of the BAV-3 genome.
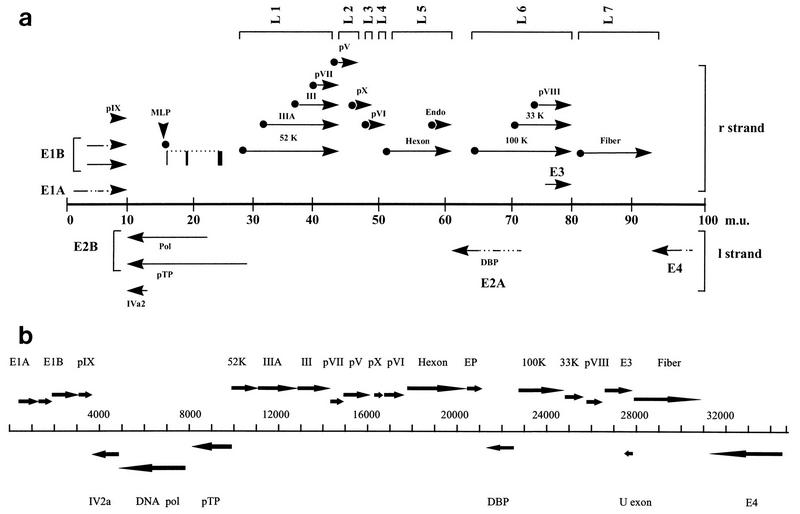
The complete nucleotide sequence of the BAV-3 genome is 34,446 bp in length and has a base composition of 26.6% G, 27.5% C, 23.3% A, and 22.6% T. Thus, the sequence of the BAV-3 genome has a G+C content of 54%, which is similar to the G+C content of the HAV type 12 (HAV-12) genome (59) but approximately 7 and 20% more than the G+C contents of the canine adenovirus type 1 (CAV-1) (45) and ovine adenovirus (OAV) (70) genomes, respectively. The size of the BAV-3 genome is very similar to that reported for the genome of HAV-12 (59) but is 3.9 and 4.9 kb longer than those reported for CAV-1 (45) and OAV (70), respectively. ORFs with coding capacities for more than 75 amino acids which could be identified by their homologies with published adenovirus proteins or by genomic locations are described below. The organization of the genes in the BAV-3 genome, as determined by analysis of ORFs and sequencing of cDNA clones, is summarized in Fig. 1.
FIG. 1.
Transcription map and genome organization of BAV-3. (a) Summary of transcriptional analysis of BAV-3. The schematic diagram of the BAV-3 genome (34,446 bp) is divided into 100 m.u. Arrows above and below the central line represent mRNAs from the r and l strands, respectively. Solid lines represent sequences found in mRNA, broken lines indicate introns, and arrowheads represent poly(A) sites and show the direction of transcription. Each mRNA in L1 to L7 has a TPL spliced to its 5′ terminus (•). Detailed structures of transcripts in E1A and E1B (50), E3 (31) and E4 (38) have been described elsewhere. (b) Summary of the genomic organization of BAV-3. Genes are named as homologs of HAV-2. Arrows show the positions of the ORFs for the indicated proteins.
The genome termini share inverted terminal repeats (ITR) of 195 bp (57). As in all adenoviruses, the ITR of BAV-3 can be divided into two regions, an AT-rich region (first 70 nucleotides [nt] of ITR) and a GC-rich region. The primary sequence of the AT-rich region shows a high degree of homology with those of other adenoviruses (57). The unusual feature of the GC-rich region of BAV-3 ITR is its very high (84%) G+C content. The blocks of sequences, such as the core origin of replication and auxiliary region, that were shown to be important for DNA replication in HAV-2 (66) are also conserved in the ITR of BAV-3. In HAV-5, the DNA packaging domain is composed of a number of A repeats between nt 194 to 358 from the left end of the genome (26). The nucleotide sequence of BAV-3 reveals similar repeats between nt 225 and 310.
E1 region.
The gene products encoded by early region 1 (E1) of HAVs are involved in transactivation of viral and cellular genes (27), transformation of cells in culture, induction of cell DNA synthesis, and mitosis (56). As in other adenoviruses, the E1 transcription unit of BAV-3 is located at the left end of the genome (24, 75) and contains the ORFs for proteins that are homologous to the E1A and E1B proteins of HAV-5. Transcriptional analysis of the E1 region has previously indicated that unlike those of HAV-2 (7), the BAV-3 E1A and E1B transcripts are 3′ coterminal (50). Transcriptional analysis has also identified transcriptional start and termination sites and splice acceptor and donor sites (50) (Table 2).
TABLE 2.
Summary of the transcriptional and translational features of the BAV-3 genome
| Region | Gene | Transcription start site | ATG | Splice donor site | Splice acceptor site | Poly(A) signal(s) | Poly(A) site(s) |
|---|---|---|---|---|---|---|---|
| E1Aa | 211R | 296–298 | 606 | 1215 | 1323 | 3577, 3612 | 3601, 3618, 3632, 3637 |
| 2073 | 3326 | ||||||
| E1Ba | 157R | 1452 | 1476 | 2073 | 3326 | 3577, 3612 | 3601, 3618, 3632, 3637 |
| 420R | 1452 | 1850 | 3577, 3612 | 3601, 3618, 3632, 3637 | |||
| pIXa | pIX | 3161 | 3200 | Nd | N | 3612 | 3632, 3737 |
| E2A | DBP | 24528cb | 22582c | 24468c | 23373c | 21202c | 21175c |
| 23297 | 22591c | ||||||
| E2B | TP | NAc | 9958c | NA | NA | 3627c | 3601c, 3604c |
| Pol | NA | 7790c | NA | NA | 3627c | 3601c, 3604c | |
| IVa2 | IVa2 | NA | 4740c | N | N | 3603c | NA |
| E3a | 284R | 26184 | 26512 | N | N | 27754 | 27776, 27777 |
| 121R | 26184 | 27377 | 26458 | 27151 | 27752 | 27776, 27777 | |
| E4a | 268R | 33950c | 32829c | 33914c | 33635c | 30952c | 30932c |
| 143R | 33950c | 32001c | 33914c | 32003c | 30952c | 30932c | |
| 219 | 33950c | 31631c | 33914c | 31651c | 30952c | 30932c | |
| L1 | 52K | 5587 | 9988 | 9162 | 9988 | 14974 | 14999 |
| IIIA | 5587 | 11098 | 9162 | NA | 14974 | 14999 | |
| III | 5587 | 12919 | 9162 | 12909 | 14974 | 14999 | |
| pVII | 5587 | 14433 | 9162 | 14419 | 14974 | 14999 | |
| L2 | pV | 5587 | 15068 | 9162 | 15016 | 16371 | 16399 |
| L3 | pX | 5587 | 16469 | 9162 | 16444 | 16738 | 16760 |
| L4 | pVI | 5587 | 16871 | 9162 | 16835 | 17679 | 17700 |
| L5 | Hexon | 5587 | 17809 | 9162 | 17763 | 21209 | 21227 |
| Protease | 5587 | 20545 | 9162 | 20537 | 21209 | 21227 | |
| L6 | 100K | 5587 | 22609 | 9162 | 22596 | 27754 | 27776, 27777 |
| 33K | 5587 | 24794 | 9162 | 24788 | 27754 | 27776, 27777 | |
| pVIII | 5587 | 25803 | 9162 | 25723 | 27754 | 27776, 27777 | |
| L7 | Fiber | 5587 | 27968 | 9162 | 27926 | 30909 | 30926 |
E2A and E2B.
The gene products encoded by the E2 region are involved in viral DNA replication. The DNA-binding protein (DBP) encoded by the E2A region of adenoviruses is a multifunctional protein, which is involved in diverse processes (12). The ORF for the BAV-3 DBP is located on the complementary strand between nt 22583 and 21285 and lies between the ORFs for viral endoproteinase (EP) and the 100,000-molecular-weight (100K) hexon assembly protein (Fig. 1b). The BAV-3 DBP contains 432 residues (Fig. 2), which is longer than the DBP of OAV (70) but shorter than the DBPs of various HAVs (34). The DBP of BAV-3 shows amino acid identities of 18 to 47% to the DBPs of other adenoviruses (Table 3). Sequence analysis of the cDNA clones representing DBP transcripts of BAV-3 suggests that two untranslated leaders are spliced to the main body of the ORF, separating the ATG from the second splice junction by 9 nt (Fig. 1a; Table 2). The poly(A) signal, which is 82 bp downstream of the termination codon, is the signal for poly(A) addition (Table 2). There is one more poly(A) signal, located 18 nt downstream from the termination codon of the BAV-3 DBP; its significance is not known. As with other adenoviruses (63), the BAV-3 DBP contains a variable amino-terminal domain and a conserved carboxy-terminal domain (Fig. 2). The variable amino-terminal domain of the HAV DBP is dispensable for DNA replication but is involved in the determination of host range (2). The conserved carboxy-terminal domain contains several stretches of highly conserved amino acids (34), which function in DNA binding and replication and transcriptional activation of the major late promoter (MLP). Recently, it has been shown that the DBP of HAV contains two zinc atoms in different coordinations (65). The first atom is coordinated by four conserved cysteine residues, whereas the second one is coordinated by three cysteine residues and one histidine residue. The zinc atoms have a structural role and are required for functional activity of the DBP (65). The first cysteine in the motif HXCX8CXH (HAV-2 residues 273 to 286), which has previously been shown to be involved in zinc binding, is conserved in the DBP of BAV-3 (HXCX9CXH; Fig. 2). A second zinc atom is bound by cysteine residues at positions 396, 398, 450, and 467 in HAV-2. All of these residues and the distances between residues are conserved in the DBP of BAV-3 (residues 302, 304, 356, and 372; Fig. 2). The glycine residues, which have previously been shown to be involved in tight β turns (65), are also conserved in the DBP of BAV-3 (residues 150, 172, 186, 193, 262, 268, and 315; Fig. 2). In HAVs, the N-terminal domain is heavily phosphorylated at serine and threonine residues and contains most of the phosphates bound by the DBP molecule (43). The DBP of BAV-3 also contains a number of serine and threonine residues in the amino-terminal domain (Fig. 2) which may serve as phosphorylation sites. The DBP of HAV-5 has a bipartite nuclear localization signal (NLS) 42PPKKR46 and 84PKKKKK89 (44). The amino-terminal region of the BAV-3 DBP contains 29PRKK32, 35RKRR38, and 53KRAK56 (Fig. 2), which could function as an NLS for the transport of the protein into the nucleus. The E2B region of HAV-2 codes for DNA polymerase (Pol) and precursor terminal protein (pTP). As in HAV-2, the E2B region of BAV-3 codes for DNA Pol and pTP (6), which show significant homologies to the homologous proteins of other adenoviruses (Table 3). Sequence analysis of cDNA clones representing the DNA Pol and pTP of BAV-3 showed that the transcripts coding for them are 3′ coterminal (Fig. 1a; Table 2). Similar observations have previously been made for HAV-2 but not for OAV (69).
FIG. 2.

Multiple-sequence alignment of DBP homologs. The amino acid sequences of BAV-3 (B3), CAV-1 (C1), and HAV-2 (H2) DBPs were aligned as indicated. Identical and conserved amino acid residues are indicated by asterisks and dots, respectively. Dashes indicate gaps. Conserved Cys residues that are involved in metal binding are shown in bold, underlined, and shaded. NLSs are shown in bold italics and shaded. Conserved glycine residues are shown in bold italics and underlined. The sequence motif HXCX8CXH is shown in bold and shaded. The alignment was done with the CLUSTAL program by using default parameters (open gap cost and unit gap cost = 10). The same parameters were used for the alignment of protein sequences in Fig. 3.
TABLE 3.
Protein sequence identities between BAV-3 and other adenovirusesa
| BAV-3 protein | Identity (%)
|
|||||
|---|---|---|---|---|---|---|
| HAV-2 | HAV-12 | HAV-40 | CAV-1 | OAV | CELO | |
| DBP | 41.9 | 43.5 | 38.7 | 47 | 26.7 | 18.1 |
| pTP | 58.1 | 59.9 | 59.4 | 57.8 | 24 | 31 |
| Pol | 58.8 | 60.1 | 61.5 | 60 | 42.4 | 29 |
| IVa2 | 68.9 | 67.8 | 69.7 | 63.6 | 34.6 | 29.8 |
| pIX | 27.2 | 16.8 | 28.2 | 16.5 | N | N |
| 52K | 61.6 | 58.6 | 59.8 | 56.8 | 24.1 | 21.4 |
| IIIA | 57 | 57.2 | 55.6 | 51.5 | 25.2 | 26.4 |
| III | 62.9 | 63.3 | 63.3 | 64.8 | 52.4 | 44.2 |
| pVII | 53.2 | Nb | 51.5 | 43.9 | 29.7 | 33.3 |
| pV | 40.9 | 38.3 | 38.3 | 28.3 | N | N |
| pX | 52.5 | 59.7 | 64.3 | 63.2 | 38 | 45 |
| pVI | 32 | 31.6 | 33.1 | 38.7 | 15.8 | 26 |
| Hexon | 65.7 | 69 | 69.1 | 70.8 | 51.1 | 45.3 |
| EP | 65.7 | 65.7 | 65.2 | 65.2 | 30.3 | 38.7 |
| 100K | 52.1 | 50.1 | 52.1 | 51.2 | 28.3 | 27.6 |
| pVIII | 52.3 | 51.4 | 51.4 | 56.5 | 19 | 19.4 |
| Fiber | 23.7 | 21.5 | 23.2 | 26.5 | 22.8 | 17 |
E3 region.
The E3 region gene products of HAVs are involved in modulating the host immune response to virus infection in vivo but are not required for virus replication in vitro (73). As in other adenoviruses, the E3 region of BAV-3 is located between the genes coding for pVIII and fiber proteins (41). It is relatively small (1.4 kb) and contains four ORFs, one of which shows significant homology to the 14.7K protein of HAV-5 (41). The E3 region contains a TATA box at nt 26154. Transcriptional analysis of E3 region identified the transcription start and termination sites and splice donor and acceptor sites (31) (Table 2).
E4 region.
The gene products encoded by the E4 region of HAVs are involved in several levels of cellular and viral gene expression (8). The E4 region of BAV-3 was identified by its corresponding genomic location relative to that in HAVs. It lies near the right end of the genome (nt 30932c to 33950c) and is transcribed from the l strand of the genome. Nucleotide sequence analysis identified five ORFs, two of which (ORF3 and ORF5) show homologies to the 34K protein (ORF6) of HAVs (38). Transcriptional analysis of the E4 region identified the transcription start and termination sites and splice donor and acceptor sites (38) (Table 2).
VA RNAs.
HAV-infected cells contain large quantities of two low-molecular-weight RNAs that are designated virus-associated (VA) RNAs, VA RNAI and VA RNAII (37). The genes for VA RNAs are located at around 30 map units (m.u.) and are situated between the genes coding for the 52/55K protein and pTP. In the case of BAV-3, the ORFs coding for the 52/55K protein and pTP are adjacent to each other. Consequently, there is no room for a VA RNA gene(s) unless the VA gene(s) overlaps both ORFs, suggesting that a VA RNA gene(s) is absent in BAV-3. Similar observations have previously been reported for CAV-1 (45) and OAV (70). However, the VA RNA gene(s) in BAV-3, OAV, and CAV-1 may be located elsewhere in the genome, as has previously been shown for CELO virus (13). If VA RNA genes are absent in adenoviruses of animal origin, it will be interesting to see what novel mechanisms these viruses use to evade the effects of protein kinase DAI (29).
Intermediate region.
In HAVs, the two genes coding for pIX and IVa2 proteins are classified as intermediate-region genes. The pIX protein of HAVs is a structural component of the virion (67) and acts as a transcriptional activator (38a). The pIX protein of BAV-3 is 125 amino acids long (6, 75), which is shorter than the pIX protein of HAV-2 (61) but longer than the pIX protein of CAV-1 (45). The BAV-3 pIX protein shows homologies of 16 to 28% to the pIX proteins of other adenoviruses (Table 3). Unlike in HAV-2 (7), BAV-3 pIX, E1A, and E1B transcripts are 3′ coterminal (50) (Table 2). In HAVs, the IVa2 protein has previously been shown to contribute to MLP activation (62). The IVa2 protein of BAV-3 is 376 amino acids long (6), which is shorter than the IVa2 proteins of other adenoviruses (45, 61, 69). The BAV-3 IVa2 protein shows amino acid identities of 29 to 69% to the IVa2 proteins of other adenoviruses (6) (Table 3). Unlike in other adenoviruses (69), the BAV-3 IVa2 transcript is not 3′ coterminal with either DNA Pol or pTP transcripts (6) (Table 2).
Late regions.
The gene products encoded by the late region of HAVs are mainly structural proteins. In HAVs, the late transcription unit is transcribed from an MLP located at 16.3 m.u. and continues to the right end of the genome. The primary transcript is processed to about 20 late mRNAs that comprise five families (L1 to L5) of mRNAs (25). In BAV-3, the late mRNAs can be divided into seven families (L1 to L7) of mRNAs, depending on which poly(A) signal is used by these mRNAs (Fig. 1; Table 2). In HAV-2, mRNAs transcribed from the MLP contain an identical 5′ noncoding segment which is 200 nt in length, the tripartite leader (TPL) (14). It enhances the translation of mRNAs in infected and transfected cells (22) and is derived from the splicing of three small exons located at 16.3, 19.0, and 26 m.u. (14). In BAV-3, the MLP (58) is located at 16.2 m.u. and has a canonical TATA box (nt 5508 to 5513), inverted CAAT box (nt 5582 to 5592), transcriptional factor USF binding site (nt 5508 to 5513), and initiator element (nt 5565 to 5572). The activity of the HAV-2 MLP increases manyfold after the onset of viral DNA replication. The sequence elements (DE1 and DE2) downstream of the HAV-2 MLP transcriptional start site have previously been shown to play an important role in this enhancement (62). Two such sequence elements are also located within the first intron between nt 5671 to 5681 and 5686 to 5699 in BAV-3. Nucleotide sequence determinations of several cDNA clones representing late transcripts of BAV-3 suggest that the MLP transcriptional start site is at nt 5587, which is 47 nt downstream of the TATA box (Table 2). The first leader of the TPL in BAV-3 lies adjacent to the MLP and is 40 nt in length (nt 5587 to 5626). The second leader lies within the gene encoding DNA Pol and is 78 nt in length (nt 6642 to 6719). The third leader lies within the gene encoding pTP and is 87 nt in length (nt 9076 to 9162). Thus, the TPL of BAV-3 is 205 nt long and derived from three exons located at 16.2, 19.3, and 26.4 m.u. (Fig. 1a). A similar organization has previously been reported for the TPL of HAV-2 (14) but not for the TPL of OAV (69). The overall length of the TPL in HAV-2 is 201 nt, with its first exon of 41 nt from the MLP region (16.2 m.u.), its second exon of 71 nt from the DNA Pol region (19.6 m.u.), and its third exon of 89 nt from the pTP region (26.3 m.u.) of the genome (14). In OAV, the length of the TPL is only 157 nt, with its first exon of 31 nt from the MLP region and its second and third exons of 63 nt each from the pTP region (69). Thus, the TPL of OAV is much shorter than is the TPL of BAV-3 and the second exon of the OAV TPL lies in the gene encoding pTP instead of DNA Pol. In addition to the three exons of the TPL, HAV-2 fiber mRNA contains three additional leader sequences (x, y, and z) derived from the E-3 region (14). No such leader sequences were detected in cDNAs representing the fiber mRNA of BAV-3 (Fig. 1a).
L1 region.
In BAV-3, four late-region genes, 52K, IIIA, III, and pVII, have common poly(A) signals and sites and thus belong to the L1 family (Fig. 1a; Table 2). However, in HAVs, only 52/55K and pIIIA proteins are encoded by the L1 region, whereas pIII and pVII proteins are encoded by the L2 region (61). The HAV-2 52/55K proteins represent two differentially phosphorylated forms of the 48K protein which are involved in virion assembly (28). The 52/55K protein homolog in BAV-3 is 331 amino acids in length, which is similar to the size reported for OAV (70) but much shorter than those of HAV-2 (61) and CAV-1 (45), mainly due to a large carboxy-terminal deletion. The 52/55K protein of BAV-3 has amino acid identities of 21 to 61% to the 52/55K proteins of other adenoviruses (Table 3). In HAV-2, the pIIIa protein is a virion phosphoprotein which is associated with the hexon polypeptide (17) and cleaved by the viral protease during virion assembly. In BAV-3, pIIIa is 568 amino acids long, which is nearly the same size as the pIIIa proteins of CAV-1 (45) and HAV-2 (61) but longer than the pIIIa protein of OAV (70). The BAV-3 IIIa protein has amino acid identities of 25 to 57% to the pIIIa proteins of other adenoviruses (Table 3). The possible protease cleavage site in the pIIIa protein of BAV-3 is located 19 residues from the carboxy terminus (LRGT↓G).
The penton base protein (pIII) is known to play an important role in virus entry into cells (72). The penton base protein of BAV-3 is 482 residues in length (Fig. 3), which is longer than the pIII protein of OAV (70) but shorter than the pIII protein of HAV-2 (51). The BAV-3 penton base protein has amino acid identities of 44 to 64% to the penton base proteins of other adenoviruses (Table 3). The following two sequence motifs have previously been identified in the HAV penton base protein: the RGD motif, which interacts with surface integrins αvβ3 and αvβ5 (72), and the LDV motif, which interacts with α4β1 integrin (35). Although both these sequence motifs are absent in the penton base protein of BAV-3, the LDV motif seems to be replaced by an MDV motif (Fig. 3). Though it is conserved in most but not all adenoviruses, the RGD motif is also absent in the penton base proteins of OAV (70), egg drop syndrome virus 76 (52), and CAV-1 (45), indicating that these viruses may use other motifs in interacting with cell surface integrins. Alternatively, animal adenoviruses may use a different entry pathway. In addition to its interactions with cell surface integrins, the penton base protein interacts with the fiber protein to form penton. The fiber-interacting domain (HSRLSNLLGIRKR) of the HAV-2 penton base protein is highly conserved in the penton base proteins of other adenoviruses (11), including BAV-3 (residues 241 and 253; Fig. 3).
FIG. 3.
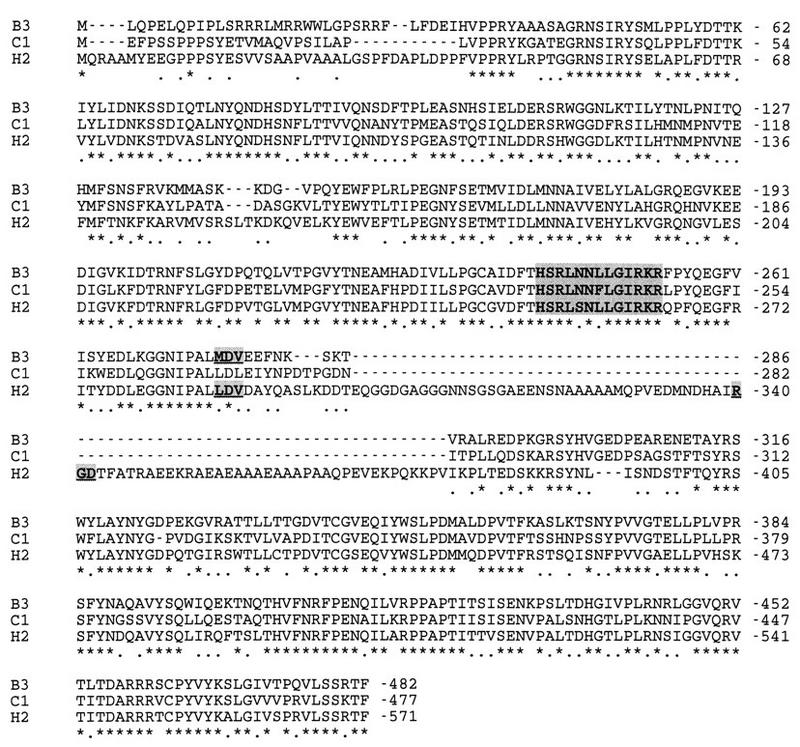
Multiple-sequence alignment of penton base protein homologs. The amino acid sequences of BAV-3 (B3), CAV-1 (C1), and HAV-2 (H2) penton base proteins were aligned as described in the legend to Fig. 2. Identical and conserved amino acids are indicated by asterisks and dots, respectively. Dashes indicate gaps. RGD, LDV, MDV motifs are shown in bold, underlined, and shaded. The putative fiber-interacting domain is shown in bold and shaded.
Polypeptide pVII is a major protein associated with adenovirus cores and alone accounts for about 10% of the protein mass of the virion (60). The ORF coding for the pVII protein of BAV-3 is 171 residues in length, which is similar in size to that of CAV-1 (45) but longer than that of OAV (70). The BAV-3 pVII protein has amino acid identities of 29 to 53% to the pVII proteins of other adenoviruses (Table 3). An analysis of the amino acid sequence of the BAV-3 pVII protein revealed a number of basic amino acids (43 of 171 residues), which is consistent with the function of pVII homolog proteins in the packaging of viral DNA. The consensus protease cleavage site (MYGG↓A) is located 19 nt from the amino terminus of the BAV-3 pVII protein.
L2 region.
The L2 region of HAV-2 codes for three proteins, including pV (61). However, in BAV-3, the L2 region codes for only the pV protein (Fig. 1a; Table 2). The ORF coding for the BAV-3 pV protein is 410 residues in length, which is similar in size to that of CAV-1 (45) but longer than that of HAV-2 (51). The pV protein of BAV-3 shows amino acid identities of 28 to 40% to the pV proteins of other adenoviruses (Table 3), except for OAV (70) and CELO (13), which do not contain homologs of the pV protein. Like pVII, the BAV-3 pV protein is rich in basic amino acids (77 of 410 residues). The pV proteins of HAVs contain a bipartite NLS sequence close to the carboxy terminus (21). A similar sequence motif (290KRGVGKVEPTIQVLASKKRR) is present in the central region of the BAV-3 pV protein.
L3 region.
In HAV-2, the precursor of pX is encoded by the L2 region (1). However, in BAV-3, the precursor of pX is encoded by the L3 region (Fig. 1a; Table 2). The precursor pX protein of BAV-3 is 80 amino acids in length, which is identical to that of HAV-2 (1) but longer than those of CAV-1 (45) and OAV (70). The pX protein of BAV-3 shows amino acid identities of 38 to 64% to the pX proteins of other adenoviruses (Table 3). The pX protein of BAV-3 is rich in basic amino acids, especially in the central domain, which in pX homologs is thought to be involved in DNA binding (1). It has a bipartite NLS (23RRTGLRGGSAYLLGRRRRR) which is conserved in the pX proteins of other adenoviruses, including HAV-2 (1). In addition, the BAV-3 pX protein contains two consensus protease cleavage sites (27LRGG↓S and 47LRGG↓F), suggesting that as in HAV-2 (54, 71), the pX protein of BAV-3 is cleaved by viral protease.
L4 region.
The BAV-3 pVI protein is coded for by the L4 region (Fig. 1a; Table 2). The homolog of the pVI protein in HAV-2 is encoded by the L3 region (61). The mature pVI protein is a minor capsid component of the virion, whereas the precursor pVI protein is involved in the transport of hexon capsomers to the nucleus (33). The length of the BAV-3 pVI sequence is predicted to be 263 residues, which is longer than those of the pVI proteins of HAV-2 (61) and OAV (70). The BAV-3 pVI protein shows amino acid identities of 15 to 38% to the pVI proteins of other adenoviruses (Table 3). In BAV-3, the precursor pVI protein contains two sequence motifs, with one near the amino terminus (30MHGG↓R) and the other at the carboxy terminus (249IVGL↓G). Both of these motifs conform to a consensus sequence motif for cleavage by endoprotease (EP) (54, 71). Previous studies with HAV-2 proteinase have indicated that it requires two cofactors for its activity (39). The first cofactor is DNA, and the second cofactor is the carboxy-terminal fragment of pVI. As the carboxy-terminal regions of the pVI proteins of BAV-3 and HAV-2 show significant homology, the carboxy-terminal fragment of the BAV-3 pVI protein may act as a cofactor for the BAV-3 EP.
L5 region.
The L3 region of HAV-2 codes for pVI, hexon, and 23K proteins (61). However, the L5 region of BAV-3 codes for only the hexon protein and proteinase (Fig. 1a; Table 2). The proteinase of BAV-3 is 204 amino acids in length (10) and shows amino acid identities of 30 to 65% to the proteinases of other adenoviruses (Table 3). The BAV-3 pIIIa, pVII, pX, pVI, pVIII, and pTP proteins have maintained protease cleavage sites, as have their homologs in other mastadenoviruses. The hexon protein of BAV-3 is 910 amino acids in length (30) and shows amino acid identities of 45 to 70% to the hexon proteins of other adenoviruses (Table 3). The hexon proteins of various adenovirus serotypes can be distinguished from each other mainly because of differences in external loops 1 and 2 (l1 and l2, respectively) of the protein (3). In BAV-3, loop l1 is shorter (30) partly because of the absence of a region which is rich in acidic amino acids in HAV-2 (3). Most mutations and deletions occur in loops l1 and l2 of the molecule. As these loops are flexible and are not involved in the structural stability of the protein, the overall structure of the BAV-3 hexon protein should be similar to that determined for HAV-2 (3). Unlike in OAV (70), the ORFs of the hexon protein and EP do not overlap in the genome of BAV-3.
L6 region.
The L4 region of HAV-2 codes for two nonstructural (100K and 33K) proteins and one structural (pVIII) protein (47, 61). However, in BAV-3, these proteins are coded by mRNAs produced from the L6 region (Fig. 1a; Table 2). Since the 100K protein is involved in folding hexon polypeptide chains into trimers (48), it is also called the hexon assembly protein. The 100K protein of BAV-3 is 850 amino acids in length, which is longer than the 100K proteins of HAV-2 (61) and OAV (68). The BAV-3 100K protein shows amino acid identities of 27 to 52% to the 100K proteins of other adenoviruses (Table 3). The gene coding for the HAV-2 33K phosphoprotein is unique among all the genes transcribed from the MLP in that it contains a 202-nt intron (47). The gene coding for the 33K phosphoprotein can encode either the 22K protein (with splicing) or 33K protein (without splicing). In BAV-3, the corresponding ORF is predicted to code for a protein of 274 amino acids in the absence of splicing and a protein of 279 amino acids with splicing. This is longer than the 33K proteins of HAV-2 (47) and OAV (68). Surprisingly, neither the 279- nor the 274-amino-acid protein shows any homology to the corresponding proteins of other adenoviruses (data not shown). Unlike in other adenoviruses (68), the coding regions of the 100K and 33K proteins do not overlap in BAV-3.
Polypeptide pVIII, which is one of the hexon-associated proteins, connects the core with the inner surface of the adenovirus capsid. The predicted amino acid sequence of the BAV-3 pVIII protein is 216 residues long, which is similar in size to the pVIII proteins of HAV-2 (61) and OAV (68). The BAV-3 pVIII protein shows amino acid identities of 19 to 56% to the pVIII proteins of other adenoviruses (Table 3). The pVIII protein of BAV-3 contains two recognizable protease cleavage sites (108IAGG↓G and 143LGGG↓S). Unlike in HAV-2 (61) and OAV (68), L6 region transcripts of BAV-3 are polyadenylated at the poly(A) site of the E3 region (Fig. 1a; Table 2). A similar observation has previously been made for the L4 region of HAV-40 (20). In addition to these proteins, an ORF of 55 amino acids (U exon) is encoded on the complementary strand; it starts 32 nt upstream of the ATG of the fiber protein and overlaps the 3′ end of E3. This ORF is conserved in all adenoviruses for which nucleotide sequence data are available, except for murine adenovirus type 1 (20, 49). However, there is no apparent TATA signal upstream of this ORF in BAV-3.
L7 region.
The fiber protein of HAV-2 is encoded by the L5 region (61). However, in BAV-3, the fiber protein is encoded by a transcript produced from the L7 region (Fig. 1a; Table 2). The BAV-3 fiber protein is 976 amino acids in length and contains a very long shaft region with 46.5 repeats, 80% of which are glycine repeats (53). The BAV-3 fiber protein shows amino acid identities of 17 to 26% to the fiber proteins of other adenoviruses (Table 3). In all HAVs, a hydrophobic sequence motif [FNPVYPY(D/E)] in the amino-terminal tail of the fiber protein is speculated to be involved in the specific interaction between the fiber and penton base proteins (11). Such a sequence element (9FNLVYPYKA) is strongly conserved in the fiber protein of BAV-3. In all adenoviruses, the head of the fiber protein, which is a carboxy-terminal domain, begins at the well-conserved TLWT motif (16). In the BAV-3 fiber protein, an identical motif is located at the junction of the head and the central shaft.
Phylogenetic analysis.
In addition to serological tests, determinations of phylogenetic relationships by comparisons of sequence data play a major role in the classification of viruses within a particular genus (4). Two-way comparisons between the predicted protein sequences of BAV-3 with those of other adenoviruses clearly demonstrated that certain regions of the genome are evolving more rapidly than are others (Table 3). This change is more clearly evident in the fiber protein and proteins encoded by the E1, E3, and E4 regions of the genome. The fiber protein carries major antigenic determinants of the virus, and changes in this protein sequence are probably due to antigenic drift produced in response to humoral immunity. The proteins encoded by the E1, E3, and E4 regions mainly interact with host cell factors in preparing infected cells for viral DNA replication. Variations in these proteins are expected, as host cell factors vary from host to host. In general, the proteins of BAV-3 that interact with DNA or other capsid proteins show their highest similarities to the corresponding proteins of HAVs and CAV-1 and their lowest homologies to those of OAV and CELO virus, indicating that BAV-3 is phylogenetically closely related to HAVs and CAVs.
In conclusion, the size and overall organization of the BAV-3 genome appear to be similar to those of HAVs for which published data are available. However, a relatively smaller and simple E3 region, seven families of late-region genes, the absence of an RGD motif in the penton base protein, the absence of additional leader (x, y, and z) sequences from fiber mRNA, and the absence of VA RNA genes from their usual location are distinctive features of the BAV-3 genome.
ACKNOWLEDGMENTS
We are grateful for the expert help of Ron MacLachlan with computer analysis of sequences.
This work was supported by grants from the National Science and Engineering Research Council of Canada, Saskatchewan Agriculture Development, Western Economic Diversification, and Alberta Agriculture Research Institute. P.S.R., A.N.Z., and M.K.B. are recipients of postdoctoral research fellowships from the Health Services Utilization and Research Commission, Saskatoon, Saskatchewan, Canada.
Footnotes
Published as VIDO journal series no. 235.
REFERENCES
- 1.Anderson C W, Young M E, Flint S J. Characterization of the adenovirus 2 virion protein, Mu. Virology. 1989;172:506–512. doi: 10.1016/0042-6822(89)90193-1. [DOI] [PubMed] [Google Scholar]
- 2.Anderson K P, Klessig D F. Altered mRNA splicing in monkey cells abortively infected with human adenovirus may be responsible for inefficient synthesis of the virion fiber polypeptide. Proc Natl Acad Sci USA. 1984;81:4023–4027. doi: 10.1073/pnas.81.13.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athappilly F K, Murali R, Rux J J, Cai Z P, Burnett R M. The refined crystal structure of hexon, the major coat protein of adenovirus type 2 at 2.9 angstrom resolution. J Mol Biol. 1994;242:430–455. doi: 10.1006/jmbi.1994.1593. [DOI] [PubMed] [Google Scholar]
- 4.Bailey A, Mautner V. Phylogenetic relationships among adenovirus serotypes. Virology. 1994;205:438–452. doi: 10.1006/viro.1994.1664. [DOI] [PubMed] [Google Scholar]
- 5.Bartha A. Proposal for subgrouping of bovine adenoviruses. Acta Vet. 1969;19:319–321. [PubMed] [Google Scholar]
- 6.Baxi, M. K., P. S. Reddy, A. N. Zakhartchouck, N. Idamakanti, C. Pyne, L. A. Babiuk, and S. K. Tikoo. Virus Genes, in press. [DOI] [PubMed]
- 7.Berk A J, Sharp P A. Structure of the adenovirus 2 early mRNAs. Cell. 1978;14:695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- 8.Bridge E, Ketner G. Redundant control of adenovirus late gene expression by early region 4. J Virol. 1989;63:631–638. doi: 10.1128/jvi.63.2.631-638.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brody S L, Crystal R G. Adenovirus-mediated in vivo gene therapy. Ann NY Acad Sci. 1994;716:90–101. doi: 10.1111/j.1749-6632.1994.tb21705.x. [DOI] [PubMed] [Google Scholar]
- 10.Cai F, Bourbonniere M, Tang D, Hu S L, Weber J M. Nucleotide and deduced amino acid sequence of the bovine adenovirus type 3 proteinase. Nucleic Acids Res. 1990;18:5568. doi: 10.1093/nar/18.18.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caillet-Boudin M L. Complementary peptide sequences in partner proteins of the adenovirus capsid. J Mol Biol. 1989;208:195–198. doi: 10.1016/0022-2836(89)90095-8. [DOI] [PubMed] [Google Scholar]
- 12.Chase J W, Williams K R. Single stranded DNA-binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 13.Chiocca S, Kurzbauer R, Schaffner G, Baker A, Mautner V, Cotton M. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J Virol. 1996;70:2939–2949. doi: 10.1128/jvi.70.5.2939-2949.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow L T, Gelinas R E, Broker T R, Roberts R J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 15.Chroboczek J, Bieber F, Jacrot B. The sequence of the genome of adenovirus type 5 and its comparisons with the genome of adenovirus type 2. Virology. 1992;186:280–285. doi: 10.1016/0042-6822(92)90082-z. [DOI] [PubMed] [Google Scholar]
- 16.Chroboczek J, Ruigrok R W H, Cusack S. Adenovirus fiber. Curr Top Microbiol Immunol. 1995;199(1):163–200. doi: 10.1007/978-3-642-79496-4_10. [DOI] [PubMed] [Google Scholar]
- 17.Cuillel M, Milleville M, D’Halluin J C. Expression of human Ad2 IIIa protein in E. coli. Gene. 1987;55:295–301. doi: 10.1016/0378-1119(87)90289-7. [DOI] [PubMed] [Google Scholar]
- 18.Darbyshire J H. Oncogenicity of bovine adenovirus type 3 in hamsters. Nature (London) 1966;211:102. doi: 10.1038/211102a0. [DOI] [PubMed] [Google Scholar]
- 19.Darbyshire J H, Dawson P S, Lamont P H, Ostler D C, Pereira H G. A new adenovirus serotype of bovine origin. J Comp Pathol. 1965;75:327–330. doi: 10.1016/0021-9975(65)90038-1. [DOI] [PubMed] [Google Scholar]
- 20.Davison A J, Telford E A R, Watson M S, McBride K, Mautner V. The DNA sequence of adenovirus type 40. J Mol Biol. 1993;234:1308–1316. doi: 10.1006/jmbi.1993.1687. [DOI] [PubMed] [Google Scholar]
- 21.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 22.Dolph P J, Huang J, Schneider R J. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J Virol. 1990;64:2669–2677. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elgadi M M, Haj-Ahmad Y. Molecular cloning and restriction enzyme analysis of bovine adenovirus type 3. Intervirology. 1992;34:113–120. doi: 10.1159/000150272. [DOI] [PubMed] [Google Scholar]
- 24.Elgadi M M, Rghei N, Haj-Ahmad Y. Sequence and sequence analysis of E1 and pIX regions of the BAV3 genome. Intervirology. 1993;36:113–120. doi: 10.1159/000150329. [DOI] [PubMed] [Google Scholar]
- 25.Frasar N W, Baker C C, Moore M A, Ziff E B. Poly(A) sites of adenovirus serotype 2 transcription units. J Mol Biol. 1982;155:207–212. doi: 10.1016/0022-2836(82)90002-x. [DOI] [PubMed] [Google Scholar]
- 26.Grable M, Hearing P. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J Virol. 1990;64:2047–2056. doi: 10.1128/jvi.64.5.2047-2056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grand R J A. The structure and function of the adenovirus early region 1 proteins. Biochem J. 1987;241:25–38. doi: 10.1042/bj2410025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasson T B, Soloway P D, Ornelles D A, Doerfler W, Shenk T. Adenovirus L1 52-kilodalton proteins are required for assembly of virions. J Virol. 1989;63:3612–3621. doi: 10.1128/jvi.63.9.3612-3621.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hovanessian A G. The double stranded RNA-activated protein kinase induced by interferon: dsRNA-PK. J Interferon Res. 1989;6:641–647. doi: 10.1089/jir.1989.9.641. [DOI] [PubMed] [Google Scholar]
- 30.Hu S L, Hays W W, Potts D E. Sequence homology between bovine and human adenoviruses. J Virol. 1984;49:604–608. doi: 10.1128/jvi.49.2.604-608.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Idamakanti, N., P. S. Reddy, L. A. Babiuk, and S. K. Tikoo. Submitted for publication.
- 32.Imler J-L. Adenovirus vectors as recombinant viral vaccines. Vaccine. 1995;13:1143–1151. doi: 10.1016/0264-410x(95)00032-v. [DOI] [PubMed] [Google Scholar]
- 33.Kaufman R S, Ginsberg H S. Characterization of a temperature-sensitive, hexon transport mutant of type 5 adenovirus. J Virol. 1976;19:643–658. doi: 10.1128/jvi.19.2.643-658.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitchingman G R. Sequence of the DNA-binding protein of a human subgroup E adenovirus (type 4) and comparisons with subgroup A (type 12), subgroup B (type 7) and subgroup C (type 5) Virology. 1985;146:90–101. doi: 10.1016/0042-6822(85)90055-8. [DOI] [PubMed] [Google Scholar]
- 35.Komoriya A, Green L H, Mervic M, Yamada S S, Yamada K M, Humphries M H. The minimum essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J Biol Chem. 1991;266:15075–15079. [PubMed] [Google Scholar]
- 36.Kurokawa T, Igarashi K, Sugino Y. Biochemical studies on bovine adenovirus type 3. III. Cleavage maps of viral DNA by restriction endonucleases EcoRI, BamHI, and HindIII. J Virol. 1978;28:212–218. doi: 10.1128/jvi.28.1.212-218.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsson S, Bellett A, Akusjarvi G. VA RNAs from avian and human adenoviruses: dramatic differences in length, sequence, and gene location. J Virol. 1986;58:600–609. doi: 10.1128/jvi.58.2.600-609.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, J. B., M. K. Baxi, N. Idamakanti, P. S. Reddy, A. N. Zakhartchouk, C. Pyne, L. A. Babiuk, and S. K. Tikoo. Submitted for publication.
- 38a.Lutz P, Rosa-Calatrava M, Kedinger C. The product of the adenovirus intermediate gene IX is a transcriptional activator. J Virol. 1997;71:5102–5109. doi: 10.1128/jvi.71.7.5102-5109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangel W F, McGrath W J, Toledo D L, Anderson C W. Viral DNA and a viral peptide act as cofactors of adenovirus virion proteinase activity. Nature (London) 1993;361:274–275. doi: 10.1038/361274a0. [DOI] [PubMed] [Google Scholar]
- 40.Mattson D E, Norman B B, Dunbar J R. Bovine adenovirus type 3 infection in feedlot calves. Am J Vet Res. 1988;49:67–69. [PubMed] [Google Scholar]
- 41.Mittal S K, Prevec L, Babiuk L A, Graham F L. Corrigendum. Sequence analysis of bovine adenovirus type 3 early region 3 and fiber protein genes. J Gen Virol. 1993;74:2825. doi: 10.1099/0022-1317-74-12-2825. [DOI] [PubMed] [Google Scholar]
- 42.Mohanty B S. Comparative study of bovine adenoviruses. Am J Vet Res. 1971;32:1899–1905. [PubMed] [Google Scholar]
- 43.Morin N, Delsert C, Klessig D F. Mutations that affect phosphorylation of the adenovirus DNA-binding protein alters its ability to enhance its own synthesis. J Virol. 1989;63:5228–5237. doi: 10.1128/jvi.63.12.5228-5237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morin N, Delsert C, Klessig D F. Nuclear localization of the adenovirus DNA-binding protein: requirement for two signals and complementation during viral infection. Mol Cell Biol. 1989;9:4372–4380. doi: 10.1128/mcb.9.10.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrison M D, Onions D E, Nicolson L. Complete DNA sequence of canine adenovirus type 1. J Gen Virol. 1997;78:873–878. doi: 10.1099/0022-1317-78-4-873. [DOI] [PubMed] [Google Scholar]
- 46.Niiyama Y, Igarashi K, Tsukamoto K, Kurokawa T, Sugino Y. Biochemical studies on bovine adenovirus type 3. I. Purification and properties. J Virol. 1975;16:621–633. doi: 10.1128/jvi.16.3.621-633.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oosterom-Dragon E A, Anderson C W. Polypeptide structure and encoding location of the adenovirus serotype 2 late, nonstructural 33K protein. J Virol. 1983;45:251–263. doi: 10.1128/jvi.45.1.251-263.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oosterom-Dragon E A, Ginsberg H S. Characterization of two temperature-sensitive mutants of type 5 adenovirus with mutations in the 100,000 dalton protein gene. J Virol. 1981;40:491–500. doi: 10.1128/jvi.40.2.491-500.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raviprakash K S, Grunhaus A, El Kholy M A, Horwitz M. The mouse adenovirus type 1 contains an unusual E3 region. J Virol. 1989;63:5455–5458. doi: 10.1128/jvi.63.12.5455-5458.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy, P. S., N. Idamakanti, L. A. Babiuk, and S. K. Tikoo. Submitted for publication.
- 51.Roberts R J, Akusjarvi G, Alestrom P, Gelinas R E, Gingeras T R, Sciaky D, Pettersson U. Adenovirus DNA. In: Doerfler W, editor. Adenovirus DNA. Boston, Mass: Martinus Nijhoff; 1986. pp. 1–51. [Google Scholar]
- 52.Rohn K, Prusas C, Monereal G, Hess H. Identification and characterization of penton base and pVIII protein of egg drop syndrome virus. Virus Res. 1997;47:59–65. doi: 10.1016/s0168-1702(96)01407-4. [DOI] [PubMed] [Google Scholar]
- 53.Ruigrok R W H, Barge A, Mittal S K, Jacrot B. The fiber of bovine adenovirus type 3 is very long but bent. J Gen Virol. 1994;75:2069–2073. doi: 10.1099/0022-1317-75-8-2069. [DOI] [PubMed] [Google Scholar]
- 54.Russell W C, Kemp G D. Role of adenovirus structural components in the regulation of adenovirus infection. Curr Top Microbiol Immunol. 1995;199(1):81–98. doi: 10.1007/978-3-642-79496-4_6. [DOI] [PubMed] [Google Scholar]
- 55.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shenk T, Flint S J. Transcriptional and transforming activities of the adenovirus E1A proteins. Adv Cancer Res. 1991;57:47–85. doi: 10.1016/s0065-230x(08)60995-1. [DOI] [PubMed] [Google Scholar]
- 57.Shinagawa M, Lida Y, Matsuda A, Tsuiyama T, Sato G. Phylogenetic relationships between adenoviruses as inferred from nucleotide sequences of inverted terminal repeats. Gene. 1987;55:85–93. doi: 10.1016/0378-1119(87)90251-4. [DOI] [PubMed] [Google Scholar]
- 58.Song B, Hu S, Darai G, Spindler K, Young C S H. Conservation of DNA sequence in the predicted major late promoter region of selected mastadenoviruses. Virology. 1996;220:390–401. doi: 10.1006/viro.1996.0327. [DOI] [PubMed] [Google Scholar]
- 59.Sprengel J, Schmitz B, Heuss-Neitzel D, Zock C, Doerfler W. Nucleotide sequence of human adenovirus type 12 DNA: comparative functional analysis. J Virol. 1994;68:379–389. doi: 10.1128/jvi.68.1.379-389.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sung M T, Lische M A, Richards J C, Hosokawa K. Adenovirus chromatin. I. Isolation and characterization of the major core protein VII and precursor pro-VIII. J Biol Chem. 1977;252:4981–4988. [PubMed] [Google Scholar]
- 61.Sussenbach J S. The structure of the genome. In: Ginsberg H S, editor. The adenovirus. New York, N.Y: Plenum Press; 1984. pp. 35–172. [Google Scholar]
- 62.Tribouley C, Lutz P, Staub A, Kedinger C. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J Virol. 1994;68:4450–4457. doi: 10.1128/jvi.68.7.4450-4457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsernoglou D, Tsugita A, Tucker A D, Van der Vliet P C. Characterization of the chymotryptic core of the adenovirus DNA binding protein. FEBS Lett. 1985;188:248–252. doi: 10.1016/0014-5793(85)80381-1. [DOI] [PubMed] [Google Scholar]
- 64.Tsukamoto K, Sugino Y. Nonproductive infection and induction of cellular deoxyribonucleic acid synthesis by bovine adenovirus type 3 in a contact-inhibited mouse cell line. J Virol. 1972;9:465–473. doi: 10.1128/jvi.9.3.465-473.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tucker P A, Tsernoglou D, Tucker A D, Coenjaerts F E J, Leenders H, Van der Vliet P C. Crystal structure of the adenovirus DNA binding protein reveals a hook-on model for cooperative DNA binding. EMBO J. 1994;13:2994–3002. doi: 10.1002/j.1460-2075.1994.tb06598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Der Vliet P C. Adenovirus DNA replication. Curr Top Microbiol Immunol. 1995;199(2):1–30. doi: 10.1007/978-3-642-79499-5_1. [DOI] [PubMed] [Google Scholar]
- 67.van Oostrum J, Burnett R M. Molecular composition of the adenovirus type 2 virion. J Virol. 1985;56:439–448. doi: 10.1128/jvi.56.2.439-448.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vrati S, Boyle D B, Kocherhans R, Both G W. Sequence of ovine adenovirus homologs for 100K hexon assembly, 33K, pVIII and fiber genes: early region E3 is not in the expected location. Virology. 1995;209:400–408. doi: 10.1006/viro.1995.1272. [DOI] [PubMed] [Google Scholar]
- 69.Vrati S, Brookes D E, Boyle D B, Both G W. Nucleotide sequence of ovine adenovirus tripartite leader sequence and homologues of IVa2, DNA polymerase and terminal proteins. Gene. 1996;177:35–41. doi: 10.1016/0378-1119(96)00266-1. [DOI] [PubMed] [Google Scholar]
- 70.Vrati S, Brookes D E, Strike P, Khatri A, Boyle D B, Both G W. Unique genome arrangement of an ovine adenovirus: identification of new proteins and proteinase cleavage sites. Virology. 1996;220:186–199. doi: 10.1006/viro.1996.0299. [DOI] [PubMed] [Google Scholar]
- 71.Weber J M. Adenovirus endopeptidase and its role in virus infection. Curr Top Microbiol Immunol. 1995;192:227–235. doi: 10.1007/978-3-642-79496-4_12. [DOI] [PubMed] [Google Scholar]
- 72.Wicham T J, Filardo E J, Cheresh D A, Nemerow G R. Integrin-alpha-V-beta 3 and integrin-alpha-V-beta-5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 73.Wold W S M, Gooding L R. Region E3 of adenovirus: a cassette of genes involved in host immunosurveillance and virus-cell interaction. Virology. 1991;184:1–8. doi: 10.1016/0042-6822(91)90815-s. [DOI] [PubMed] [Google Scholar]
- 74.Xu Z Z, Hyatt A, Boyle D B, Both G W. Construction of ovine adenovirus recombinants by gene insertions or deletions of related terminal region sequences. Virology. 1997;230:62–71. doi: 10.1006/viro.1997.8452. [DOI] [PubMed] [Google Scholar]
- 75.Zheng B, Mittal S K, Graham F L, Prevec L. The E1 sequence of bovine adenovirus type 3 and complementation of human adenovirus type E1a function in bovine cells. Virus Res. 1994;31:163–186. doi: 10.1016/0168-1702(94)90002-7. [DOI] [PubMed] [Google Scholar]