Abstract
Deletion mutants of the pathogenic clone of simian immunodeficiency virus isolate 239 (SIVmac239) were derived that are missing nef, vpr, and upstream sequences (US) in the U3 region of the LTR (SIVmac239Δ3), nef, vpx, and US (SIVmac239Δ3x), and nef, vpr, vpx, and US (SIVmac239Δ4). These multiply deleted derivatives replicated well in the continuously growing CEMx174 cell line and were infectious for rhesus monkeys. However, on the basis of virus load measurements, strength of antibody responses, and lack of disease progression, these mutants were highly attenuated. Measurements of cell-associated viral load agreed well with assays of plasma viral RNA load and with the strengths of the antibody responses; thus, these measurements likely reflected the extent of viral replication in vivo. A derivative of SIVmac239 lacking vif sequences (SIVmac239Δvif) could be consistently grown only in a vif-complementing cell line. This Δvif virus appeared to be very weakly infectious for rhesus monkeys on the basis of sensitive antibody tests only. The weak antibody responses elicited by SIVmac239Δvif were apparently in response to low levels of replicating virus since they were not elicited by heat-inactivated virus and the anti-SIV antibody responses persisted for greater than 1 year. These results, and the results of previous studies, allow a rank ordering of the relative virulence of nine mutant strains of SIVmac according to the following order: Δvpr > Δvpx > ΔvprΔvpx ≅ Δnef > Δ3 > Δ3x ≥ Δ4 > Δvif > Δ5. The results also demonstrate that almost any desired level of attenuation can be achieved, ranging from still pathogenic in a significant proportion of animals (Δvpr and Δvpx) to not detectably infectious (Δ5), simply by varying the number and location of deletions in these five loci.
Attempts to develop a vaccine for the prevention of AIDS in humans have relied heavily on macaque monkey models that utilize simian immunodeficiency virus (SIV) (9). SIV closely parallels its human counterpart, human immunodeficiency virus (HIV), in genetic makeup and biological properties. We have used SIVmac239 derived from infectious cloned DNA of defined sequence (16) and uncloned, early-passage SIVmac251 (5, 21) for our vaccine studies. These viruses produce consistently high virus loads that can be readily measured in rhesus monkeys, and they induce AIDS in rhesus monkeys in a time frame suitable for laboratory investigation. These strains resemble primary HIV type 1 (HIV-1) isolates in that they are difficult to neutralize even with sera from infected animals (24).
Trials in rhesus monkeys have shown that vaccine protection against SIVmac239 and SIVmac251 is very difficult to achieve even under idealized laboratory conditions. Vaccine trials that have used inactivated whole virus, envelope protein, vaccinia virus recombinants, multivalent vaccinia virus recombinants, and multivalent vaccinia virus recombinants followed by particle boosting have shown little or no protection against challenge by these viruses (7, 10, 22, 26, 30, 31). These vaccine failures have occurred despite extensive attempts to match the strain of challenge virus closely to the vaccine and to time the challenge at or near the peak of vaccine-induced immune responses.
In contrast to these vaccine failures, the live attenuated vaccine approach has afforded impressive protection against challenge by SIVmac251 and SIVmac239 (1, 4, 35). Some studies have used vaccine strains with defined attenuating mutations (1, 4, 35), while others have used attenuated or partially attenuated strains whose attenuating mutations have not been defined (2, 23). In our studies, we have demonstrated strong protective immunity by vaccination with SIVmac239Δnef (lacking nef sequences) and SIVmac239Δ3 (lacking nef, vpr, and US [upstream sequences of U3]) (4, 35). The live attenuated vaccine approach has not, however, been universally successful in protecting against SIV (2, 23, 35). The length of time between vaccination and challenge appears to be one variable that influences protective efficacy with the live attenuated approach (35); other factors that determine protective efficacy have not been defined.
Most viral vaccines currently in use in humans are live attenuated strains of virus. Extensive experience with the development and testing of these viral vaccines in humans has demonstrated the importance of seeking a critical balance between safety and potency (28). The vaccine strain should be attenuated enough to ensure relative safety in the target population but potent enough to induce good protective immunity. Whether this concept also holds for live attenuated vaccination against SIV and HIV remains to be demonstrated.
We have targeted five regions of the SIV genome for attenuating mutations. These five regions are the nef gene, the vpx gene, the vpr gene, the vif gene, and sequences in the upstream region of U3 in the long terminal repeat (US). In all cases, we have used the infectious, pathogenic SIVmac239 clone as the starting parental strain and have introduced large deletions in order to prevent reversion at the targeted locus (12). For deletions involving vpr and vpx, we have demonstrated the normal expression of adjacent open reading frames (11). While US is probably nothing more than nef coding sequence (13, 18, 19), it will be treated as a discrete entity for the purpose of this report. Previous publications have described the properties of Δnef, Δvpr, Δvpx, ΔvprΔvpx, and Δ3 constructs in rhesus monkeys (11, 17, 35). Here we describe the properties of even more highly attenuated strains Δ3x (missing nef, vpx, and US), Δ4 (missing nef, vpr, vpx, and US), and Δvif (missing vif) as they relate to Δ3 and the other strains studied previously.
MATERIALS AND METHODS
vif-complementing CEMx174 cell line.
The amphotropic Moloney murine leukemia virus packaging cell line gpenvAm12 was propagated in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS) and 2 mM glutamine (Gibco). Hypoxanthine, xanthine, and mycophenolic acid (Gibco) were added on a weekly basis to select for cells expressing retrovirus packaging components. The SIV vif gene was amplified by PCR from 10 ng of pSIVSphSph5′ plasmid template DNA with oligonucleotides 5′SIVvifEcoRI and 3′SIVvifBamHI. Amplified SIV vif DNA was molecularly cloned into bacterial plasmid pCRII, using a TA cloning kit (Promega) according to the manufacturer’s directions. The TA vector intermediate was purified, verified by DNA sequencing across the vif gene, and digested with EcoRI and BamHI. Similarly digested pLXSN Moloney murine leukemia virus bacterial plasmid was purified by electrophoresis in low-melting-point agarose (FMC) and cut from the gel. Melted gel fragments containing appropriate amounts of DNA were ligated, thereby molecularly cloning the SIV and HIV-1 vif genes between the EcoRI and BamHI restriction sites of pLXSN. The final plasmid, pLXSIVvif, and the empty vector pLXSN were transfected into gpenvAm12 cells by using DEAE-dextran as previously described (12). Transfected cells were selected with G418 (0.6 mg/ml; Gibco) beginning 24 h posttransfection. The cell lines were then named gpenvSIVvif and pgenvLXSN. Three weeks postselection, 5 ml of supernatant was drawn off these cell lines and used to infect CEMx174 cells, which were selected with G418 (0.6 mg/ml) beginning 24 h postinfection. Resultant selected cell lines were designated CEMxSIVvif and CEMxLXSN.
Virus inoculation into rhesus monkeys.
In all cases except Δvif virus, virus stocks were prepared by transfection of cloned DNA into CEMx174 cells by using DEAE-dextran (12). For SIVΔvif, the vif-complementing CEMx174 cell line was used. Virus was harvested from the cell-free supernatant at or near the peak of virus production, usually 8 to 11 days after transfection. All animals were inoculated intravenously. Inocula were normalized according to the content of p27gag antigen, determined with a commercial antigen capture kit (Coulter, Hialeah, Fla.). Inocula containing 100 ng of p27 of SIVΔ3, SIVΔ3x, or SIVΔ4 were used for each animal in experiment A (Table 1). Viruses containing 34, 396, and 325 ng of p27 were used for experiments B, D, and F, respectively, in Table 1. We noticed no effect of inoculum dose on virus load. SIVΔvif-inoculated animals 166-89 and 427-92 received virus containing 360 ng of p27, animal 151-93 received SIVΔvif containing 180 ng of p27, and animal 180-93 received heat-inactivated SIVΔvif containing 180 ng of p27. Peripheral blood samples were taken at intervals and used to assay the numbers of infectious cells in peripheral blood mononuclear cells (PBMC) (4, 35) and to determine anti-SIV antibody responses by enzyme-linked immunosorbent assay (ELISA) (6, 7, 11, 17, 35); PBMC were stored for DNA and PCR (4, 35), and plasma was stored for RNA quantitation.
TABLE 1.
Summary of attenuated mutants of SIVmac239
| Strain | Gene(s) deleted | No. of monkeys analyzed | Expt(s) (no. of animals studied) | AIDS | Relative virulencea |
|---|---|---|---|---|---|
| SIVmac239 | None | NAb | Yes | 1.0 | |
| SIVmac239Δvpr | vpr | NA | Yes | 0.5 | |
| SIVmac239Δvpx | vpx | NA | Yes | 0.1 | |
| SIVmac239ΔvpxΔvpr | vpx, vpr | NA | No | 0.01 | |
| SIVmac239Δnef | nef | NA | No | 0.01 | |
| SIVmac239Δ3 | nef, vpr, US | 9 | A (4), B (5) | No | 0.005 |
| SIVmac239Δ3x | nef, vpx, US | 6 | A (4), C (2) | No | 0.001 |
| SIVmac239Δ4 | nef, vpx, vpr, US | 6 | A (4), D (2) | No | 0.0001 |
| SIVmac239Δvif | vif | 3 | E (3) | No | 0.00005 |
| SIVmac239Δ5 | nef, vpx, vpr, US, vif | 2 | F (2) | No | 0 |
The extent of viral replication in vivo as reflected by cell-associated viral loads, plasma viral RNA loads, and strengths of antibody responses.
NA, not applicable.
Plasma RNA quantification.
Virion-associated SIV RNA in plasma samples was quantified as an index of ongoing viral replication by using a real-time reverse transcription-PCR assay on an Applied Biosystems Prism 7700 sequence detection system as described in detail elsewhere (32). For each test specimen, three reactions that each used 20% of the total extracted RNA were performed. Duplicate aliquots were separately reverse transcribed and amplified, and the amplification cycle during which detectable PCR product was first observed (threshold cycle) was determined from real-time kinetic analysis of fluorescent product generation as a consequence of specific amplification (32). One reaction was processed and amplified without addition of reverse transcriptase. Nominal copy numbers for test samples were then automatically calculated by interpolation of the experimentally determined threshold cycle values onto a regression curve derived from control transcript standards, followed by normalization for the volume of the extracted plasma specimen. Results shown are averages of duplicate determinations. The average interassay coefficient of variation was 25% on replicate plasma aliquots.
Western blot analysis of sera from infected monkeys.
One milligram (Lowry protein) of sucrose density gradient-purified SIV (Mne)/HUT78 clone E11S was subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (20) using a 12- by 14-cm gel. A 5-mm-wide lane of prestained molecular weight standards (Kaleidoscope; Bio-Rad) was included on each gel. A single 12.2-cm-wide lane was used for the virus. The separated proteins were electroblotted (34) to an Immobilon membrane (Millipore) for 1 h at 1.5 A in transfer buffer (0.025 M Tris base, 0.192 M glycine, 15% methanol, and 0.01% SDS in Milli-Q water). The membrane was blocked with 3% gelatin in Tris-buffered saline (TBS; 20 mM Tris-HCl [pH 7.5] and 0.5 M NaCl in Milli-Q water), air dried, and sliced into 4-mm-wide strips (strips represent ∼30 μg of SIV). Plasma samples (10 μl) were preincubated with 90 μl of heat-inactivated FBS for 1 h at 37°C in 15-ml tubes in an attempt to adsorb any antibodies that bind to FBS proteins. (FBS proteins are contaminants of the purified SIV used to make the strips). After addition of 5 ml of TBS containing 1% gelatin, strips were added and incubated overnight at room temperature on a rocking platform. Each strip was decanted, transferred to an incubation tray, and washed five times for 10 min each with TBS containing 1% Tween 20 and 0.1% SDS. Three milliliters of 1:10,000 peroxidase-labeled goat anti-human immunoglobulin G (Sigma) in TBS containing 1% gelatin was added to each strip and incubated for 1 h at room temperature on a rocking platform. After five washes as before, 2 ml of enhanced chemiluminescence (ECL) reagent (Amersham) was added to each strip. After 1 min, strips were arranged on a clear sheet protector and placed on X-ray film (Lumi-Film; Boehringer Mannheim) for 1 to 10 min. Films were developed with an automated processor (Konica SRX-101).
Western blot analysis of virus preparations.
Wild-type SIVmac239open and SIVmac239Δvpr virus stocks were prepared by transfection of cloned DNAs into CEMx174 cells as described previously (12). For SIVmac239Δvif, the vif-complementing CEMx174 cell line was used. Cells were removed from 250 ml of culture by centrifugation at 17,500 rpm for 3 h in a type 19 rotor. Virus pellets were resuspended in 50 μl of phosphate-buffered saline (PBS) containing 0.5% Triton X-100, and p27gag antigen concentrations of the virus stocks were determined by using a commercial antigen capture kit (Coulter).
Polyclonal Vpr-specific and Vif-specific antisera were raised in rabbits by using α-β-galactosidase-Vpr and -Vif fusion proteins, respectively. Monoclonal antibody to SIV p27gag was harvested from the FA2 hybridoma cell line (33). Virus preparations containing 200 ng of p27gag were treated with Laemmli sample buffer, electrophoresed through an SDS–15% polyacrylamide gel, and electroblotted onto Immobilon-P membranes (Millipore, Bedford, Mass.). Membranes were first blocked with 8% skim milk in PBS–0.05% Tween 20 (PBST) for 1 h and then incubated with a 1:500 dilution of the corresponding antiserum in the same blocking solution for 1 h at room temperature. Primary antibodies were removed by washing the membranes three times with PBST at room temperature. Dilutions of the secondary antibodies and detection were performed according to the protocol for the Amersham ECL system.
RESULTS
Stocks of SIVmac239Δ3, -Δ3x, and -Δ4 were produced by transfection of cloned DNA into CEMx174 cells and harvesting virus from the cell-free supernatant at or near the peak of virus production. The precise size and limit of deletions in these strains can be found in reference 12. Virus stocks were stored in the vapor phase of liquid nitrogen at approximately −160°C. All three of these multiply deleted viruses replicated well in CEMx174 cells (12). In one experiment (experiment A [Table 1]), four rhesus monkeys were inoculated intravenously with normalized amounts of each strain, Δ3, Δ3x, and Δ4, containing 100 ng of p27 antigen. Other monkeys were infected with these same strains at different times (experiments B to D [Table 1]). We observed no unusual variations in the behavior of these viruses with infections initiated at different times.
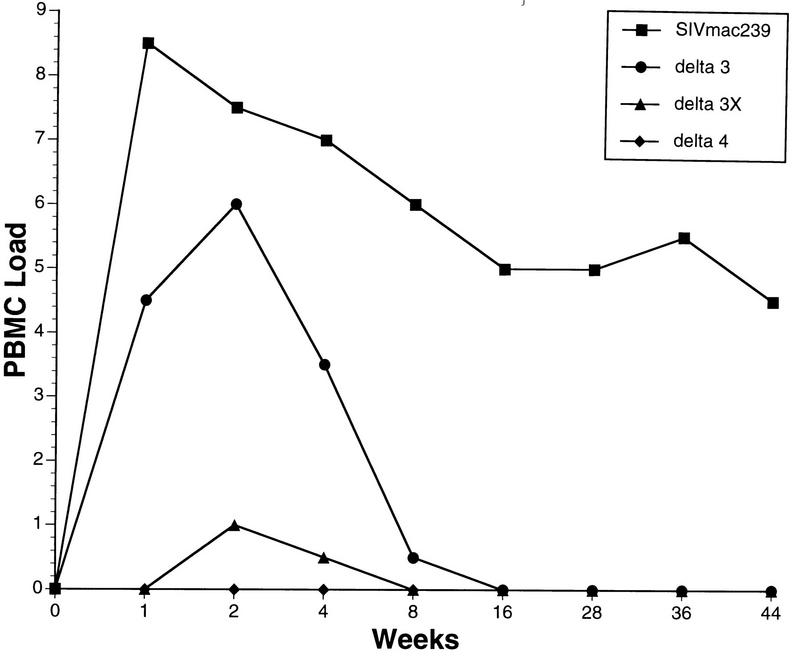
Cell-associated virus loads were measured by limiting dilution culture of PBMC (Fig. 1). Peak virus loads in this assay around week 2 decreased stepwise in the order Δ3→Δ3x→Δ4. This result is consistent with previous virus load measurements in animals infected with Δvpr, Δvpx, and ΔvprΔvpx in which deletion of vpx had a greater effect than deletion of vpr and deletion of vpx and vpr had a much greater effect than deletion of either element alone (11). Peak viral loads occurred around week 2 postinoculation. With parental SIVmac239, an average of 305 PBMC were required to recover virus at peak, compared to 4,115 PBMC for Δ3, 106 PBMC for Δ3x, and >106 PBMC for Δ4. Thus, peak loads in this assay for the parental wild type were 13.5 times higher than those obtained with Δ3, which in turn were 243 times higher than those obtained with Δ3x. Peak virus loads with this assay were thus approximately 3,000 times higher with parental SIVmac239 than with Δ3x and even greater than with Δ4. While not evident in Fig. 1, SIVΔ4 was occasionally recovered from inoculated animals in bulk cultures when 5 × 106 or more PBMC were used in both experiments A and D. SIVΔ4 was recovered from one animal at week 1 only, one animal at weeks 2 and 4 only, and one animal at week 40 only, all with ≥5 × 106 PBMC. SIV was never recovered from two of the six SIVΔ4-infected animals that were studied. Virus was more frequently recovered in these bulk cultures with SIVΔ3x-infected animals in experiments A and C.
FIG. 1.
Cell-associated virus loads. The numbers of infectious cells in PBMC are expressed in code on the y axis as a function of weeks after inoculation of virus. Each curve represents the average of multiple animals. For SIVmac239, averages of 16 and 18 animals were used for weeks 1 and 2; averages of 5 to 9 animals were used for the subsequent times due to the lack of sampling of individual animals at those times or due to the planned sacrifice of the animal. Five animals from experiment B were used for Δ3, two animals from experiment C were used for Δ3x, and two animals from experiment D were used for Δ4. Four animals that received SIVmac239Δ4 at TSI Mason Laboratories yielded very similar results (22a). Code for PBMC load: 0, virus was not recovered even when 106 PBMC were used; 1, virus was recovered with an average of 106 but not fewer PBMC; 2, 333,333 PBMC; 3, 111,111 PBMC; 4, 37,037 PBMC; 5, 12,345 PBMC; 6, 4,115 PBMC; 7, 1,371 PBMC; 8, 457 PBMC.
DNA was prepared from selected PBMC samples obtained at 2 and 16 weeks postinfection and used in a sensitive nested PCR assay for the detection of viral DNA (4, 35). The results overall were in good agreement with the cell-associated viral load measurements. Viral DNA was detected in two of three animals infected with Δ3 at 2 weeks after infection and in three of three animals infected with Δ3 at 16 weeks after infection. In contrast, viral DNA was detected in none of four animals infected with Δ4 at both 2 and 16 weeks after infection.
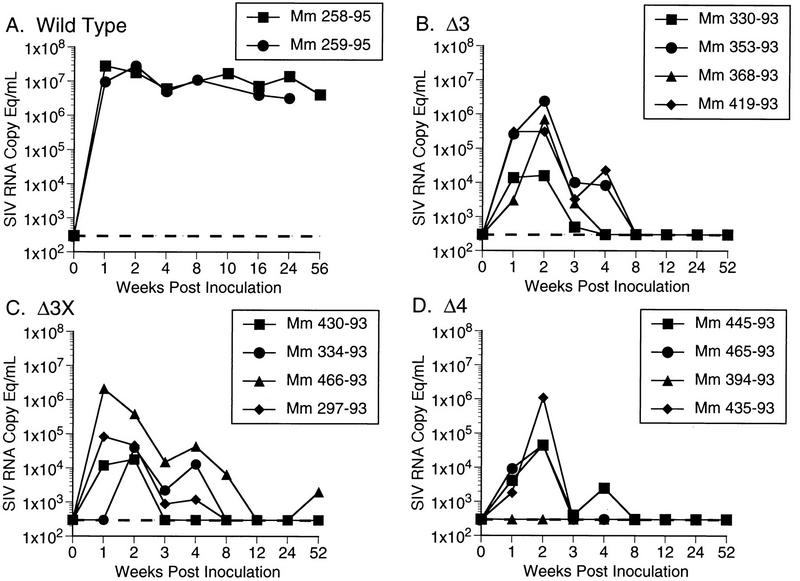
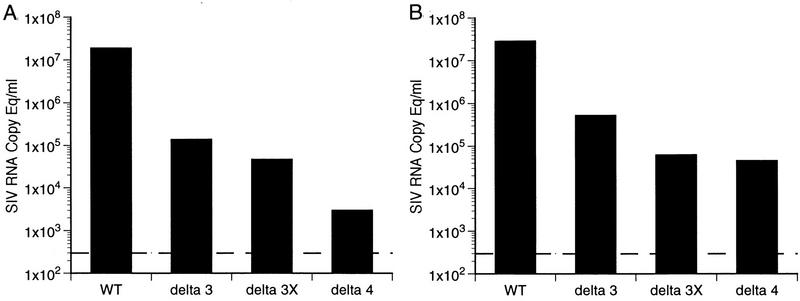
Plasma samples from all 12 animals infected with Δ3, Δ3x, and Δ4 in experiment A were used to quantify viral RNA burdens (Fig. 2). While there was some overlap in peak levels for individual animals infected with different viruses, the results overall agreed quite nicely with the cell-associated viral load measurements shown in Fig. 1. For all of the viruses evaluated, plasma SIV RNA levels peaked within the first 2 weeks following inoculation (Fig. 2). Early plasma RNA viral loads exhibited a similar rank order, i.e., wild type ≫ Δ3 > Δ3x > Δ4 (Fig. 3). For animals infected with all viruses other than wild type, plasma SIV RNA levels decreased to below the threshold sensitivity of the assay used (300 copy eq/ml) between 8 and 12 weeks postinoculation (Fig. 2).
FIG. 2.
Plasma SIV RNA levels at the indicated weeks postinoculation for animals infected with wild-type SIVmac239 (A), Δ3 (B), Δ3x (C), and Δ4 (D) for the 12 animals in experiment A and two controls analyzed separately. Dashed lines indicate threshold sensitivity of the assay, 300 copy eq/ml.
FIG. 3.
Early plasma SIV RNA loads. The median plasma RNA values at week 1 postinoculation (A) and at peak (B) for each group of infected animals are derived from the data shown in Fig. 2. The dashed lines indicate the threshold sensitivity of the assay.
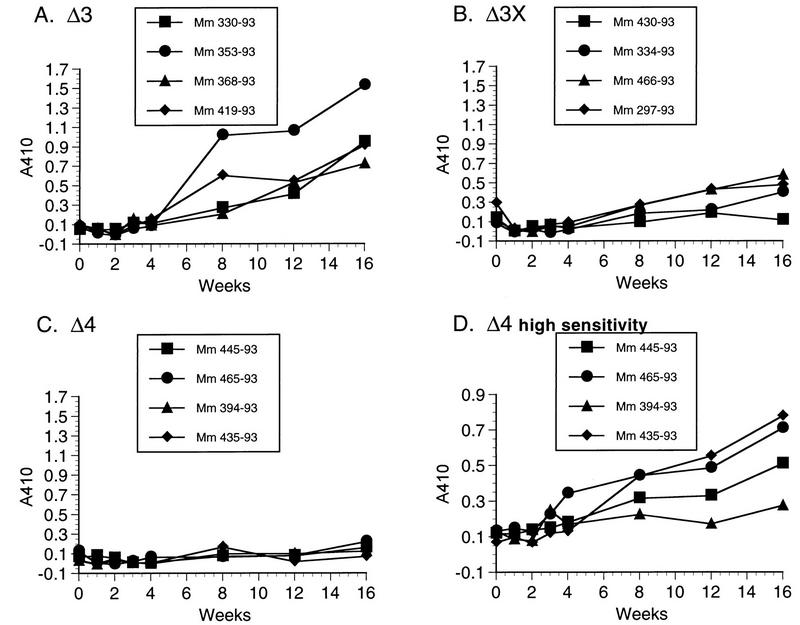
The strengths of the anti-SIV antibody responses varied with each mutant strain, with the same rank ordering as the levels of replicating virus and plasma RNA that were detected. This is best illustrated by the 12 animals infected in parallel in experiment A, whose antibody responses are shown in Fig. 4. SIVmac239Δ3x infection induced anti-SIV antibodies that were readily detected by ELISA, but they were considerably lower in strength than those detected in the animals infected with Δ3 (Fig. 4A and B). Under the identical assay conditions and parallel testing, anti-SIV antibodies were not evident in the Δ4-inoculated monkeys (Fig. 4C). However, in a much more sensitive ELISA testing format, the induction of anti-SIV antibodies could be readily measured in the Δ4-infected animals (Fig. 4D). The ultrasensitive assay conditions included more virus antigen per well (lysed virus containing 91 ng of p27 antigen per well) and testing of test serum at a dilution of 1:10 and the alkaline phosphatase-conjugated goat anti-human detection serum at 1:40. The levels of anti-SIV antibodies resulting from Δ3 infection were similar to those resulting from parental SIVmac239 infection (data not shown).
FIG. 4.
Antibody responses. Plasma samples from the 12 rhesus monkeys infected in parallel in experiment A were tested for the development of anti-SIV antibody responses by ELISA reactivity to whole lysed virus. All of the samples in panels A to C were tested on the same day under identical conditions. The samples from the Δ4-infected monkeys were retested under the high-sensitivity ELISA conditions (D).
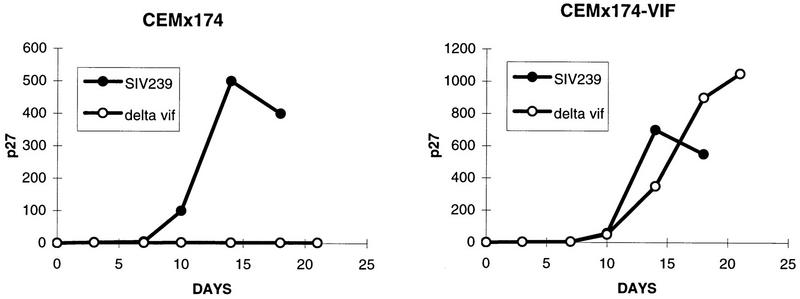
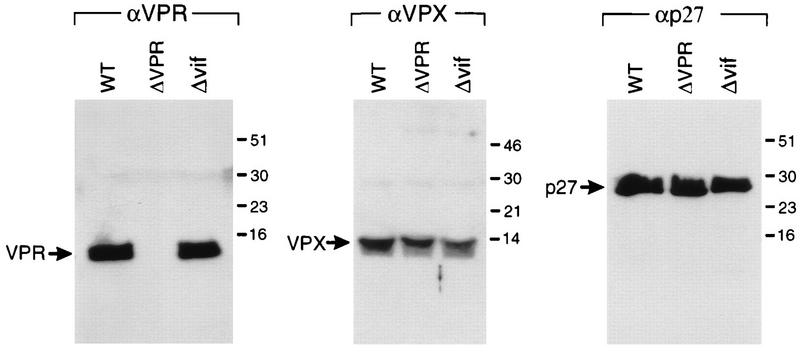
The properties of virus from which a large segment of vif sequences had been deleted were investigated next. The deletion in Δvif encompasses bp 5421 to 5655 (12, 29). The only way we have been able to grow the Δvif virus is in vif-complementing cell lines. Transfection of Δvif proviral DNA into CEMx174 cells did not yield detectable levels of replicating virus, but transfection into a vif-complementing CEMx174 cell line yielded high levels of replicating virus. SIVmac239Δvif derived from the vif-complementing cell line did not replicate appreciably in CEMx174 cells but replicated to high levels in the vif-complementing CEMx174 cells (Fig. 5). The Δvif virus also did not replicate detectably in lectin-stimulated rhesus monkey PBMC cultures in the presence of interleukin-2 (data not shown). The deletion in Δvif was demonstrated not to affect the normal expression of vpr and vpx by Western blot analysis (Fig. 6).
FIG. 5.
Replication of SIVmac239Δvif. Stock viruses containing 10 ng of p27 were tested in parallel for replication in CEMx174 cells and in the vif-completing CEMx174 cell line.
FIG. 6.
Synthesis of vpx and vpr by SIVmac239Δvif. Virion proteins of SIVmac239open (wild type [WT]), SIVmac239Δvpr (ΔVPR), and SIVmac239Δvif (Δvif) containing 200 ng of p27gag were separated in an SDS–15% polyacrylamide gel and electroblotted onto membrane filters. Vpr, Vpx, and p27gag proteins were detected by anti-Vpr (αVPR), anti-Vpx (αVPX), and anti-p27gag (αp27) antisera, respectively, using the Amersham ECL detection system. Sizes are indicated in kilodaltons.
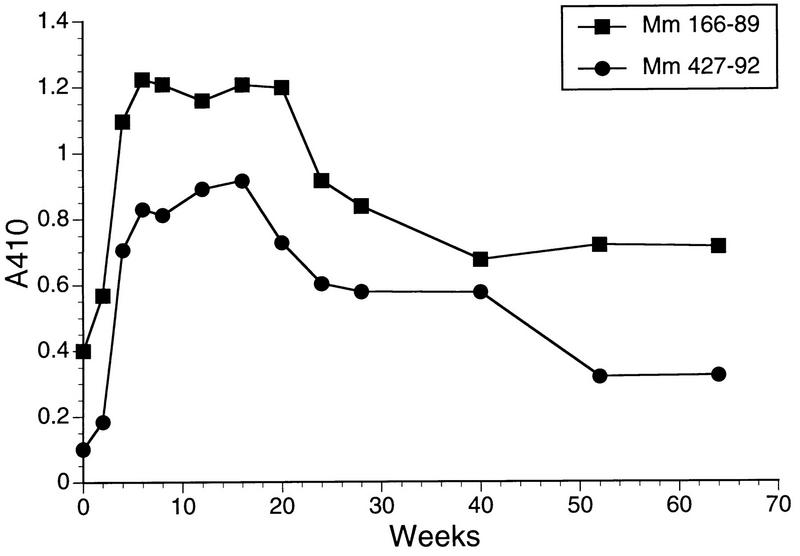
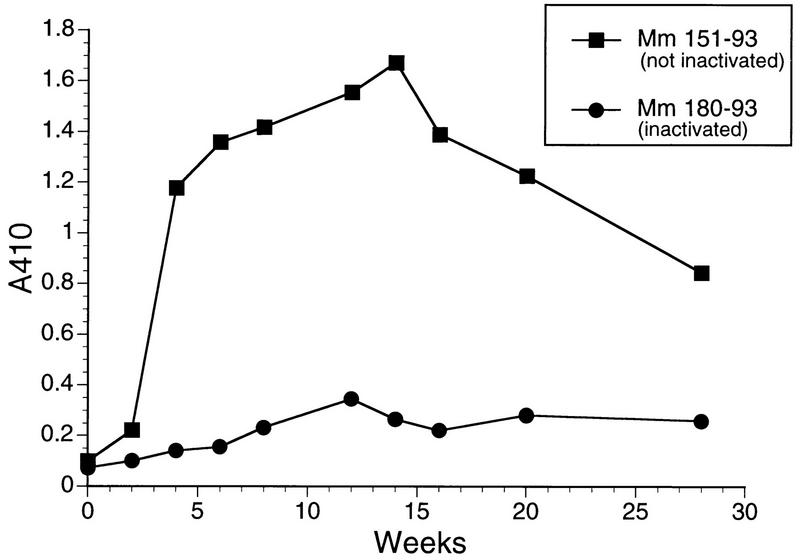
Rhesus monkeys 166-89 and 427-92 were inoculated intravenously with SIVmac239Δvif containing 360 ng of p27 prepared in the vif-complementing CEMx174 cell line. Attempts to recover SIV from PBMC were repeatedly negative, even with bulk cultures containing 106, 5 × 106, or 107 PBMC and the vif-complementing CEMx174 cell line. Analysis of sequential sera from these animals in the usual ELISA format did not reveal evidence for the emergence of anti-SIV antibodies in these animals. However, with the ultrasensitive ELISA format, anti-SIV antibodies clearly were demonstrable over time following the inoculation of the Δvif virus, and these antibodies persisted over the starting baseline levels for more than 1 year (Fig. 7).
FIG. 7.
Antibody responses to SIV in rhesus monkeys inoculated with SIVmac239Δvif. Sequential plasma samples from the rhesus monkeys inoculated with SIVmac239Δvif containing 360 ng of p27 were tested for the presence of antibodies to SIV under high-sensitivity conditions.
To investigate further whether the antibody response elicited by Δvif was indeed due to replicating virus, a vial of the Δvif stock virus produced in the vif-complementing cell line and containing 180 ng of p27 was thawed and split evenly into two tubes. One tube was heat inactivated at 56°C for 30 min, and the other tube was kept on ice. One rhesus monkey (180-93) was inoculated intravenously with the heat-inactivated Δvif virus, and another rhesus monkey (151-93) was inoculated with the Δvif virus that had not been heat inactivated. While 151-93 showed the same sort of anti-SIV antibody response with the sensitive ELISA system as was seen previously with 166-89 and 427-92, 180-93 had no such anti-SIV antibody response (Fig. 8). Repeated attempts to recover SIV with CEMx174 and the vif-complementing CEMx174 cell lines were again negative. These results provided further evidence that the anti-SIV antibody responses to Δvif virus were due to low levels of replicating virus.
FIG. 8.
Effect of heat inactivation of SIVmac239Δvif on antibody responses in inoculated rhesus monkeys. Sequential plasma samples were analyzed as described in the legend to Fig. 7.
We also attempted to detect SIV DNA by PCR in the animals inoculated with Δvif. SIV sequences were not detected in DNA of PBMC obtained at weeks 2 and 16 from animals 166-89, 427-92, 151-93, and 180-93 in the same nested PCR assays that readily detected SIV DNA in Δ3-inoculated animals (see above).
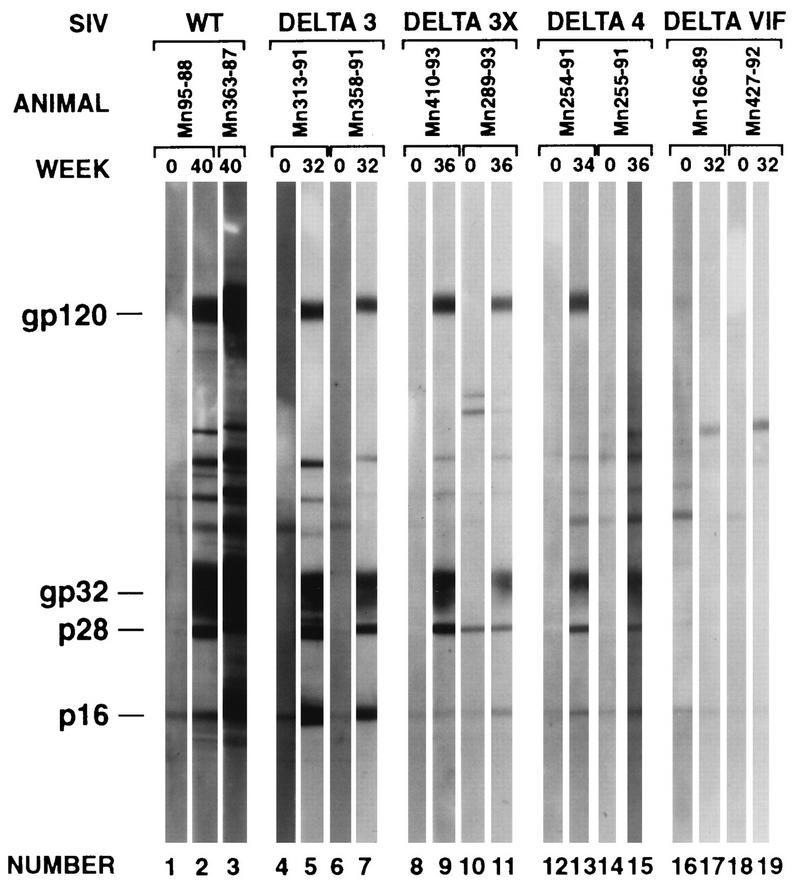
We analyzed plasma samples from selected animals for Western blot reactivity to viral antigens (Fig. 9). Plasma samples taken at week zero (preinoculation) and weeks 32 to 40 (postinoculation) were used. The results agreed very nicely with the quantitative ELISA measurements shown in Fig. 4 and in Fig. 7. Plasma from the Δvif-infected animals showed little or no reactivity under the standard conditions used (Fig. 9). Week 34–36 plasma samples from Δ4-infected animals showed weak but definite antibody levels that were considerably lower than those in the Δ3- and Δ3x-infected animals (Fig. 9).
FIG. 9.
Western blot reactivity of selected sera. WT, wild type.
DISCUSSION
Our results together with those of previous studies (11, 17, 35) demonstrate that almost any desired level of attenuation can be achieved simply by varying the number and location of deletion mutations in the five loci described in this report. Other loci, such as the NF-κB and Sp1 transcriptional control elements, can also be targeted singly and in combinations for attenuating deletion mutations (14, 15). The SIVmac239Δ4 and -Δvif mutant strains described in this report are extremely attenuated yet are still apparently infectious. However, animals infected with these attenuated strains routinely score negative in standard assays for infection, including routine antibody tests. A strain with deletions in all five loci (12) was not detectably infectious for rhesus monkeys in our hands. Our results allow ranking of the relative virulence of deletion mutants of SIVmac239 according to the following order: Δvpr > Δvpx > Δvpr Δvpx = Δnef > Δ3 > Δ3x > Δ4 Δvif > Δ5. We have shown good protective efficacy in a vaccine format with Δnef and Δ3 (4, 35). The vaccine capabilities of other strains remain to be determined.
To what extent can the relative virulence of these SIV strains be extrapolated to HIV-1? A number of long-term nonprogressing humans who are naturally infected with nef-deleted HIV-1 have been identified (8, 18). The level of attenuation of HIV-1Δnef in humans appears to be quite similar to the level of attenuation of SIVmac239Δnef in rhesus monkeys. However, there is no information on the attenuating effects of loss of other HIV-1 genes. There is no reason to presume a priori that the effects of loss of a corresponding gene or genes will necessarily be the same. Answers to this can probably only be derived from identification of rare individuals harboring nonrevertible defects at individual genomic locations.
While our studies to date have characterized the overall level of attenuation of these mutant strains, it will also be important to determine whether any of these mutations affect specific aspects, features, or characteristics of the infection. For example, mutation of specific genetic elements could conceivably alter cell type or tissue targeting, the primary site of replication within an individual lymphoid tissue, or viral dependence on the state of cellular activation. It is also possible that none of these mutations alters what the virus does or where it goes but simply results in the virus doing it less efficiently. Along these lines, it is worth noting that none of the infectious mutants that we have studied resulted in lack of persistence as a consistent phenotype. Several groups (3, 25, 27, 35) have noted rare animals with apparently transient infections, sometimes even with wild-type virus, in which anti-SIV antibodies increase after infection but subsequently decline to basal levels below the limit of deletion; none of the mutants exhibited these features of a nonpersisting infection as a consistent phenotype.
ACKNOWLEDGMENTS
We thank T. Wiltrout and G. Vasquez for plasma SIV RNA measurements and D. L. Xia, A. McPhee, D. Silva, R. Imig, J. Bess, and M. G. Moll for technical assistance. We also thank Prabhat Sehgal and Elaine Roberts for animal care, blood sampling, and clinical care.
This work was supported by PHS grants AI 35365, AI 25328, and RR 00168.
REFERENCES
- 1.Almond N, Kent K, Cranage M, Rud E, Clarke B, Stott E J. Protection by attenuated simian immunodeficiency virus in macaques against challenge with virus-infected cells. Lancet. 1995;345:1342–1344. doi: 10.1016/s0140-6736(95)92540-6. [DOI] [PubMed] [Google Scholar]
- 2.Clements J E, Montelaro R C, Zink M C, Amedee A M, Miller S, Trichel A M, Jagerski B, Hauer D, Martin L N, Bohm R P, Murphey-Corb M. Protective immune responses induced in rhesus macaques by immunization with attenuated macrophage-tropic simian immunodeficiency virus. J Virol. 1995;69:2737–2744. doi: 10.1128/jvi.69.5.2737-2744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clerici M, Clark E A, Polacino P, Axberg I, Kuller L, Casey N I, Morton W R, Shearer G M, Benveniste R E. T-cell proliferation to subinfectious SIV correlates with lack of infection after challenge of macaques. AIDS. 1994;8:1391–1395. doi: 10.1097/00002030-199410000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live-attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 5.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 6.Daniel M D, Letvin N L, Sehgal P K, Hunsmann G, Schmidt D K, King N W, Desrosiers R C. Long-term persistent infection of macaque monkeys with the simian immunodeficiency virus. J Gen Virol. 1987;68:3183–3189. doi: 10.1099/0022-1317-68-12-3183. [DOI] [PubMed] [Google Scholar]
- 7.Daniel M D, Mazzara G P, Simon M A, Sehgal P K, Kodama T, Panicali D L, Desrosiers R C. High titer immune responses elicited by recombinant vaccinia virus priming and particle boosting are ineffective in preventing virulent SIV infection. AIDS Res Hum Retroviruses. 1994;10:839–851. doi: 10.1089/aid.1994.10.839. [DOI] [PubMed] [Google Scholar]
- 8.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, Lawson V A, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan J S, Cunningham A, Dwyer D, Downton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 9.Desrosiers, R. C. 1995. Non-human primate models for AIDS vaccines. AIDS 9(Suppl. A):S137–S141. [PubMed]
- 10.Giavedoni L D, Planelles V, Haigwood N L, Ahmad S, Kluge J D, Marthas M L, Gardner M B, Luciw P A, Yilma T D. Immune response of rhesus macaques to recombinant simian immunodeficiency virus gp130 does not protect from challenge infection. J Virol. 1993;67:577–583. doi: 10.1128/jvi.67.1.577-583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs J S, Lackner A A, Lang S M, Simon M A, Sehgal P K, Daniel M D, Desrosiers R C. Progression to AIDS in the absence of genes for vpr or vpx. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of SIVmac mutants with deletions in the “nonessential” genes. AIDS Res Hum Retroviruses. 1994;10:607–616. doi: 10.1089/aid.1994.10.607. [DOI] [PubMed] [Google Scholar]
- 13.Ilyinskii P O, Daniel M D, Simon M A, Lackner A A, Desrosiers R C. The role of upstream U3 sequences in the pathogenesis of simian immunodeficiency virus-induced AIDS in rhesus monkeys. J Virol. 1994;68:5933–5944. doi: 10.1128/jvi.68.9.5933-5944.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilyinskii P O, Desrosiers R C. Efficient transcription and replication of simian immunodeficiency virus in the absence of NF-κb and Sp1 binding elements. J Virol. 1996;70:3118–3126. doi: 10.1128/jvi.70.5.3118-3126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilyinskii P O, Simon M A, Czajak S C, Lackner A A, Desrosiers R C. Induction of AIDS by simian immunodeficiency virus lacking NF-κB and SP1 binding elements. J Virol. 1997;71:1880–1887. doi: 10.1128/jvi.71.3.1880-1887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 17.Kestler H W, III, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for the development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 18.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 19.Kirchhoff F, Kestler III H W, Desrosiers R C. Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene. J Virol. 1994;68:2031–2037. doi: 10.1128/jvi.68.3.2031-2037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lewis M G, Bellah S, McKinnon K, Yalley-Ogunro J, Zack P M, Elkins W R, Desrosiers R C, Eddy G E. Titration and characterization of two rhesus derived SIVmac challenge stocks. AIDS Res Hum Retroviruses. 1994;10:213–220. doi: 10.1089/aid.1994.10.213. [DOI] [PubMed] [Google Scholar]
- 22.Lüke W, Coulibaly C, Dittmer U, Voss G, Oesterle R, Makoschey B, Sauermann U, Jurkiewicz E, Stahl-Hennig C, Petry H, Hunsmann G. Simian immunodeficiency virus (SIV) gp130 oligomers protect rhesus macaques (Macaca mulatta) against the infection with SIVmac32H grown on T-cells or derived ex vivo. Virology. 1996;216:444–450. doi: 10.1006/viro.1996.0082. [DOI] [PubMed] [Google Scholar]
- 22a.Manson, K., and M. Wynand. Personal communication.
- 23.Marthas M L, Sutjipto S, Higgins J, Lohman B, Torten J, Luciw P A, Marx P A, Pedersen N C. Immunization with a live, attenuated simian immunodeficiency virus (SIV) prevents early disease but not infection in rhesus macaques challenged with pathogenic SIV. J Virol. 1990;64:3694–3700. doi: 10.1128/jvi.64.8.3694-3700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Means R E, Greenough T, Desrosiers R C. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J Virol. 1997;71:7895–7902. doi: 10.1128/jvi.71.10.7895-7902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller C J, Marthas M, Torten J, Alexander N J, Moore J P, Doncel G F, Hendrickx A G. Intravaginal inoculation of rhesus macaques with cell-free simian immunodeficiency virus results in persistent or transient viremia. J Virol. 1994;68:6391–6400. doi: 10.1128/jvi.68.10.6391-6400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills K H G, Page M, Kitchin P, Chan L, Jones W, Silvera P, Corcoran T, Flanagan B, Ling C, Thiriart C, DeWilde M, Bruck C, Rud E, Clark B, Stott E J. Immunization of macaques with SIV env recombinants: specificity of T cell and antibody responses and evaluation of protective efficacy. J Med Primatol. 1993;22:104–109. [PubMed] [Google Scholar]
- 27.Pauza D, Trivedi P, Johnson E, Meyer K K, Streblow D N, Malkovsky M, Emau P, Schultz K T, Salvato M S. Acquired resistance to mucosal SIV infection after low dose intrarectal inoculation: the roles of virus selection and CD8-mediated T cell immunity. In: Girard M, Valette L, editors. Retroviruses of human A.I.D.S. and related animal diseases. Lyon, France: Foundation Marcel Merieux; 1993. pp. 151–156. [Google Scholar]
- 28.Plotkin S A, Mortimer E A. Vaccines. Philadelphia, Pa: The W. B. Saunders Company; 1988. [Google Scholar]
- 29.Regier D A, Desrosiers R C. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1990;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- 30.Schlienger K, Montefiori D C, Mancini M, Riviere Y, Tiollais P, Michel M. Vaccine-induced neutralizing antibodies directed in part to the simian immunodeficiency virus (SIV) V2 domain were unable to protect rhesus monkeys from SIV experimental challenge. J Virol. 1994;68:6578–6588. doi: 10.1128/jvi.68.10.6578-6588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl-Hennig C, Voss G, Dittmer U, Coulibaly C, Petry H, Makoschey B, Cranage M P, Aubertin A M, Lüke W, Hunsmann G. Protection of monkeys by a split vaccine against SIVmac depends upon biological properties of the challenge virus. AIDS. 1993;7:787–795. doi: 10.1097/00002030-199306000-00005. [DOI] [PubMed] [Google Scholar]
- 32.Suryanarayana, K., T. A. Wiltrout, G. M. Vasquez, V. M. Hirsch, and J. D. Lifson. 1997. Unpublished data.
- 33.Sutjipto S, Kodama T, Yee J, Gettie A, Jennings M, Desrosiers R C, Marx P A. Characterization of monoclonal antibodies that distinguish simian immunodeficiency virus isolates from each other and from human immunodeficiency virus types 1 and 2. J Gen Virol. 1990;71:247–249. doi: 10.1099/0022-1317-71-1-247. [DOI] [PubMed] [Google Scholar]
- 34.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyand M S, Manson K H, Garcia-Moll M, Montefiori D, Desrosiers R C. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]