Abstract
The NS1 protein (139 amino acids) is one of the two nonstructural proteins of human respiratory syncytial virus (RSV) and is encoded by a very abundant mRNA transcribed from the promoter-proximal RSV gene. The function of NS1 was unknown and was investigated here by using a reconstituted transcription and RNA replication system that involves a minireplicon and viral proteins (N, P, L and M2-1) expressed from separate cotransfected plasmids. Coexpression of the NS1 cDNA strongly inhibited transcription and RNA replication mediated by the RSV polymerase, even when the level of expressed NS1 protein was substantially below that observed in RSV-infected cells. The effect depended on synthesis of NS1 protein rather than NS1 RNA alone. Transcription and both steps of RNA replication, namely, synthesis of the antigenome and the genome, appeared to be equally sensitive to inhibition. The efficiency of encapsidation of the plasmid-derived minigenome was not altered by coexpression of NS1, indicating that the inhibition occurs at a later step. In two different dicistronic minigenomes, transcription of each gene was equally sensitive to inhibition by NS1. This suggested that the gradient of transcriptional polarity was unaffected and that the effect of NS1 instead probably involves an early event such as polymerase entry on the genome. NS1-mediated inhibition of transcription and RNA replication was not affected by coexpression of the M2 mRNA, which has two open reading frames encoding the transcriptional elongation factor M2-1 and the putative negative regulatory factor M2-2. The potent nature of the NS1-mediated inhibition suggests that negative regulation is an authentic function of the NS1 protein, albeit not necessarily the only one.
Human respiratory syncytial virus (RSV) is an important etiologic agent of severe lower respiratory tract disease in infants and young children worldwide (7, 10). It is the prototype member of the genus Pneumovirus of the family Paramyxoviridae and has a single-stranded, nonsegmented, negative-sense RNA genome of 15,222 nucleotides (nt).
RSV encodes 10 mRNAs and at least 10 (probably 11) distinct viral proteins. Three RSV proteins are transmembrane surface glycoproteins: the attachment G protein, the fusion F protein, and the small hydrophobic SH protein of unknown function. An unglycosylated matrix M protein is present as an inner virion protein. Three proteins associate with genomic RNA to form the nucleocapsid: the major nucleocapsid N protein, the P phosphoprotein, and the large L polymerase subunit. The M2-1 protein, which is encoded by the upstream translational open reading frame (ORF) of the M2 mRNA, has been shown to be a transcription elongation factor and therefore probably also is nucleocapsid associated (8, 9). The M2 mRNA contains a second, conserved ORF called M2-2 that overlaps the first, which encodes a putative negative regulatory activity and which therefore appears to be an 11th RSV gene (9). Here, the two M2 ORFs and their encoded proteins are referred to as M2-1 and M2-2 and the mRNA containing both ORFs in their native configuration is called M2(1+2). Finally, RSV encodes two nonstructural proteins, NS1 and NS2, of unknown function. The RSV gene order is 3′-(leader)-NS1-NS2-N-P-M-SH-G-F-M2(1+2)-L-(trailer)-5′ (7, 10). The M2-1, M2-2, NS1, and NS2 proteins lack obvious counterparts in other nonsegmented negative-strand viruses.
The general features of RSV transcription and RNA replication resemble those of the model nonsegmented negative-strand viruses Sendai virus and vesicular stomatitis virus (24, 29). The genomic RNA is tightly encapsidated both in the virion and intracellularly. The polymerase enters the template at the 3′-terminal extragenic leader region (21). Transcription involves a sequential stop-start mechanism that produces subgenomic mRNAs. Transcription is guided by short conserved signals that flank each mRNA coding unit, namely, a transcription gene start (GS) signal and a termination/polyadenylation gene end (GE) signal (21, 23). RNA replication occurs when the polymerase somehow switches to a readthrough mode in which the transcription signals are not recognized. This results in the synthesis of a positive-sense replicative intermediate, or antigenome, which also is encapsidated. The N, P, and L proteins constitute the replicase, whereas the transcriptase also requires the M2-1 elongation factor (9, 17).
The NS1 and NS2 proteins are expressed from separate mRNAs encoded by the first and second genes, respectively, that follow the 44-nt leader region (6). Because of their promoter-proximal location, these two mRNAs are the most abundant of the RSV transcripts (5). The NS1 protein is 139 amino acids long and is slightly acidic, but the predicted amino acid sequence provides no obvious clues to its function. The NS1 and NS2 proteins each contain the same four carboxy-terminal amino acids but otherwise are unrelated. Of the various RSV proteins, NS1 (and NS2) exhibits an intermediate degree of sequence conservation between the two human antigenic subgroups and ovine or bovine RSV; it is more highly conserved than the SH or G proteins but is less well conserved than any of the other proteins (1, 20, 25).
We recently described a system in which RSV transcription and RNA replication are reconstituted by the intracellular coexpression of an RSV minireplicon and RSV proteins from separate transfected plasmids (9, 14, 17). Here, we present evidence from this system that the NS1 protein is a potent inhibitor of intracellular RSV transcription and RNA replication.
MATERIALS AND METHODS
cDNAs.
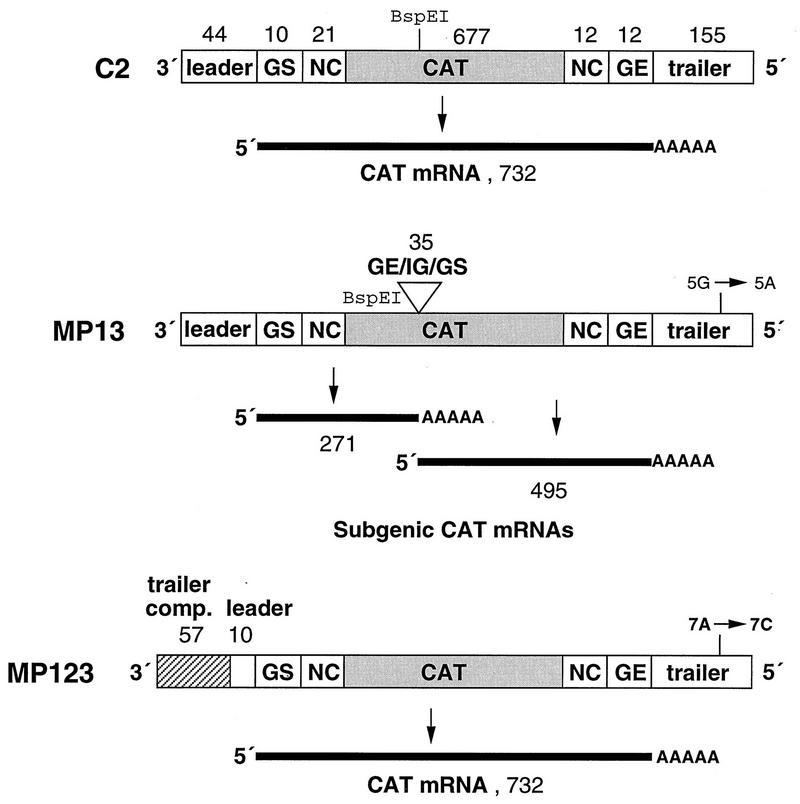
We previously described cDNAs encoding the C2 and C4 RSV-CAT minireplicons (17). The C2 RNA is 931-nucleotide negative-sense minigenome analog of the RSV genome in which the viral genes have been replaced by a negative-sense copy of the chloramphenicol acetyltransferase (CAT) ORF flanked by GS and GE signals and by the extragenic leader and trailer regions (Fig. 1) (17, 21–23). The C4 RNA is a positive-sense mini-antigenome that is the complement of the C2 minigenome.
FIG. 1.
Schematic representation (not to scale) of the cDNA-encoded, negative-sense RSV-CAT C2 minigenome RNA and its modified derivatives, MP13 and MP123. The C2 minigenome contains the 3′-terminal 75 nt and 5′-terminal 179 nt of the genome of RSV strain A2, including the leader, GS, non-protein-coding (NC), GE, and trailer regions (open boxes). These sequences flank a negative-sense copy of the CAT ORF (shaded box). MP13 is a version of C2 in which a 35-nt sequence containing a GE signal, a single-nucleotide intergenic region (IG), and GS signal has been inserted in the CAT ORF at its BspEI site. This converts the minigenome into one which is dicistronic, encoding two subgenic CAT mRNAs of 271 and 495 nt, exclusive of poly(A). MP123 is a version of C2 in which the first 34 nt of the leader region have been replaced with a sequence that is complementary to the 5′-terminal 57 nt of the trailer region (Tc) and thus contains the antigenome promoter. In addition, MP13 and MP123 differ from C2 in each having a single-nucleotide substitution in the trailer region that greatly inhibits amplification by the RSV polymerase: in MP13 nt 5 from the 5′ end has been changed from G to A (TrG5A), and in MP123 nt 7 from the 5′ end has been changed from A to C (TrA7C). Nucleotide lengths of the various segments are indicated.
cDNA encoding minigenome MP13 (Fig. 1) was constructed from the C2 cDNA by insertion, into the unique BspEI site within the CAT sequence, of a synthetic DNA containing a GE motif, the single-nucleotide N-P intergenic region of the authentic genome, and a GS motif (Fig. 1) (21–23). The synthetic DNA contained the sequence TCCGGgAGTTAATAAAAAATGGGGCAAATAGGATCcCCGGA, which is shown in the positive sense with the GS and GE motifs underlined, the BspEI-flanking sites italicized, and a single nucleotide substitution which destroyed each site in lowercase. This insertion makes MP13 into a dicistronic minigenome which encodes two subgenic CAT mRNAs of 271 (upstream mRNA) and 495 (downstream mRNA) nt. In addition, the MP13 minigenome was constructed to contain a point mutation (G to A in the negative-sense genome) in the trailer region at position 5 relative to the 5′ end. This substitution (called TrG5A) strongly inhibits the synthesis of progeny minigenomes from a minigenome template by the RSV polymerase and thus restricts the reconstituted system to the synthesis of positive-sense RNA. The reason for using this mutation is described below.
MP96 (not shown) is a version of C2 which has two differences: (i) the hammerhead ribozyme is replaced by that of hepatitis delta virus (26), and (ii) it contains an A-to-C substitution in the trailer region at position 7 relative to the 5′ end (TrA7C), which, like the TrG5A mutation mentioned above, strongly inhibits the synthesis of a progeny minigenome from a minigenome template.
MP123 is a version of MP96 in which the 3′-terminal 34 nt of the leader region have been replaced with the complement of the last 57 nt of the trailer region, the 3′ end of the antigenome. Thus, this substitution replaces the genomic promoter with that of the antigenome and makes a copyback-type replicon.
cDNA containing the NS1 ORF was cloned into the NcoI-BamHI window of plasmid pTM-1 (13), in which the foreign ORF is under the transcriptional control of the promoter for T7 RNA polymerase and the translational control of the internal ribosome entry site of encephalomyocarditis virus. The N, P, and L cDNAs were placed in the same vector in a previous study (17). The M2(1+2) cDNA included nt 10 to 909 of the complete 957-nt mRNA and thus contained both ORFs in their original configuration, whereas the M2-1 cDNA contained only the upstream ORF encoding the transcription elongation factor (9). In addition, a mutant of the NS1 plasmid was prepared in which a 2-nt deletion was placed in codon 8 to ablate translation by shifting the reading frame to a register which terminates shortly thereafter. This mutation was inserted by PCR with plasmid pTM1-NS1 as the template and with the forward primer AATACCATGGGCAGCAATTCATTGAGTGATAAAAGT (positive sense, with the NcoI site containing the NS1 translational start codon underlined) and the reverse primer CTCGAGCTGCAGGGATCCTCTAGAGTC (BamHI site underlined). This resulted in a complete copy of the NS1 ORF containing the 2-nt deletion, which was then cloned into the NcoI-BamHI window of pTM1 as indicated above for the parental cDNA. The sequence of the mutant ORF was confirmed in its entirety.
Transfections.
Confluent monolayers of 293 or HEp-2 cells (as indicated in the figure legends) in six-well dishes (each well contained 1.5 × 106 cells) were infected with recombinant vaccinia virus vTF7-3 expressing T7 RNA polymerase (15) at a multiplicity of infection of 10 PFU per cell. After 45 min of adsorption, the inoculum was replaced with 1 ml of Opti-MEM per well containing 10 μl of Lipofectamine or 12 μl of Lipofectace (Life Technologies) and amounts of minireplicon and N, P, and L plasmids as indicated in the figure legends. In some experiments, the standard plasmid mix also included the M2(1+2) or M2-1 (i.e., ORF 1 only) plasmid. The concentration of the NS1 plasmid was varied as indicated. In all experiments, pTM-1 vector plasmid lacking an insert was added to keep the total amount of plasmid transfected per well of cell monolayer constant. After 16 h of transfection, the transfection medium was replaced with 1.5 ml of Eagle’s minimal essential medium containing 10% fetal bovine serum for 293 cells or Opti-MEM containing 2% fetal bovine serum for HEp-2 cells. The cells were typically harvested 24 h after the change of medium, i.e., 40 h postinfection.
Analysis of protein expression by Western blotting.
Typically, one-fifth of the volume of the total cells harvested from one well was lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer containing β-mercaptoethanol, and about 1/150 of total cellular equivalent from a single well was run in each lane of precast 4 to 20% gradient minigels (Novex) as specified by the supplier. The gels were transferred onto Optitran (Schleicher & Schuell) nitrocellulose membranes with the buffer system of Towbin et al. (28) at 250 mA for 2 h. The blots were washed in TTBS (0.1 M Tris-HCl [pH 7.5], 0.5 M NaCl, 0.01% Tween 20) and blocked in Blotto (5% [wt/vol] nonfat dry milk in TTBS) for 1 to 2 h before the specific antibody was added for overnight binding. For detecting the N, P, and other structural proteins, a rabbit antiserum against gradient-purified RSV virions was used (17). This antibody detected the G, F, N, P, M, M2-1, and SH proteins but not the NS1 and NS2 proteins. The NS1 and NS2 proteins were detected with a rabbit antiserum directed against a 10-amino-acid C-terminal peptide of NS2 protein. This antibody efficiently recognized both the NS1 and NS2 proteins in a Western blot, presumably because the two proteins are identical for the C-terminal 4 amino acids. After overnight incubation with the antibody, the blots were washed three times for 10 min each in TTBS and then given a further incubation for 2 h in a secondary antibody solution prepared in Blotto containing 1 μg of goat anti-rabbit immunoglobulin conjugated to alkaline phosphatase per ml. The blots were washed three times for 10 min each in TTBS lacking Tween 20 and then subjected to color development with Nitro Blue Tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) as specified by the supplier (Promega).
CAT assay.
Typically, 1/10 of the cells harvested from a single well was used to make 200 μl of cell lysate, and an aliquot of either 5 or 10 μl of lysate was used for the CAT assay. The CAT assay was based on the conversion of [14C]chloramphenicol to acetylated forms in the presence of acetyl coenzyme A as described before (16). The acetylated forms were separated by ascending thin-layer chromatography and visualized by autoradiography.
RNA isolation and Northern blot analysis.
Total intracellular RNA was extracted by lysing the cell pellets in Trizol reagent (Life Technologies) as specified by the supplier, except that the RNAs were extracted twice with phenol-chloroform following isopropanol precipitation. The RNA samples were quantitated by spectrophotometry as well as by electrophoresis in agarose gels with ethidium bromide staining to ensure that the amounts of samples analyzed were equivalent. Total intracellular RNA samples (8 to 10 μg, representing one-fifth of the total amount from each well) were subjected to Northern blot analysis as described previously (4, 17). The radioactive signals on the blots were quantitated with a PhosphorImager (Molecular Dynamics) and were also exposed to X-ray film (Kodak X-AR).
Micrococcal nuclease treatment of intracellular RNA.
Cells which had been infected with vTF7-3 and transfected with plasmids were harvested 40 h postinfection by scraping, washed once in phosphate-buffered saline, and lysed in 100 μl of 10 mM Tris-HCl (pH 7.5)–10 mM NaCl–1.5 mM MgCl2–10 mM CaCl2–1% Triton X-100–0.5% deoxycholate per well. An aliquot of each sample was treated with micrococcal nuclease (Nuclease S7; Boehringer Mannheim) at a final concentration of 20 μg/ml and incubated for 30 min at 30°C (2). RNA was isolated with Trizol reagent as described above.
RESULTS
Expression of the NS1 protein in a reconstituted transcription and RNA replication system.
We previously described a system in which RSV transcription and RNA replication are reconstituted intracellularly from transfected plasmids (9, 14, 17). The plasmids encode (i) minireplicon RNA, which can be a negative-sense analog of genomic RNA (called a minigenome) or a positive-sense analog of antigenomic RNA (a mini-antigenome), and (ii) the subset of RSV proteins necessary to reconstitute the RSV nucleocapsid and polymerase (9, 14, 17).
In the reconstituted system, coexpression of the N, P, and L proteins is necessary and sufficient for RSV RNA replication whereas processive transcription also requires expression of the M2-1 protein (9, 14, 17). The purpose of this study is to describe the effect of coexpression of the NS1 protein on RSV transcription and RNA replication. The system used conditions under which the levels of plasmid-expressed viral proteins were similar to those expressed by RSV infection, which should be an important factor for re-creating authentic effects. This is illustrated in Fig. 2 (upper panel), in which the levels of N and P expressed in RSV-infected cells (lane 1) were shown by Western blot analysis to be similar to those expressed from transfected N and P plasmids (lane 3). Similar findings have been described elsewhere, based on both Western blot analysis and radiolabeling (references 9, 14, 17 and unpublished data). Coexpression of incrementally increasing amounts of the NS1 plasmid resulted in concomitant increases in the expression of the NS1 protein, as detected by Western blot analysis (Fig. 2, lower panel, lanes 4 to 6) with an antipeptide rabbit serum that cross-reacts with the NS1 and NS2 proteins (see Materials and Methods) or by metabolic labeling with [35S]methionine, immunoprecipitation, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). In RSV-infected cells, the NS1 protein could be readily detected by Western blot analysis at 8 h postinfection and was expressed at maximal levels between 16 and 24 h postinfection (data not shown). The maximum level of NS1 protein expressed during RSV infection was similar to the amount expressed in response to 100 ng of transfected plasmid (Fig. 2). The expression of the NS1 protein from the transfected plasmid had no effect on the expression of the N, P, or M2-1 proteins from cotransfected plasmids (Fig. 2 and data not shown).
FIG. 2.
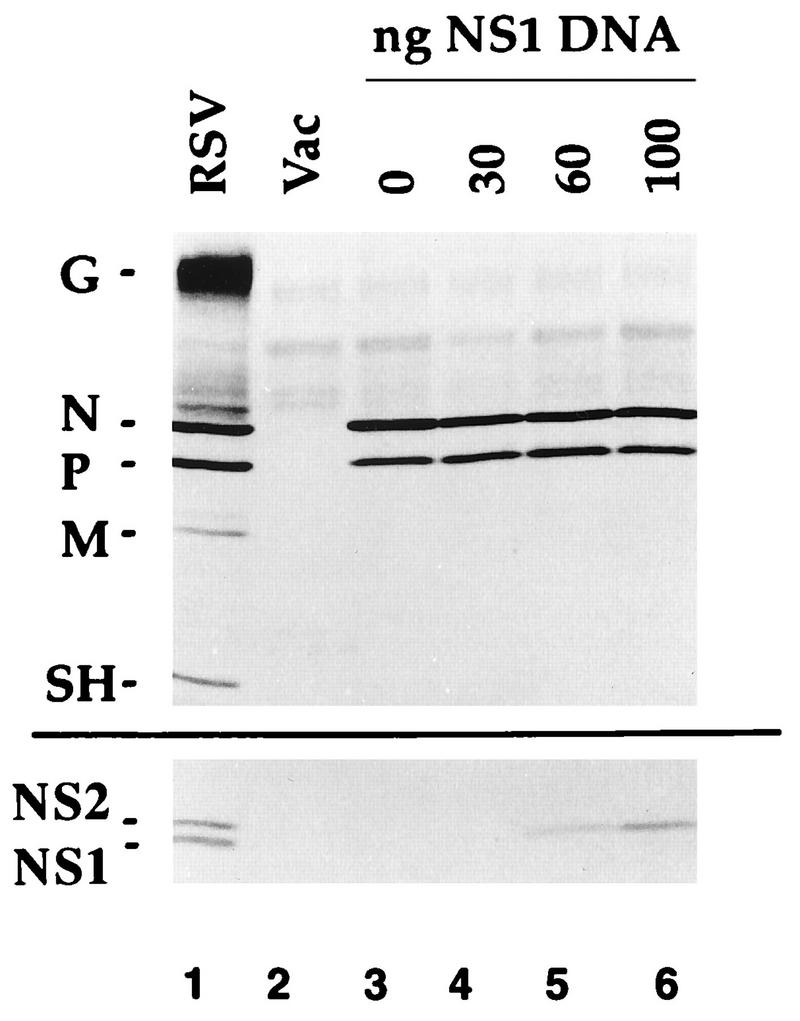
Western blot analysis of proteins synthesized in 293 cells which had been infected with 10 PFU of RSV per cell (lane 1) or infected with the vaccinia virus recombinant vTF7-3 (lanes 2 to 6) and mock transfected (lane 2) and then transfected with 200 ng of N plasmid and 100 ng of P plasmid (lane 3) or the same amounts of N and P plasmids supplemented with 30 ng (lane 4), 60 ng (lane 5), or 100 ng (lane 6) of NS1 plasmid. Cells were harvested 18 h (lane 1) or 40 h (lanes 2 to 6) postinfection, and lysates were electrophoresed on a 4 to 20% gradient gel and transferred to nitrocellulose. The upper blot was probed with a polyclonal antiserum against gradient-purified, disrupted RSV virions (17), which is reactive with most of the RSV structural proteins including G, N, P, M, and SH. The lower blot was probed with an antibody against a C-terminal peptide of the NS2 protein, which recognizes both the NS1 and NS2 proteins in RSV-infected cells (lane 1) and NS1 in the plasmid-transfected cells (lanes 4 to 6).
Effect of NS1 on RNA synthesis from cDNA-encoded C2 minigenome or C4 mini-antigenome.
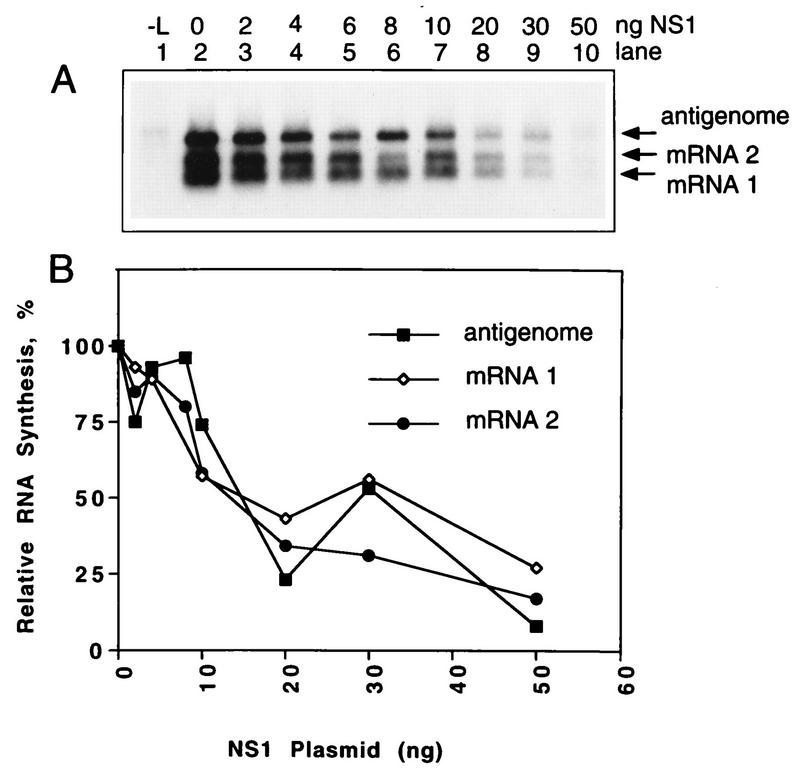
We evaluated the effect of coexpression of the NS1 protein on the synthesis of positive-sense RNA from the C2 minigenome (Fig. 3A and B) or the synthesis of negative-sense RNA from the C4 mini-antigenome (Fig. 3C and D). The plasmid-supplied C2 or C4 minireplicon was complemented by the N, P, and L plasmids with or without the M2(1+2) plasmid (i.e., that containing both ORFs in their natural configuration). In previous work, the transcription elongation activity of ORF1 predominated in situations where smaller amounts of this plasmid (no more than 20% of that of the N plasmid) were transfected whereas the inhibitory activity associated with ORF2 predominated at higher plasmid levels (9). The amount of M2(1+2) plasmid supplied here was 5% of that of the N plasmid.
FIG. 3.
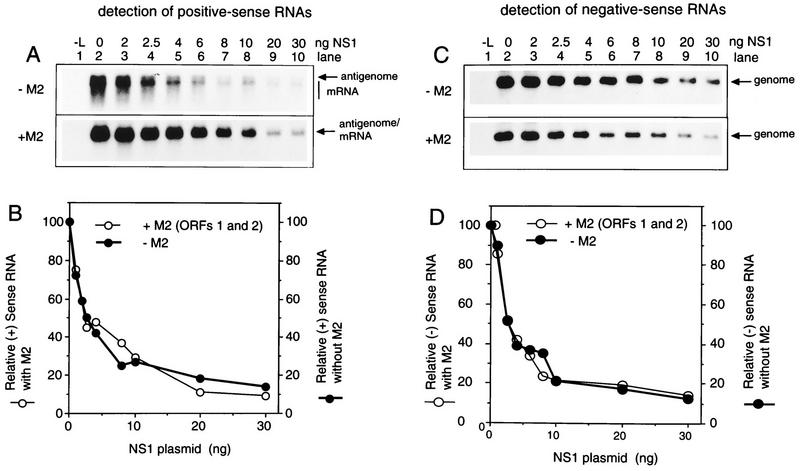
Northern blot (A and C) and PhosphorImager (B and D) analysis showing the effects of increasing expression of NS1 plasmid on the synthesis of positive-sense RNA from the C2 minigenome (A and B) and the synthesis of negative-sense RNA from the C4 mini-antigenome (C and D). 293 cells were infected with vaccinia virus vTF7-3 and transfected with 50 ng of C2 minigenome (A) or C4 mini-antigenome (C) plasmid, 200 ng of N plasmid, 100 ng of P plasmid, 0 ng (lane 1) or 25 ng (lanes 2 to 10) of L plasmid, 0 ng (top panel) or 10 ng (bottom panel) of M2 plasmid containing both ORFs, and the indicated amount (0 to 30 ng [lanes 2 to 10]) of NS1 plasmid. Intracellular RNA was harvested 40 h postinfection and analyzed by Northern blot hybridization with a negative-sense (A) or positive-sense (C) RSV-CAT riboprobe. Hybridized radioactivity was quantitated by PhosphorImager analysis: quantitation of panels A and C is shown in panels B and D, respectively.
The positive-sense RNAs synthesized by the plasmid-supplied C2 minigenome in the absence of M2 include the mini-antigenome, a small amount of full-size CAT mRNA, and a smear of incomplete mRNAs resulting from premature termination of transcription (Fig. 3A, upper panel, lane 2), as previously described (9, 17). Because the amount of full-length mRNA is reduced, the antigenome and mRNA could be distinguished. With the addition of a small amount of M2(1+2) plasmid, all of the mRNA accumulated as full-length molecules and the broadness of the mRNA band obscured the mini-antigenome (Fig. 3A, lower panel, lane 2).
The NS1 plasmid was then added in increasing amounts (Fig. 3A, lanes 3 to 10). This resulted in incremental decreases in the accumulation of positive-sense RNA. The presence or absence of the M2(1+2) plasmid did not affect this decrease (Fig. 3A and B). Effects on the levels of mini-antigenome and mRNA could be observed in the absence of M2(1+2) plasmid (Fig. 3A, upper panel); this showed that the syntheses of the two RNAs appeared to be affected equally, a point which is addressed again below.
Complementation of the plasmid-supplied C4 mini-antigenome with the N, P, and L proteins resulted in the synthesis of negative-sense minigenome (Fig. 3C, lane 2), as described previously (9, 17). The inclusion of increasing amounts of NS1 plasmid resulted in incremental decreases in the synthesis of this RNA (Fig. 3C, lanes 3 to 10). Coexpression of the M2(1+2) plasmid had no apparent effect on this inhibitory activity. Furthermore, the syntheses of positive-sense and negative-sense RNAs appeared to be equally sensitive to inhibition (Fig. 3B and D). The inhibition was observed with very small amounts of NS1 plasmid and was associated with levels of protein synthesis below the limit of detection by Western blot analysis (Fig. 2).
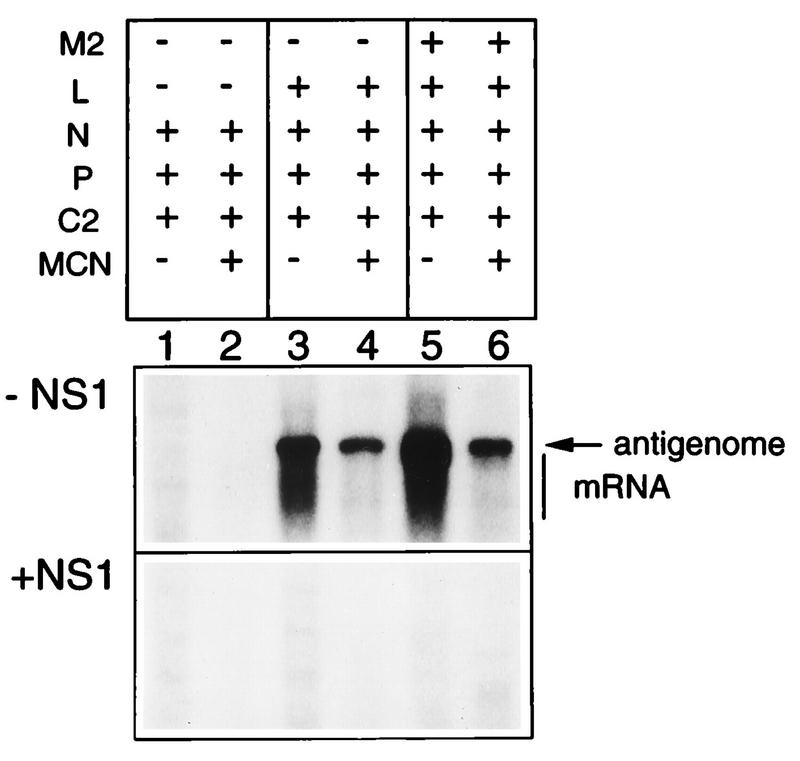
Because the mini-antigenome was partly obscured by the mRNA band (Fig. 3A), it was of interest to confirm that synthesis of the mini-antigenome RNA was indeed sensitive to NS1. Cells were transfected with plasmids expressing the C2 minigenome and the N, P, and L proteins, in the presence or absence of the M2(1+2) plasmid. Cell lysates were prepared and treated with micrococcal nuclease prior to RNA isolation, a treatment which degrades free RNA including mRNA whereas encapsidated genome and antigenome are protected (2). Samples which had not been exposed to micrococcal nuclease contained both mRNA and mini-antigenome (Fig. 4, panel −NS1, lanes 3 and 5). Treatment of the cell lysates with micrococcal nuclease prior to RNA isolation degraded most of the mRNA, whereas the level of the mini-antigenome was undiminished (lanes 4 and 6). This confirmed the identity of the antigenome, allowed it to be detected unambiguously, and confirmed that it was efficiently encapsidated in the reconstituted system. When NS1 plasmid was included at 50 ng per well, the synthesis of positive-sense RNA, including the mini-antigenome, was reduced almost to background levels (Fig. 4, panel +NS1).
FIG. 4.
Northern blot analysis showing the synthesis and encapsidation of the mini-antigenome from the plasmid-supplied C2 minigenome in the absence (−NS1) or presence (+NS1) of 50 ng of NS1 plasmid. 293 cells were infected with vTF7-3 and transfected with C2 plasmid together with the support plasmids indicated at the top, added in the same amounts as in Fig. 3. The M2 plasmid was that containing both ORFs. Lysates were prepared 40 h postinfection and were either mock treated (lanes 1, 3, and 5) or treated (lanes 2, 4, and 6) with micrococcal nuclease (MCN), after which the remaining RNA was isolated and analyzed by Northern blot hybridization with a negative-sense RSV-CAT riboprobe. The positions of the mini-antigenome and mRNA are shown.
The effect of the NS1 plasmid is dependent upon an intact NS1 ORF.
It was necessary to determine whether the inhibition associated with the NS1 plasmid depended upon its ability to express NS1 protein, as might be expected, or whether the inhibitory effect might be an artifact of transfection of the NS1 plasmid or synthesis of NS1 mRNA. Regarding this last possibility, it should be noted that although the C2 minigenome does contain the upstream nontranslated region of the NS1 mRNA (Fig. 1), this sequence is not present in the NS1 support plasmid because the initiation codon is joined directly to the internal ribosome entry site (13). It therefore seemed unlikely that the inhibition associated with expression of the NS1 plasmid was due to hybrid arrest of the minigenome by NS1 RNA, but it was important to directly rule out possible artifactual effects.
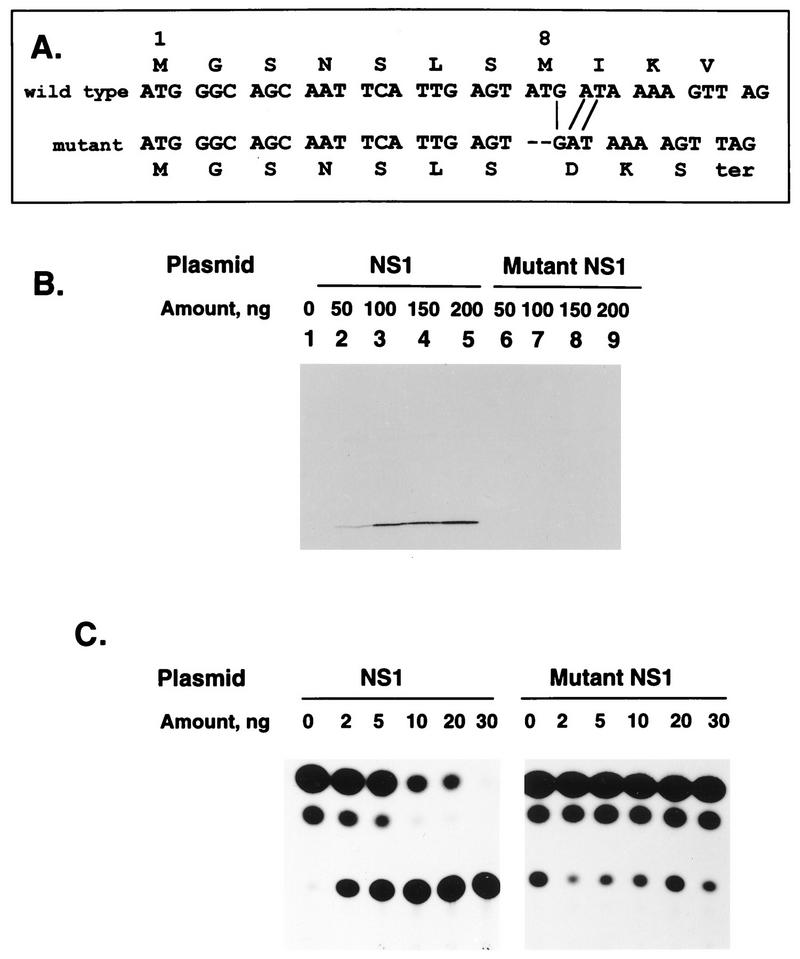
The first and eighth codons of the NS1 ORF are potential AUG translational start sites, whereas the next AUG is at codon 80. To preclude synthesis of the NS1 protein from either upstream start site, 2 nt were deleted from codon 8, which shifted the reading frame such that the mutant ORF would terminate 3 codons later (Fig. 5A). Western blot analysis of transfected cells (Fig. 5B) confirmed that while the original plasmid produced NS1 protein in a dose-dependent manner (lanes 1 to 5), the mutant plasmid was silent for its synthesis (lanes 6 to 9). Increasing amounts of the wild-type or mutant plasmid were evaluated for their effect on the expression of CAT enzyme by the C2 minigenome complemented by the N, P, L, and M2(1+2) plasmids (Fig. 5C). The wild-type plasmid was strongly inhibitory to CAT expression, and the use of this sensitive assay system showed that the inhibitory effect was complete in this particular experiment at 30 ng of input NS1 plasmid. In contrast, cotransfection of the mutant plasmid had no effect on the expression of CAT.
FIG. 5.
The inhibitory effect of the NS1 plasmid is ablated by a frameshift in the NS1 ORF. (A) The NS1 cDNA was mutagenized to delete 2 nt from codon eight, which shifts the reading frame and terminates the ORF three codons later. This is illustrated in the sequence of codons 1 to 11 of the NS1 ORF. This mutation would preclude translation from either the first or eighth codons, which are AUG. The next AUG in the 139-codon ORF is at codon 80. (B) Western blot analysis showing the expression of the NS1 protein from the wild-type (lanes 1 to 5) or mutant (lanes 6 to 9) plasmid. The indicated amounts of plasmid were transfected into 293 cells infected with vTF7-3. Cell lysates were prepared 40 h later and analyzed with a Western blot probed with an antiserum against a synthetic peptide of the NS2 protein. (C) Expression of CAT by the C2 minigenome complemented by the N, P, L, and M2 (both ORFs) plasmids together with the indicated amount of wild-type or mutant NS1 plasmid. Cells were harvested 40 h postinfection, and lysates were assayed for the ability to acetylate [14C]chloramphenicol as visualized by thin-layer chromatography and autoradiography (the unreacted chloramphenicol comprises the bottom row of spots, and the acetylated forms comprise the two higher rows).
NS1 does not interfere with encapsidation of the plasmid-supplied minigenome.
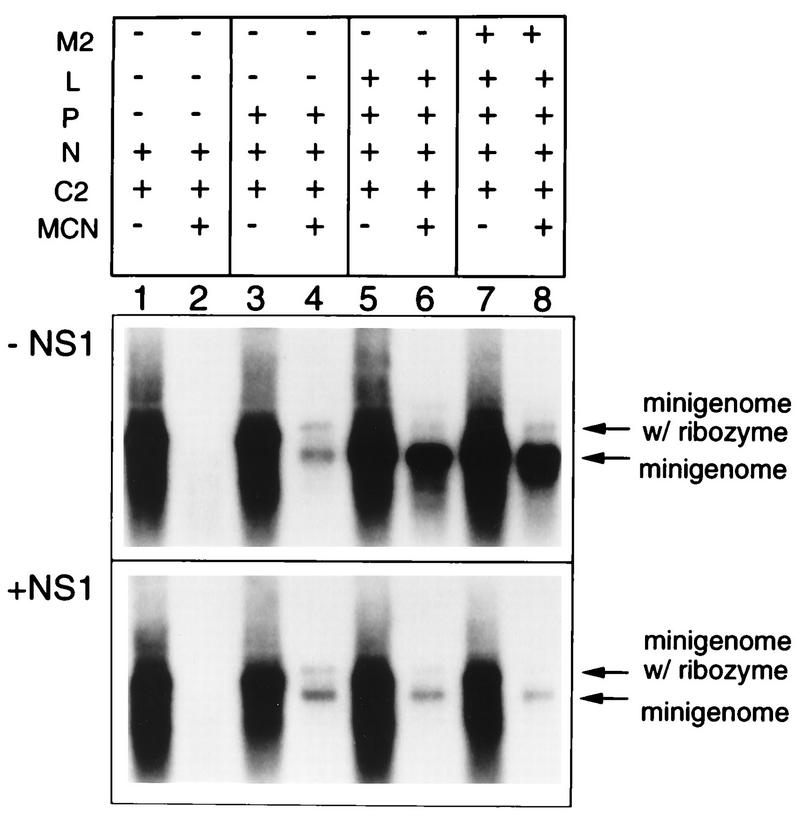
We then examined whether the action of the NS1 protein was at the level of encapsidation by examining the ability of N and P proteins to encapsidate the plasmid-supplied minigenome (Fig. 6). Cells were transfected with plasmids encoding the C2 minigenome and various combinations of support plasmids with or without the addition of NS1 plasmid (Fig. 6, panels +NS1 and −NS1, respectively). Cell lysates were prepared, incubated with micrococcal nuclease or mock treated, and processed for RNA purification. RNA which was encapsidated would be protected from degradation and thus detected. The RNA was analyzed by Northern blot hybridization with a positive-sense probe, which would detect plasmid-encoded minigenome produced by T7 RNA polymerase as well as progeny minigenome produced by the RSV polymerase.
FIG. 6.
The NS1 protein does not affect encapsidation of plasmid-supplied or RSV-amplified C2 minigenome. 293 cells were infected with vTF7-3 and transfected with 50 ng of C2 minigenome, 0 (−NS1) or 50 ng (+NS1) of NS1, and the indicated support plasmids in the same amounts as in Fig. 3. The M2 plasmid was that containing both ORFs. Lysates were prepared 40 h postinfection and were either mock treated (lanes 1, 3, 5 and 7, both panels) or treated (lanes 2, 4, 6, and 8, both panels) with micrococcal nuclease (MCN), after which the remaining RNA was isolated. The RNAs were analyzed by Northern blot hybridization with a positive-sense RSV-CAT riboprobe, which would detect minigenome that is plasmid encoded as well as that which is generated by RSV-mediated replication. Some of the plasmid-encoded minigenome accumulates in a form (labeled minigenome w/ ribozyme) that is slightly larger than the minigenome because it contains uncleaved ribozyme at its 3′ end.
When the support proteins did not include the complete RSV replicase (for example, when the P or L protein was omitted), the only source of minigenome RNA was T7 transcription of the plasmid. When N alone was supplied as the support protein, all of the minigenome was sensitive to micrococcal nuclease, indicating that it was not encapsidated (Fig. 6, lanes 1 and 2, panels −NS1 and +NS1). Incidently, the ability of unencapsidated minigenome RNA to accumulate intracellularly appears to be a property of 293 cells, because in HEp-2 cells it accumulates only when protected by encapsidation (14). Inclusion of P in addition to N resulted in the encapsidation of a fraction of the total plasmid-encoded minigenome RNA (lane 4, both panels). Some of the plasmid-supplied minigenome was in a somewhat larger form, which appears to be stabilized by the presence of uncleaved ribozyme at the 3′ end (14, 17). Thus, while encapsidation of the plasmid-encoded minigenome may or may not be a good model for authentic RSV encapsidation, it does require both N and P, as is thought to be the case in the authentic situation. Importantly, the amount of micrococcal nuclease-resistant genomic RNA was approximately the same in both the absence and presence of the NS1 protein (lane 4, both panels), indicating that NS1 did not prevent the formation of the encapsidated template. When L was included in addition to N and P and in the absence of NS1, thus providing all of the components of the RSV replicase, the encapsidated RNA was amplified 10- to 50-fold, depending on the experiment (panel −NS1, compare lane 6 with lane 4). Inclusion of the NS1 plasmid blocked this amplification (panel +NS1, lane 6), and the presence of the M2(1+2) plasmid made no difference (lane 8). These experiments showed that NS1 did not prevent the formation of the encapsidated template but prevented its subsequent utilization by the polymerase.
Effect of NS1 on the sequential transcription of successive small genes of a minigenome restricted to a single step in RNA replication.
The experiment shown in Fig. 6 showed that reconstituted RNA replication involved extensive amplification of the plasmid-supplied minireplicon by the RSV polymerase. This complicated the analysis of the effects of NS1. For example, since most of the encapsidated minigenome which is available as a template for transcription is the product of the reconstituted RSV polymerase, it is difficult to know whether reduced accumulation of mRNA was due to a direct effect on transcription or was an indirect effect of reduced template availability because of reduced RNA replication. With regard to RNA replication, it is difficult to know whether inhibition was due to an effect on the synthesis of the genome or the antigenome or both.
We recently found that the ability of the minigenome to execute multiple rounds of RNA replication can be blocked by certain nucleotide substitutions in the trailer region, such as changing the fifth nucleotide from the 5′ end from G to A (mutation TrG5A), or the seventh nucleotide from the 5′ end from A to C (mutation TrA7C) (14, 25a). These mutations probably inactivate the promoter at the 3′ end of the encoded mini-antigenome. Consistent with this, they did not affect the encapsidation of the T7-produced minigenome and did not reduce the amount of positive-strand RNA made per amount of encapsidated minigenome template. However, the synthesis of progeny minigenome was severely inhibited. Therefore, we analyzed the effect of NS1 on RNA synthesis by one of these mutants, MP13, which is a version of the C2 minigenome which contains the above-mentioned TrG5A mutation (Fig. 1) and would allow a direct examination of the effects on transcription. In addition, MP13 contains the insertion of a gene junction cassette into the BspEI site in the CAT ORF such that it encodes two subgenic CAT mRNAs of 271 and 495 nt (see Materials and Methods (Fig. 1). This has two advantages: (i) the mini-antigenome can be clearly distinguished from mRNA, and (ii) it was possible to investigate whether NS1 affected the gradient of transcription of these two small genes.
Complementation of MP13 with N, P and L resulted in the synthesis of antigenome and the two subgenic CAT mRNAs (Fig. 7A, lane 2). The inclusion of increasing amounts of the NS1 plasmid resulted in a dose-dependent decrease in the synthesis of the mini-antigenome and mRNAs 1 and 2 (lanes 3 to 10). The presence of the M2(1+2) plasmid made little difference (data not shown). PhosphorImager analysis indicated that the effect of NS1 was similar for antigenome, mRNA 1 or mRNA 2 (Fig. 7B). This provided evidence that (i) the synthesis of all positive-sense RNAs was directly inhibited by NS1, (ii) the synthesis of mRNA (i.e., transcription) and the synthesis of the mini-antigenome (i.e., replication) were equally affected, and (iii) the gradient of transcriptional polarity was not altered detectably.
FIG. 7.
The NS1 protein inhibits the synthesis of positive-sense RNA from the minigenome MP13. MP13 is an analog of genomic RNA that encodes two short mRNAs from the upstream and downstream ends of the CAT ORF (mRNAs 1 and 2, which are 271 and 495 nt exclusive of poly[A]), as illustrated in Fig. 1. In addition, MP13 is restricted from amplification by the RSV polymerase due to a point mutation (TrG5A) in the trailer region. (A) 293 cells were infected with vTF7-3 and transfected with MP13 minigenome and the N, P, and L support plasmids in the same amounts as in Fig. 3 and the indicated amount (0 to 50 ng) of NS1 plasmid. L plasmid was omitted from lane 1 as a negative control. RNA was isolated 40 h postinfection and analyzed by Northern blot hybridization with negative-sense RSV-CAT riboprobe. (B) The hybridized radioactivity was quantitated on a PhosphorImager (B). Each RNA species was normalized separately with respect to the sample that received L, N, P, but no NS1 (lane 2) as 100%.
We also examined the effect of NS1 on a larger, previously described (22) dicistronic minigenome, RSV-CAT-LUC(CAT). This minigenome contains two genes, CAT and LUC(CAT), each under the control of a separate set of GS and GE signals and separated by the naturally occurring NS1/NS2 intergenic region. The second gene is designated LUC(CAT) because the LUC gene has been modified by the insertion of a nearly complete copy of the CAT gene so that it too is detected by a CAT-specific probe (22). The CAT and LUC(CAT) transcription cassettes are 730 and 1,876 nt long, respectively, making it possible to examine possible effects on the gradient of polar transcription over a gene pair of greater than 2.6 kb. The effect of NS1 on this larger transcriptional unit was the same as with the shorter MP13 minigenome; specifically, the two mRNAs were equally sensitive to the effect of NS1 and the gradient of transcriptional polarity was unaffected (data not shown).
Effect of NS1 on the antigenomic promoter.
As described above, the use of the mutant minigenome MP13, which is restricted to the synthesis of positive-sense RNA, made it possible to directly analyze the effect of NS1 on the genomic promoter. It also was of interest to test whether NS1 has an effect on synthesis from the antigenomic promoter. Therefore, we constructed a minigenome, MP123, in which the 3′-terminal 34 nt of the leader region were removed and replaced with the 3′-terminal 57 nt of the antigenome (Fig. 1), resulting in a copyback-type minireplicon. Since the 3′ end of the antigenome is encoded by the minigenome trailer, it is referred to as the trailer complement. In addition, MP123 contained the A-to-C substitution at position 7 relative to the 5′ end (TrA7C), a change which has an effect similar to that of the TrG5A mutation described above for MP13 and MP25 and greatly restricts amplification by the RSV polymerase. As a control, MP123 was compared with MP96 (see Materials and Methods), which is a version of the standard C2 minigenome which also contains the TrA7C mutation.
When complemented by N, P, L, and M2-1, MP123 directed the synthesis of the antigenome and polyadenylated subgenomic mRNA (Fig. 8C, lane 2). It is noteworthy that a copyback RSV minireplicon can direct the synthesis of mRNA, and the characterization of this activity will be described in greater detail elsewhere (25b). The identification of the antigenome was complicated by the fact that a background band of that size accumulated in the absence of the L protein (Fig. 8C, lane 1). However, when the cell lysates were treated with micrococcal nuclease before RNA purification, the background band was destroyed (Fig. 8D, lane 1), as was most of the mRNA (Fig. 8D, lane 2). Similar results were obtained with the MP96 minigenome analyzed in parallel (Fig. 8A and B), except that a background band was not observed in the absence of L protein (lanes 1). The addition of increasing amounts of NS1 inhibited the synthesis of positive-sense RNA by MP123 and MP96, and the magnitude of the effect appeared to be similar for the two minigenomes, with regard to both mRNA and the mini-antigenome. The effect on the mini-antigenome was seen most clearly with the micrococcal nuclease-treated samples (Fig. 8B and D). Thus, the synthesis of RNA from the genome and antigenome promoters was equally sensitive to the NS1 protein.
FIG. 8.
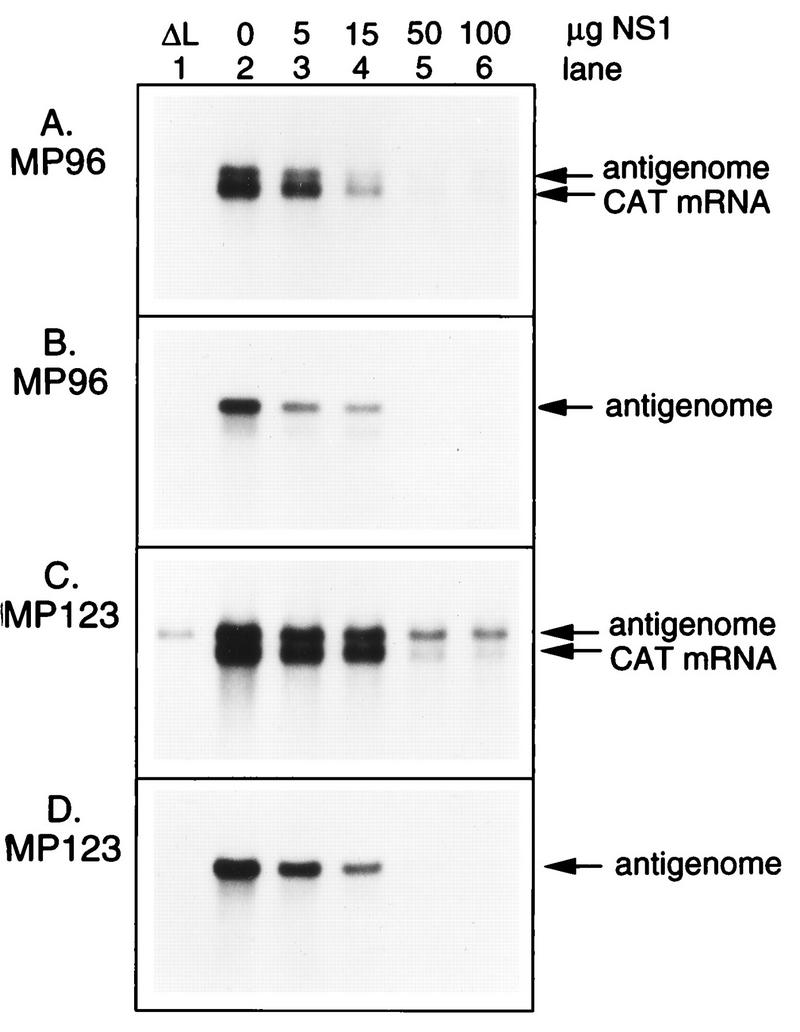
The NS1 protein inhibits RNA synthesis from the antigenome promoter. MP123, a copyback-type minigenome in which the 3′-leader region was replaced by the complement of the last 57 nucleotides of the trailer region, which corresponds to the 3′ end of the antigenome, was constructed (Fig. 1). The TrA7C trailer mutation was included to block minigenome amplification by the RSV polymerase. MP123 was analyzed in parallel with MP96, which is a version of the prototype C2 minigenome which also contains the TrA7C point mutation. HEp-2 cells were infected with vTF7-3 and transfected with 100 ng of the MP96 (A and B) or MP123 (C and D) minigenome; the N, P, and L support plasmids in the amounts as described in the legend to Fig. 3; 50 ng of M2-1 plasmid; and the indicated amount (0 to 100 ng [lanes 2 to 6]) of NS1 plasmid. Lane 1 is a negative control in which the L plasmid was omitted. Cells were harvested 40 h postinfection, and lysates were prepared and processed for RNA purification directly (A and C) or following treatment with micrococcal nuclease (B and D). RNA was analyzed by Northern blot hybridization with a negative-sense CAT riboprobe.
The inhibitory effect of NS1 on transcription is not affected by the expression of other RSV proteins.
We considered the possibility that the inhibitory effect of coexpression of NS1 in the reconstituted transcription-replication system was an artifact due to the absence of one or more other viral proteins. We therefore tested whether various combinations and concentrations of the M2-1, M, SH, G, F, and NS2 plasmids might relieve the inhibitory activity of NS1. None of these, alone or in combination, did so (data not shown).
These studies did show that coexpression of the NS2 protein in the reconstituted system also inhibited RSV transcription and RNA replication whereas none of the remaining RSV proteins had an effect (data not shown). However, compared to NS1, much larger amounts of the input NS2 plasmid were needed for the equivalent level of inhibition of transcription and replication. Western blotting showed that substantial inhibition of transcription and RNA replication was possible only with amounts of expressed NS2 protein that were in considerable excess of that found in RSV-infected cells. The requirement for high levels of NS2 protein expression raised doubts about the authenticity of the NS2 effect, and it is being characterized further. The effects of NS1 and NS2 together were additive rather than synergistic, suggesting that the two proteins do not function together (unpublished data).
DISCUSSION
In this study, we investigated the effect of the RSV NS1 protein on transcription and RNA replication of the RSV minigenome and mini-antigenome analogs executed by plasmid-encoded proteins. Coexpression of the NS1 protein was highly inhibitory to both transcription and RNA replication. Both processes were affected equally, with 7 to 10 ng of input NS1 plasmid resulting in a reduction to 33% activity (Fig. 3). This level of expression of NS1 protein was well below the limit of detection by Western blot analysis. In RSV-infected cells, in comparison, the expression of NS1 was readily detected by 8 h postinfection. A 100-ng sample of transfected NS1 plasmid was required to achieve levels comparable to the levels observed in RSV-infected cells 20 h postinfection, when viral protein synthesis is maximal (Fig. 2). That the inhibitory effect of NS1 was associated with levels of protein equal or below those of RSV-infected cells suggested that the effect is an authentic one which could also operate during RSV infection.
Somewhat unexpectedly, the effect was not completely specific to RSV. Coexpression of NS1 also inhibited CAT expression by a minigenome analog of parainfluenza virus type 3 (PIV3) complemented by plasmid-expressed PIV3 N, P, and L proteins (12, 27a). However, the magnitude of the inhibition was at least 10-fold lower than that observed for RSV and thus occurred only at relatively high levels of NS1 expression. It is difficult to directly compare the two heterologous systems: each involves the expression of a number of components, none of which are in common, except for the CAT marker enzyme. Thus, there is no basis for standardization except that the two systems are similar in activity with regard to the production of CAT RNA and enzyme (reference 12 and data not shown). The finding that the magnitude of the inhibitory effect of NS1 was more than 10-fold greater with RSV than with PIV3 suggests that most of the effect is specific to RSV. There also might be some aspect that is common to RSV and PIV3. For example, the NS1 protein might bind to an RSV RNA or protein with a specificity that has some overlap with a PIV3 component, or part of its action might involve cellular components that both viruses use. Therefore, the inhibitory action of NS1 against PIV3 is not necessarily an artifact.
The results shown here did rule out certain trivial possibilities as causing the effect of NS1. The inhibitory effect of the NS1 plasmid was shown to depend on the presence of an intact NS1 ORF, evidence that the effect was mediated by NS1 protein rather than plasmid or RNA (Fig. 5). The synthesis of other RSV proteins from T7 expression plasmids was unaffected by coexpression of NS1 (Fig. 2 and unpublished data). The synthesis and encapsidation of plasmid-derived minigenome also were unaffected (Fig. 6). Thus, the effect of NS1 cannot be ascribed to perturbation or inhibition of plasmid transfection, synthesis of T7 polymerase by vaccinia virus, T7-mediated transcription, or translation and stability of plasmid-encoded RNA and protein. The inhibition mediated by NS1 also was not mitigated by coexpression of other RSV proteins.
We investigated which steps in RSV transcription and RNA replication might be affected by the NS1 protein. The finding that encapsidation of the plasmid-supplied minigenome was unaffected (Fig. 6) implied that the effect was at a later step. To determine whether the effect was at the level of RNA synthesis from the genome or antigenome, we used minireplicons that were engineered to contain either the genomic (MP13 and MP96) or the antigenomic (MP123) promoter at the 3′ end. An important feature of these minireplicons is that they also contain a point substitution, either TrG5A or TrA7C, in the trailer region. Both mutations blocked most of the amplification by the RSV polymerase, limiting the available template to that supplied from the plasmid. These trailer point mutations will be described in greater detail elsewhere (25a). They do not interfere with the ability of the plasmid-supplied minigenome to be encapsidated and function as a template for the synthesis of mRNA and the mini-antigenome. The mini-antigenome which is produced is also encapsidated but is impaired as template for the synthesis of progeny minigenome. Thus, RNA synthesis is limited to the promoter at the 3′ end of the plasmid-supplied template, allowing it to be examined in isolation. The results confirmed that NS1 affects both transcription and RNA replication and showed that the genomic and antigenomic promoters are equally affected.
Transcription by nonsegmented negative-strand viruses is polar, so that promoter-proximal genes are transcribed more frequently than are promoter-distal ones. This appears to be due to polymerase fall-off, which was shown to take place at the intergenic regions for the four shorter genes of vesicular stomatitis virus (19). The possibility that NS1 affected the gradient of transcriptional polarity was investigated by using two dicistronic minigenomes, MP13 and RSV-CAT-LUC(CAT), which contain dicistronic transcriptional units of 0.73 and 2.6 kb, respectively (Fig. 7) (data not shown). In each case, the upstream and downstream mRNAs were equally sensitive to inhibition by NS1, indicating that the polarity of gene transcription was unaffected. Thus, NS1 did not appear to affect chain elongation by the polymerase or its recognition and utilization of transcription signals. Also, the effect of NS1 protein on transcription was not altered by the presence or absence of the M2 plasmid, supplied either in the form of the first ORF alone, which encodes the transcription elongation factor, or in the form with the two ORFs in their natural configuration.
Taken together, these observations indicate that the NS1 protein acts at a stage after encapsidation but before sequential transcription. Since transcription and replication are equally strongly affected and since the two processes are thought to use the same promoter, it seems likely that NS1 acts at a common, early stage such as initiation at the genomic (and antigenomic) promoter. It remains to be determined whether NS1 acts on the encapsidated template or the polymerase. The latter possibility is suggested by the observation that 50% inhibition occurred with a concentration of the NS1 plasmid which is equimolar with the L plasmid but approximately 34- and 24-fold, respectively, lower than the molar concentrations of the N and P plasmids. However, 100% inhibition was observed only at a 20- to 25-fold molar excess of the NS1 plasmid relative to that of L.
The regulation of RNA synthesis by nonsegmented negative-strand RNA viruses is more complex than was previously appreciated. For example, (i) the RSV M2 mRNA encodes both a transcription elongation factor from its upstream ORF and a negative regulatory factor from its downstream ORF (9); (ii) the Sendai virus V protein inhibits the replication of a defective interfering (DI) genome, probably by interfering with either formation or use of the NP0-P encapsidation substrate (18); and (iii) the Sendai virus C proteins strongly inhibit the amplification of an internal deletion type of DI genome as well as of a complete infectious genome. However, C had no effect on the replication of a copyback-type DI genome or of an infectious genome that had been engineered to contain the antigenome promoter in place of the genome promoter (3). Hence, the C proteins appear to have a negative regulatory activity specific to the genomic promoter. Other studies also suggested that the Sendai virus C protein might be involved in down-regulating viral mRNA synthesis (11). Somewhat surprisingly, Sendai virus appears to require the expression of at least some form of C protein to be infectious (3), whereas the C protein of measles virus can be silenced without ablating infectivity (27). The inhibition caused by RSV NS1 seems to be different from that caused by the Sendai virus V and C proteins, because RSV NS1 did not interfere with encapsidation and was not promoter-specific.
The data described here strongly suggest that NS1 plays a role as a negative regulatory protein. The NS1 mRNA, encoded in the promoter-proximal gene, is the most abundant of the 10 mRNAs. A careful quantitation of the relative levels of NS1 protein expression during RSV infection has not yet been reported, but it is among the most highly expressed proteins. The potent inhibitory effects of NS1 would seem to be counterproductive for viral replication, especially during the early stages of infection, and thus it is difficult to envision what role this function might play in the RSV growth cycle. But the same comment can be made for the Sendai virus C protein, for example. It is likely that the apparent anomaly simply reflects the incomplete state of our understanding of the functions of these putative regulatory proteins. In addition, there is evidence that the NS1 protein is subject to posttranslational modification (10), which might temper its negative regulatory activity. The constant production of an unstable negative regulatory activity could be part of the reason why RSV replication and gene expression are slower and less productive than is the case for many other nonsegmented negative-strand RNA viruses.
Whether or not NS1 is a potent negative regulatory factor in a natural RSV infection, the finding that it inhibits minigenome RNA synthesis suggests that it interacts with components involved in that process. It will be of interest to examine whether RSV RNA synthesis and growth are affected in cell lines expressing the NS1 protein and to determine whether expression of the NS1 gene can be ablated or reduced in cDNA-encoded infectious virus (8) without compromising viral infectivity. Additional functional roles might also exist for the NS1 protein, such as in virion morphogenesis, interaction with the host cell, or interaction with innate or induced aspects of the host immune system.
ACKNOWLEDGMENTS
We thank Ena Camargo for performing the cell culture, Myron Hill for synthesizing and sequencing oligonucleotides, Juan Cristina for synthesizing riboprobes, and Michael Teng, Mario Skiadopoulos, and Anna Durbin for testing the effect of NS1 on the PIV3 minigenome. We also thank Siba Samal and Rachel Fearns for helpful discussions.
REFERENCES
- 1.Alansari H, Potgeiter L N D. Nucleotide and predicted amino acid sequence analysis of the ovine respiratory syncytial virus nonstructural 1C and 1B genes and the small hydrophobic protein gene. J Gen Virol. 1994;75:401–404. doi: 10.1099/0022-1317-75-2-401. [DOI] [PubMed] [Google Scholar]
- 2.Baker S C, Moyer S A. Encapsidation of Sendai virus genome RNAs by purified NP protein during in vitro replication. J Virol. 1988;62:834–838. doi: 10.1128/jvi.62.3.834-838.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski P. One hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal Biochem. 1992;20:134–139. doi: 10.1016/0003-2697(92)90185-a. [DOI] [PubMed] [Google Scholar]
- 5.Collins P L, Wertz G W. cDNA cloning and transcriptional mapping of nine polyadenylated RNAs encoded by the genome of human respiratory syncytial virus. Proc Natl Acad Sci USA. 1983;80:3208–3212. doi: 10.1073/pnas.80.11.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins P L, Wertz G W. Nucleotide sequences of the 1B and 1C nonstructural protein mRNAs of human respiratory syncytial virus. Virology. 1985;143:442–451. doi: 10.1016/0042-6822(85)90384-8. [DOI] [PubMed] [Google Scholar]
- 7.Collins P L. The molecular biology of human respiratory syncytial virus (RSV) of genus Pneumovirus. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 103–162. [Google Scholar]
- 8.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the M2 (ORF1) transcription elongation factor in gene expression and provides a new capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1313–1352. [Google Scholar]
- 11.Curran J, Marq J-B, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 12.Durbin A P, Siew J W, Murphy B R, Collins P L. Minimum requirements for transcription and RNA replication of a minigenome of human parainfluenza virus type 3. Virology. 1997;234:74–83. doi: 10.1006/viro.1997.8633. [DOI] [PubMed] [Google Scholar]
- 13.Elroy-Stein O, Fuerst T R, Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid system. Proc Natl Acad Sci USA. 1989;86:6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fearns R, Peeples M E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 15.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosfeld H, Hill M G, Collins P L. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins: transcription also occurs under these conditions but required RSV superinfection for efficient synthesis of full-length mRNA. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horikami S, Smallwood S, Moyer S. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology. 1996;222:383–390. doi: 10.1006/viro.1996.0435. [DOI] [PubMed] [Google Scholar]
- 19.Iverson L E, Rose J K. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 20.Johnson P R, Collins P L. The 1B (NS2), 1C (NS1) and N proteins of human respiratory syncytial virus (RSV) of antigenic subgroups A and B: sequence conservation and divergence within RSV genomic RNA. J Gen Virol. 1989;70:1539–1547. doi: 10.1099/0022-1317-70-6-1539. [DOI] [PubMed] [Google Scholar]
- 21.Kuo L, Grosfeld H, Cristina J, Hill M G, Collins P L. Effect of Mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol. 1996;70:6892–6901. doi: 10.1128/jvi.70.10.6892-6901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo L, Fearns R, Collins P L. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J Virol. 1996;70:6143–6150. doi: 10.1128/jvi.70.9.6143-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo L, Fearns R, Collins P L. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J Virol. 1997;71:4944–4953. doi: 10.1128/jvi.71.7.4944-4953.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1177–1204. [Google Scholar]
- 25.Patsey M, Samal S K. Nucleotide sequence analysis of the non-structural NS1 (1C) and NS2 (1B) protein genes of bovine respiratory syncytial virus. J Gen Virol. 1995;76:193–197. doi: 10.1099/0022-1317-76-1-193. [DOI] [PubMed] [Google Scholar]
- 25a.Peeples, M. E., R. Fearns, J. Cristina, and P. L. Collins. Unpublished data.
- 25b.Peeples, M. E., R. Fearns, and P. L. Collins. Unpublished data.
- 26.Perotta A T, Been M D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 27.Radecke F, Billeter M A. The nonstructural C protein is not essential for multiplication of Edmonston B strain measles virus in cultured cells. Virology. 1996;217:418–421. doi: 10.1006/viro.1996.0134. [DOI] [PubMed] [Google Scholar]
- 27a.Teng, M., A. Durbin, and M. Skiadopoulos. Unpublished data.
- 28.Towbin H, Staehelin R, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner R R, Rose J K. Rhabdoviridae and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1121–1135. [Google Scholar]