Abstract
The human immunodeficiency virus type 1 transmembrane protein gp41 oligomer anchors the attachment protein, gp120, to the viral envelope and mediates viral envelope-cell membrane fusion following gp120-CD4 receptor-chemokine coreceptor binding. We have used mutation-directed chemical cross-linking with bis(sulfosuccinimidyl)suberate (BS3) to investigate the architecture of the gp41 oligomer. Treatment of gp41 with BS3 generates a ladder of four bands on sodium dodecyl sulfate-polyacrylamide gels, corresponding to monomers, dimers, trimers, and tetramers. By systematically replacing gp41 lysines with arginine and determining the mutant gp41 cross-linking pattern, we observed that gp41 N termini are cross-linked. Lysine 678, which is close to the transmembrane sequence, was readily cross-linked to Lys-678 on other monomers within the oligomeric structure. This arrangement appears to be facilitated by the close packing of membrane-anchoring sequences, since the efficiency of assembly of heterooligomers between wild-type and mutant Env proteins is improved more than twofold if the mutant contains the membrane-anchoring sequence. We also detected close contacts between Lys-596 and Lys-612 in the disulfide-bonded loop/glycan cluster of one monomer and lysines in the N-terminal amphipathic α-helical oligomerization domain (Lys-569 and Lys-583) and C-terminal α-helical sequence (Lys-650 and Lys-660) of adjacent monomers. Precursor-processing efficiency, gp120-gp41 association, soluble recombinant CD4-induced shedding of gp120 from cell surface gp41, and acquisition of gp41 ectodomain conformational antibody epitopes were unaffected by the substitutions. However, the syncytium-forming function was most dependent on the conserved Lys-569 in the N-terminal α-helix. These results indicate that gp160-derived gp41 expressed in mammalian cells is a tetramer and provide information about the juxtaposition of gp41 structural elements within the oligomer.
The human immunodeficiency virus type 1 (HIV-1) envelope (Env) glycoproteins are synthesized as a precursor, gp160, which undergoes posttranslational cleavage to yield the noncovalently associated receptor-binding gp120 and transmembrane gp41 subunits (21, 70). Attachment to the majority of target cells is mediated through a specific interaction between gp120 and cellular CD4 molecules (14, 37, 40). Entry of HIV-1 into susceptible cells also requires the presence of fusion cofactors which have recently been identified as the chemokine receptors CXCR4, CCR2b, CCR3, and CCR5 (2, 12, 15–17, 24, 35). Viral attachment via gp120-CD4-chemokine coreceptor interactions causes conformational changes within the Env complex (3, 53, 61, 62, 72) which presumably trigger gp41-mediated fusion between viral and cellular membranes, resulting in viral entry (27).
Oligomerization of the Env precursor, gp160, occurs in the endoplasmic reticulum (21) through an amphipathic α-helical sequence close to the N terminus of gp41 (9, 50, 66) and precedes the structural maturation of the gp41 domain (44). Whereas gp41 has a stable oligomeric structure (47), the association between gp120 and gp41 is labile, and gp120 dissociates into monomers when released from gp41 (20, 46, 64). Env oligomerization is likely to be important for receptor binding and entry, because multiple gp120 molecules are required for CD4-dependent infection (39) and hetero-oligomers comprising wild-type and fusion-incompetent Env monomers have a fusion-negative phenotype (26). Env oligomerization also has important implications for Env immunogenicity and antigenicity, because primary HIV isolates are potently neutralized by monoclonal antibodies (MAbs) that are able to bind to oligomeric Env, whereas neutralization by MAbs that recognize only monomeric gp120 is limited (25, 61). This is presumably due to occlusion or alteration of antibody epitopes upon Env oligomerization (54, 59).
Studies aimed at determining the quaternary structure of the HIV-1 Env glycoproteins have yielded conflicting results, with trimeric and tetrameric structures being reported. Chemical cross-linkage of viral gp41 or polyacrylamide gel electrophoresis (PAGE) of viral gp41 in low concentrations of sodium dodecyl sulfate (SDS) indicate a tetrameric structure (47, 49, 56), and this is supported by chemical cross-linking studies of gp160, gp120, and gp41 expressed in mammalian cells (19, 20, 46, 48). However, one study indicates that virion-associated gp120 is cross-linked in a trimeric form (64). Circular dichroism and sedimentation equilibrium analyses of a synthetic peptide analog of the α-helical oligomerization domain, Asn-448 to Gln-585 (the HIV-1LAI BH8 clone numbering system is used throughout this report [52]), indicates that it forms a four-stranded coiled coil in solution (51). Furthermore, fusion of the gp41 α-helical sequence to the C terminus of the normally monomeric Escherichia coli maltose-binding protein results in the formation of tetramers (57). In contrast, various peptide analogs of the N-terminal α-helical sequence form stable α-helical trimers when complexed with peptide fragments derived from a C-terminal amphipathic α-helical sequence (9, 66). The latter structures may be different from tetrameric gp41 cleaved from gp160 in mammalian cells.
Structural elements in the gp41 ectodomain were originally modeled (31) on the structure of the influenza virus hemagglutinin (HA) transmembrane protein, HA2 (71), with an N-terminal hydrophobic fusogenic sequence (30), an amphipathic α-helical/leucine zipper-like oligomerization domain (50), a disulfide-bonded loop (73), a glycan cluster (23), and a C-terminal ectodomain amphipathic α-helix (9, 66, 68). The hydrophobic transmembrane sequence is located further C terminally (38) and precedes an unusually long cytoplasmic tail that is important for incorporation of Env into virions through an interaction with matrix protein (13, 28). Chan et al. (9) and Weissenhorn et al. (66) recently reported the crystal structures of complexes between segments of the N-terminal amphipathic α-helical oligomerization domain and the C-terminal α-helical sequence. The N-terminal α-helix forms a three-stranded parallel coiled coil with C-terminal α-helices packing in an oblique antiparallel manner into highly conserved, hydrophobic grooves on the surface of the coiled coil. Those authors propose that these structures represent the core of fusion-active gp41, because they share structural features with the fusion-active form of influenza virus HA2.
We have now used mutation-directed chemical cross-linking to obtain information about the architecture of oligomeric gp41 derived from mammalian cell-expressed gp160. Lysines in the gp41 ectodomain were systematically replaced with arginine to eliminate potential sites for intermonomer chemical cross-links mediated by bis(sulfosuccinimidyl)suberate (BS3). By determining the effects of these mutations on the gp41 cross-linking pattern, we show that N termini are closely juxtaposed, as are monomer Lys-678 residues, which are adjacent to the transmembrane sequence. Close positioning of heterologous intermonomer residues was also observed; Lys-596 and/or Lys 612 could be cross-linked with lysines located within both the N- and C-terminal gp41 ectodomain amphipathic α-helical sequences.
MATERIALS AND METHODS
Cells and virus.
The vaccinia virus-T7 RNA polymerase transient-expression system (41) was used for expression of wild-type and mutant Env glycoproteins. The recombinant vaccinia virus vTF7-3 was obtained from T. M. Fuerst and B. Moss (41), and HeLa-T4 cells were obtained from P. J. Maddon (40) through the AIDS Research and Reference Reagents Program, National Institute of Allergy and Infectious Diseases (Bethesda, Md.). HeLa-T4 cells were maintained in Dulbecco’s modification of minimal essential medium containing 10% fetal calf serum and 1 mM glutamine (DMEMF10) plus 500 μg of G418 (Life Technologies, Gaithersburg, Md.) per ml, while HeLa cells were cultured in the absence of G418. Wild-type and mutant Env glycoproteins were expressed following infection of cells with vTF7-3 and transfection with 10 μg of plasmid DNA by using Lipofectin (Life Technologies).
Plasmid constructs.
The construction of pTMenv.2, which directs expression of the full-length env gene (BH8 clone of HIV-1LAI [52]), is described elsewhere (48). An EcoRI-SalI fragment from pTMenv.2 (env nucleotides 1141 to 2553) was ligated into M13mp18 for use as a single-stranded template for construction of gp41 Lys-to-Arg mutants by the Sculptor oligonucleotide-directed in vitro mutagenesis procedure (Amersham International, Little Chalfont, United Kingdom). The sequences of mutant oligonucleotide primers will be provided upon request. In some cases, multiple Lys-to-Arg substitutions were generated by exchanging EcoRI-HindIII (env nucleotides 1141 to 1902) or HindIII-BamHI (env nucleotides 1902 to 2236) fragments between mutants. The dual T7 promoter-driven expression plasmid pPT7− (50) was used for coexpression of gp160 and gp160 truncation mutants. The construction of pP672, which coexpresses gp160 with the gp160(1-672) mutant, was described previously (50). The vector pP701, which coexpresses gp160 with gp160(1-701), was constructed by replacing the HindIII-StuI fragment (env nucleotides 1902 to 2016) of pP672 with a PCR fragment carrying env nucleotides 1902 to 2103 followed by a 3′ StuI site. PCR was performed with Pfu polymerase (Stratagene, La Jolla, Calif.), the oligonucleotide primers 5′-GGAGCAGGATCCAGCACTATGGGCGCAGCGTCA and 5′-GCCTAAAGGCCTGCACTATAGAAAGTAC, and pPT7− as the template. Inclusion of the 3′ StuI site (in bold type) required an Asn-701-to-Gln substitution followed by Ala at the mutant C terminus. DNA sequences were confirmed by the dideoxy chain termination procedure with the Sequenase method (U.S. Biochemical, Cleveland, Ohio).
Sucrose density gradient centrifugation.
HeLa cells (1.1 × 106 total cells in two 9-cm2 wells) were infected with vTF7-3 and transfected with plasmid DNA as described above. Cells were lysed at 24 h posttransfection with phosphate-buffered saline (PBS) containing 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1% Triton X-100 for 10 min on ice. Lysates were clarified by centrifugation at 10,000 × g for 10 min and then layered onto 5 to 20% sucrose gradients containing PBS and 1% Triton X-100. Centrifugation was performed in a Beckman SW41Ti rotor at 35,000 rpm for 19 h at 4°C. Following centrifugation, 0.5-ml fractions were obtained, proteins therein were cross-linked with 0.5 mM BS3 (Pierce Chemical Company, Rockford, Ill.) for 1 h on ice, and the reactions were quenched with 100 mM glycine (pH 7.8) for 1 h on ice. Proteins were precipitated with trichloroacetic acid and separated by SDS-PAGE with 5 to 15% polyacrylamide gradient gels under reducing conditions. Proteins were transferred to nitrocellulose, immunoblotted with antibodies directed against gp41 residues Gly-588 to Cys-599 (32, 49), and then probed with radioiodinated protein A. Immunoblots were visualized by autoradiography. Sucrose gradients were calibrated with the molecular size markers catalase (11.3S), aldolase (7.3S), and ovalbumin (3.55S) (Pharmacia Biotech, Uppsala, Sweden). Protein A (Pharmacia Biotech) was radioiodinated by the chloramine T procedure (33).
HeLa-T4 syncytium assay.
vTF7.3-infected HeLa-T4 cells (3.5 × 105 in 9-cm2 wells) were transfected with 10 μg of plasmid DNA as described above. At 24 h posttransfection, cells were washed with PBS and stained by the May-Grünwald-Giemsa technique (18). Syncytium formation was quantitated by direct counting of 12 fields per well at a magnification of ×200 and scoring only those multinucleated cells with six or more nuclei.
Biosynthetic labeling of Env glycoproteins.
HeLa cells expressing wild-type or mutant Env were incubated in cysteine- and methionine-deficient DMEMF10 (ICN, Costa Mesa, Calif.) for 30 min at 37°C. The cells were then pulsed with 150 μCi of Tran35S-label (ICN), washed with DMEMF10, and then chased with complete DMEMF10 (0.6 ml/well) for 4 h at 37°C. Following the chase period, culture supernatants were clarified by centrifugation at 10,000 × g for 90 s, and then 0.2 ml of quadruple-strength lysis buffer (200 mM Tris HCl [pH 7.4] containing 2.4 M KCl, 4 mM EDTA, 4 mM phenylmethylsulfonyl fluoride, and 4% Triton X-100) was added and samples were held on ice. Monolayers were lysed with 0.8 ml of single-strength lysis buffer for 10 min on ice and then clarified by centrifugation at 10,000 × g for 10 min prior to radioimmunoprecipitation.
Radioimmunoprecipitation.
Samples were precleared overnight at 4°C with 10% (vol/vol) bovine serum albumin coupled to CNBr-activated Sepharose CL4B (Pharmacia Biotech). Radioimmunoprecipitation was performed with protein A-Sepharose (Pharmacia Biotech) and either human antibodies or murine MAbs plus rabbit anti-mouse immunoglobulin G (IgG). CD4-binding assays were performed in the presence of MAb OKT4 (6 μg) and soluble recombinant CD4 (sCD4) (2 μg) obtained through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, from R. Sweet, SmithKline Beecham. Immunoprecipitated glycoproteins were electrophoresed on 5 to 15% gradient gels under reducing conditions and then visualized by autoradiography or PhosphorImager SF (Molecular Dynamics, Sunnyvale, Calif.) analysis.
sCD4-induced gp120 shedding.
HeLa cells expressing wild-type or mutant Env glycoproteins were biosynthetically labeled for 1 h as described above. Following a 4-h chase period, culture supernatants were removed, and monolayers were washed and overlaid with 0 or 15 μg of sCD4 in 0.6 ml of DMEM containing 2% fetal calf serum. Following a 90-min incubation at 37°C, the medium was removed, clarified by centrifugation (10,000 × g, 90 s), and immunoprecipitated with a cocktail of human anti-recombinant gp120 (human α-rgp120) antibody plus IgG purified from pooled HIV-1-positive human plasma. Immunoprecipitates were subjected to reducing SDS-PAGE and analyzed by autoradiography.
Antibodies.
Human α-588-599 antibodies specific for the gp41 immunodominant epitope Gly-588 to Cys-599 (GIWGCSGKLIC) (32) were purified from pooled HIV-positive human plasma by peptide affinity chromatography with a gp41(588-599) peptide-Sepharose CL4B column as described previously (49). Human α-rgp120 antibodies were purified from pooled HIV-1-positive human plasma by affinity chromatography on an rgp120 (HIV-1SF2 strain)-Sepharose CL4B column as described previously (49). MAbs obtained through the AIDS Research and Reference Reagent Program, National Institute for Allergy and Infectious Diseases, include 126-6 from S. Zoller Pazner (73), MD-1 from R. Myers (43), 902 from B. Chesebro (11), and chessie 8 (C8) from G. Lewis (1). MAb OKT4 (36) was obtained from the American Type Culture Collection (Rockville, Md.). MAb IC3/86 directed to human glycophorin (7) was obtained from Agen Biomedicals, Brisbane, Australia, and used as a control antibody.
RESULTS
Identification of near-neighbor lysines within the gp41 oligomer by using mutation-directed chemical cross-linking.
Chemical cross-linking agents have been widely used to assess the oligomeric valency of viral Env proteins. Studies of gp41 ectodomain fragments and influenza virus HA indicate that the molecular weight of the highest-order cross-linked species determined by chemical cross-linking and SDS-PAGE experiments corresponds to the oligomeric valency as determined definitively by X-ray crystallography or by equilibrium sedimentation. For example, influenza virus HA, which has been shown by X-ray crystallography to be a trimer in both the pH 7 and pH 5 forms (5, 71), migrates in SDS-polyacrylamide gels with molecular weights corresponding to monomers, dimers, and trimers following cross-linkage through primary amines with dimethyl suberimidate (11-Å spacer [69]) or BS3 (11.4-Å spacer [10]). The crystal structures of complexes formed between two α-helical peptides derived from the gp41 ectodomain indicate that they pack into a trimer of peptide heterodimers (9, 66). Cross-linkage of a cysteine-mutated version of this peptide complex with bismaleimidohexane (16.1-Å spacer) also indicated a trimeric structure (65). In agreement with this finding, a trimeric quaternary structure is observed for a gp41 ectodomain fragment lacking the N-terminal fusion peptide, transmembrane domain, and cytoplasmic tail expressed in insect cells when treated with ethylene glycolbis(succinimidylsuccinate), which has a 16.1-Å spacer arm (67). In contrast, a chimera comprising the gp41 α-helical oligomerization domain fused to the C terminus of the E. coli maltose-binding protein forms a tetramer as assessed by equilibrium sedimentation (57), and treatment of this protein with ethylene glycolbis(succinimidylsuccinate) gave rise to dimers, trimers, and tetramers. The results of these studies indicate that the cross-linking profiles generated for these proteins reflect their oligomeric status and that gp41 has the potential to pack into both trimers and tetramers, depending on which sequences are expressed.
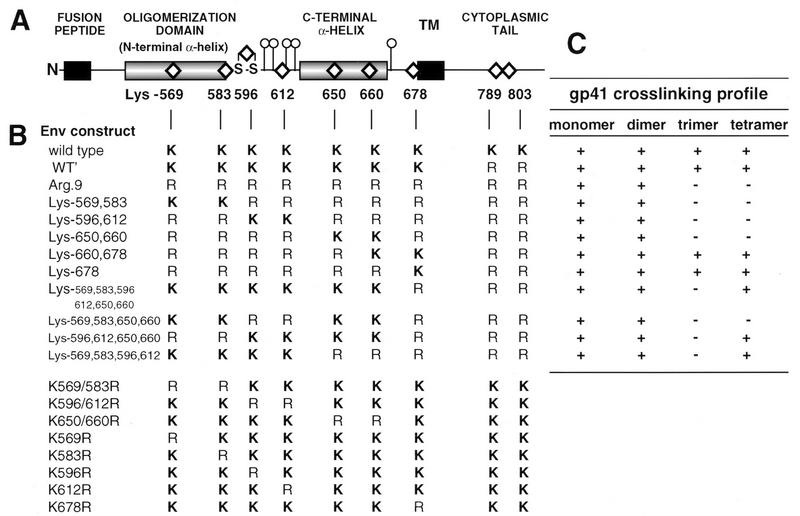
Chemical cross-linkage of mammalian cell-expressed gp160 and gp41 oligomers results in their migration as dimers, trimers, and tetramers on SDS-polyacrylamide gels, suggesting tetrameric structures for these glycoproteins (19, 20, 48, 56). There are nine lysine cross-linking targets within HIV-1LAI gp41, and lysine pairs are conveniently positioned within structure-function elements (Fig. 1A). Lys-569 and Lys-583 are within the N-terminal amphipathic α-helical oligomerization domain between Ala-536 and Gln-586 (9, 50, 66). Lys-596 and Lys-612 are within an immunodominant region containing the disulfide-bonded loop, Cys-593 to Cys-599, and glycan cluster, while Lys-650 and Lys-660 are in a C-terminal amphipathic α-helical sequence between Asn-619 and Leu-658 (9, 66, 68). Lysine-678 is adjacent to the transmembrane sequence (38). Lysine-569 is strictly conserved among clade B isolates, while the remaining lysines are present in at least 50% of clade B isolates (42). We reasoned that information about the architecture of the gp41 oligomer could be obtained by identifying which lysine residues are responsible for intermolecular cross-links and are therefore closely juxtaposed.
FIG. 1.
(A) Linear diagram of gp41 depicting regions assigned as structural and/or functional domains. The positions of the nine lysine residues targeted with Arg substitutions are indicated by diamonds. Also shown are the N-linked glycosylation sites (○|) and the disulfide loop (S-S). TM, transmembrane sequence. (B) Lys-Arg profiles of gp41 mutants. (C) Summary of mutant gp41 cross-linking profiles (see Fig. 2 to 5) indicating the presence (+) or absence (−) of cross-linked gp41 oligomeric species on immunoblots probed with human α-588-599 antibody.
Lysine residues within gp41 were systematically replaced with arginine (Fig. 1B), and the effect of these mutations on the gp41 cross-linking pattern was determined. The water-soluble chemical cross-linker BS3 specifically cross-links the NH2 of lysine residues or the protein N-terminal NH2 provided that the reactive groups are within 11.4 Å (60). This agent was used to covalently stabilize the gp41 oligomer following sedimentation over sucrose density gradients. Cross-linked gp41 species were visualized by SDS-PAGE and immunoblot analysis with human α-588-599 antibodies. Figure 2 indicates that following sedimentation over sucrose density gradients, wild-type gp41 peaks in fractions corresponding to 4.4 to 7S. BS3 treatment of gradient fractions resulted in a ladder containing four species in 4.9 to 6S fractions which correspond to gp41 monomers (40.7 kDa), dimers (84.1 kDa), trimers (111 kDa), and tetramers (157 kDa). In some experiments with wild-type Env [see, for example, Fig. 2, WT (BS3) panel], gp41 tetramers and dimers predominated in fraction 15 whereas fraction 17 contained mostly trimers and dimers, raising the possibility that gp41 sediments as a mixture of tetramers and trimers in this system. Nevertheless, our results are consistent with the tetramer being the highest-order structure for gp41 and agree with previous studies on the oligomerization of mammalian-cell-expressed gp41 and gp160 (19, 20, 47, 48, 56). An ∼240-kDa band was observed in BS3-treated fractions 13 and 15 (Fig. 2) and cannot be assigned an oligomeric structure. This may represent a cross-linked complex of monomeric gp160 irreversibly associated with BiP/GRP78 (44). A similar gp160 and gp41 cross-linking profile was obtained with Env proteins expressed in HeLa cells infected with the recombinant vaccinia virus vPE16 (19), which directs lower-level expression of BH8 Env from the vaccinia virus P7.5 early-late promoter (data not shown). The four species corresponding to cross-linked gp41 were not detected by the anti-gp120 V3 loop MAb 902, indicating that they were not gp120-containing cross-linked complexes.
FIG. 2.
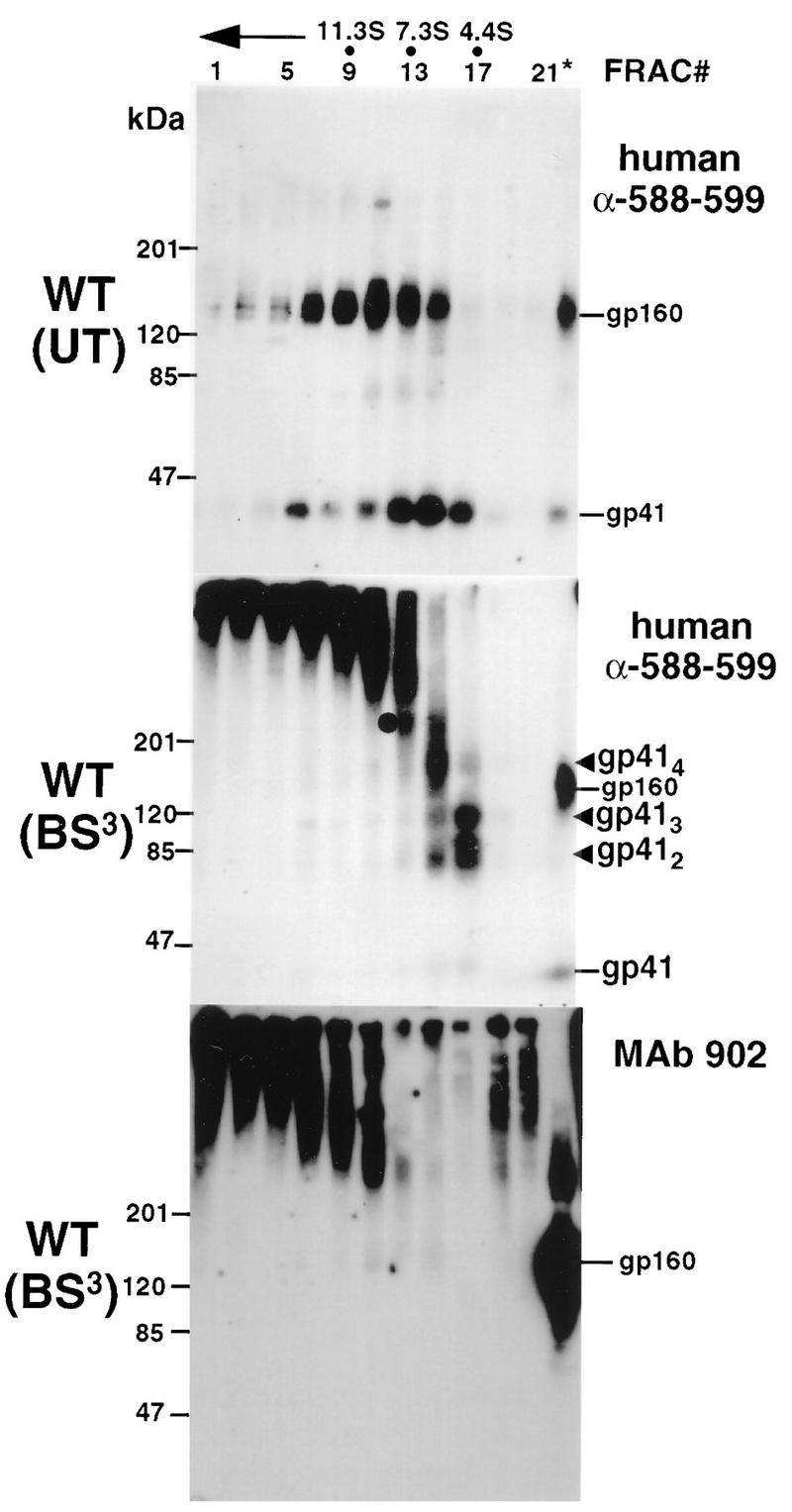
Sedimentation and cross-linking profiles of wild-type (WT) Env. vTF7-3-infected HeLa cells were transfected with a wild-type-Env-encoding plasmid and then lysed at 24 h posttransfection. Cell lysates were subjected to centrifugation through 5 to 20% sucrose gradients. Gradients were fractionated and treated with 0.5 mM BS3 where indicated. Trichloroacetic acid-precipitated proteins were separated by SDS-PAGE on 5 to 15% gradient gels under reducing conditions. Proteins were transferred to nitrocellulose, immunoblotted with human α-588-599 antibody or MAb 902, and detected by addition of 125I-protein A and autoradiography. gp41 species are indicated by arrowheads. The circle in the panel for BS3 treatment with α-588-599 antibody indicates a 240-kDa band that cannot be assigned an oligomeric structure (see text). The direction of sedimentation is right to left and is indicated by an arrow. Unfractionated cell lysates (∗) were also subjected to SDS-PAGE, and the position of monomeric gp160 is indicated. FRAC, fraction.
Results from a number of studies suggest that two amphipathic α-helical sequences within the gp41 cytoplasmic tail are membrane associated (34, 58, 74). In order to prevent intermonomer cross-linkage through Lys-789 and/or Lys-803 within the cytoplasmic tail following membrane disruption with Triton X-100, both residues were replaced with arginine. This double mutant had a sedimentation profile and cross-linking pattern similar to those observed for wild-type Env (Fig. 3, WT′ panel) and produced syncytia in HeLa-T4 cells that were indistinguishable from wild type (see Table 1). We refer to this mutant as WT′, and subsequent Lys-to-Arg gp41 ectodomain mutants were prepared on this background for cross-linking studies. An Arg.9 mutant, in which all gp41 lysine residues were replaced with arginine, was prepared. While the gp41 sedimentation profile for this mutant was similar to that for the wild type, BS3 treatment yielded gp41 dimers (85 kDa) with no evidence of trimers or tetramers (Fig. 3). This results from one cross-link site per monomer and indicates that the N termini of gp41 monomers are within 11.4 Å of each other in the oligomer.
FIG. 3.
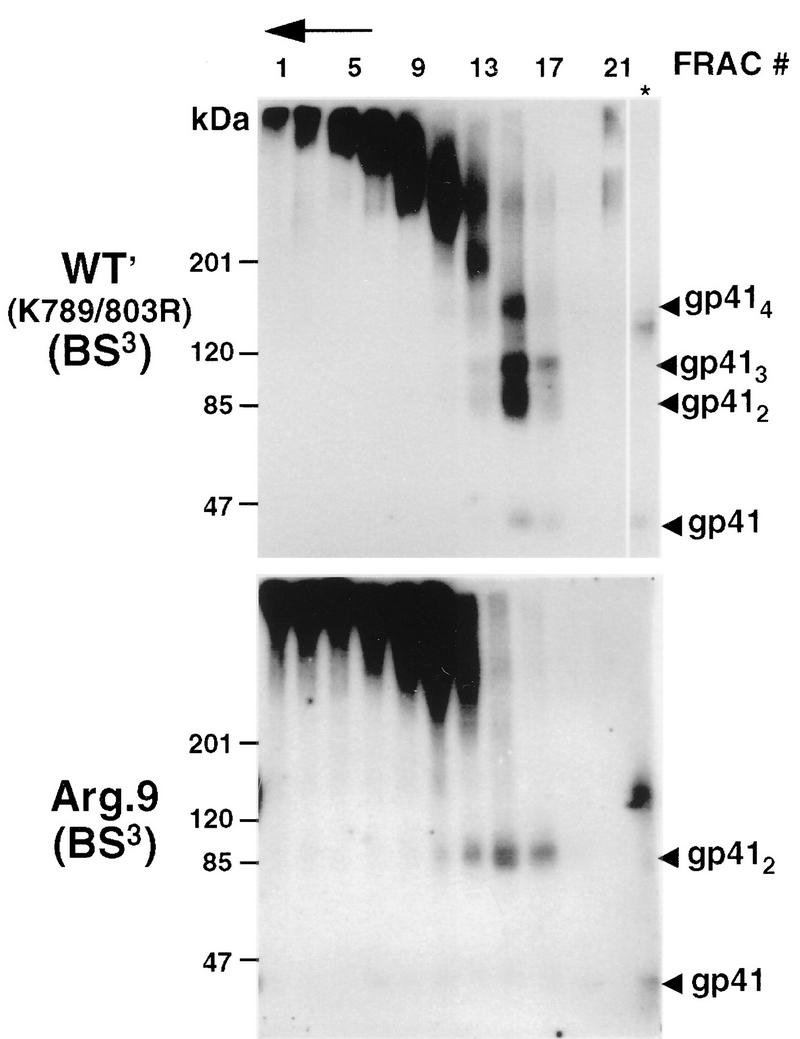
Sedimentation and BS3-cross-linking profiles of WT′ and Arg.9 Lys-to-Arg mutants. HeLa cells expressing the mutants WT′ (Lys-789,803 to Arg) or Arg.9 (a mutant in which all gp41 Lys residues were replaced with Arg) were lysed and treated as described in the legend to Fig. 2. Env glycoproteins were visualized by immunoblotting with human α-588-599 antibody and 125I-protein A. The direction of sedimentation is right to left and is indicated by an arrow. ∗, unfractionated cell lysates; FRAC, fraction.
TABLE 1.
Syncytium-forming function of Lys-to-Arg mutants
| Env protein | Relative syncytium-forming function (mean ± SD)a |
|---|---|
| Wild type | 100 |
| WT′ | 106 ± 15 |
| Arg.9 | 2 ± 2 |
| Lys-569,583 | 3 ± 3 |
| Lys-596,612 | 3 ± 2 |
| Lys-650,660 | 3 ± 1 |
| Lys-660,678 | 4 ± 1 |
| Lys-569,583,650,660 | 8 ± 5 |
| Lys-596,612,650,660 | 4 ± 1 |
| Lys-569,583,596,612 | 50 ± 3 |
| K569/583Rb | 19 ± 8 |
| K596/612Rb | 50 ± 11 |
| K650/660Rb | 87 ± 7 |
| K569Rb | 20 ± 12 |
| K583Rb | 113 ± 9 |
| K596Rb | 97 ± 11 |
| K612Rb | 117 ± 7 |
| K678Rb | 103 ± 4 |
| Control plasmid (pTM.1) | 6 ± 4 |
vTF7.3-infected HeLa T4 cells (3.5 × 105) were transfected with 10 μg of vector DNA and then stained by the May-Grünwald procedure at 24 h posttransfection. Syncytia were defined as multinucleated cells containing six or more nuclei per field at a magnification of ×200. All data represent the means from at least three experiments.
Prepared on a Lys-789,803 background. All other mutants were prepared on an Arg-789,803 background.
To identify lysine residues contributing to the gp41 trimer and tetramer cross-linking profile, a series of mutants with lysine pair combinations on the Arg.9 mutant background were prepared. The mutants Lys-569,583, Lys-596,612, and Lys-650,660 were covalently stabilized as gp41 dimers, but not trimers or tetramers, following BS3 treatment (Fig. 4). This indicates that maintenance of lysine pairs at positions 569 plus 583,596 plus 612, and 650 plus 660 does not facilitate trimer or tetramer cross-links, indicating that these lysines are out of range of their homologous positions or heterologous partners in neighboring monomers. In contrast, the gp41 cross-linking profile of mutant Lys-660,678 resembled that of wild-type gp41, because dimers, trimers, and tetramers were visible following BS3 cross-linking of gradient fractions. BS3 treatment of mutant Lys-678, which contains only Lys-678 on an Arg.9 background, resulted in gp41 tetramers, trimers, and dimers, which is similar to the case for mutant Lys-660,678 (Fig. 5). In contrast, the mutant Lys-569,583,596,612,650,660, which has a Lys-678-to-Arg substitution on a background where the other six gp41 ectodomain lysines are maintained, was cross-linked in dimeric and tetrameric forms, with trimers absent (Fig. 5). Thus, Lys-678 is sufficient for the generation of cross-linked gp41 tetramers and trimers. However, additional intermonomer cross-links are also mediated by the remaining ectodomain lysines to generate tetramers.
FIG. 4.
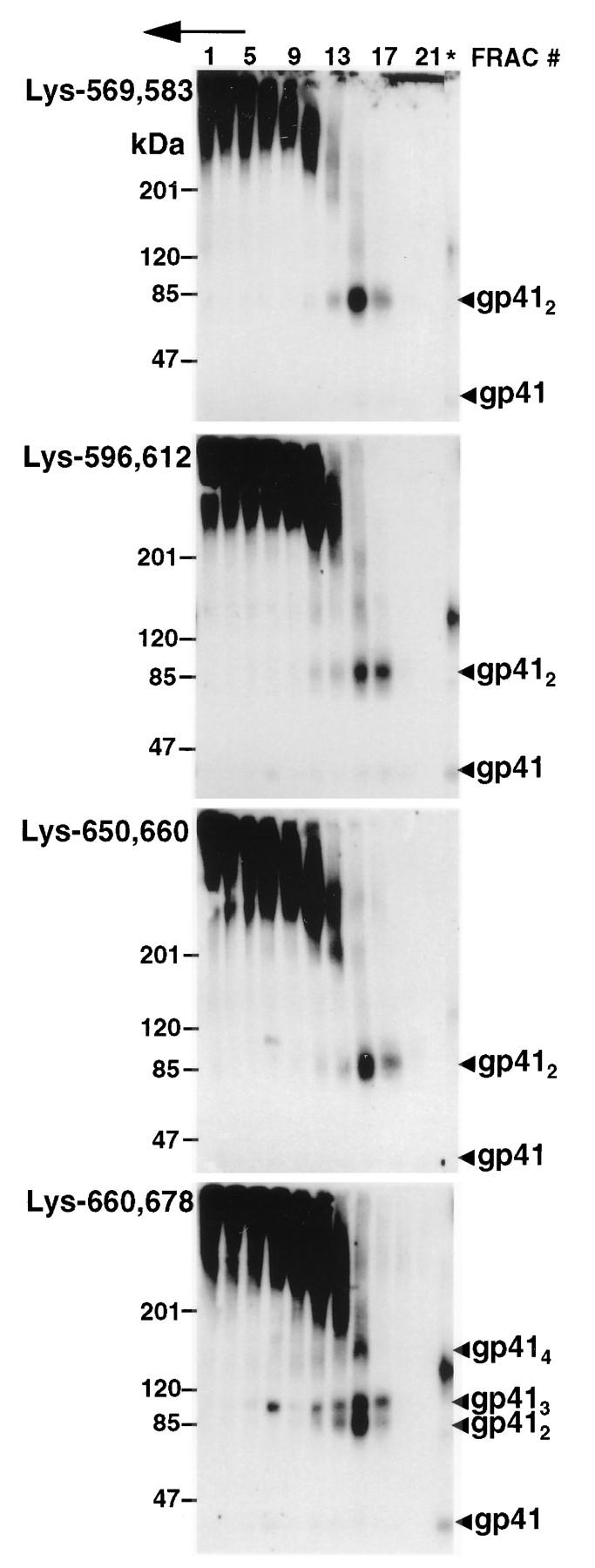
Sedimentation and BS3-cross-linking profiles of gp41 mutants containing two ectodomain lysines. Lysates from HeLa cells expressing the indicated Lys-to-Arg Env mutants were treated as described in the legend to Fig. 2. Env glycoproteins were visualized by immunoblotting with human α-588-599 antibody and 125I-protein A. The direction of sedimentation is right to left and is indicated by an arrow. ∗, unfractionated cell lysates; FRAC, fraction.
FIG. 5.
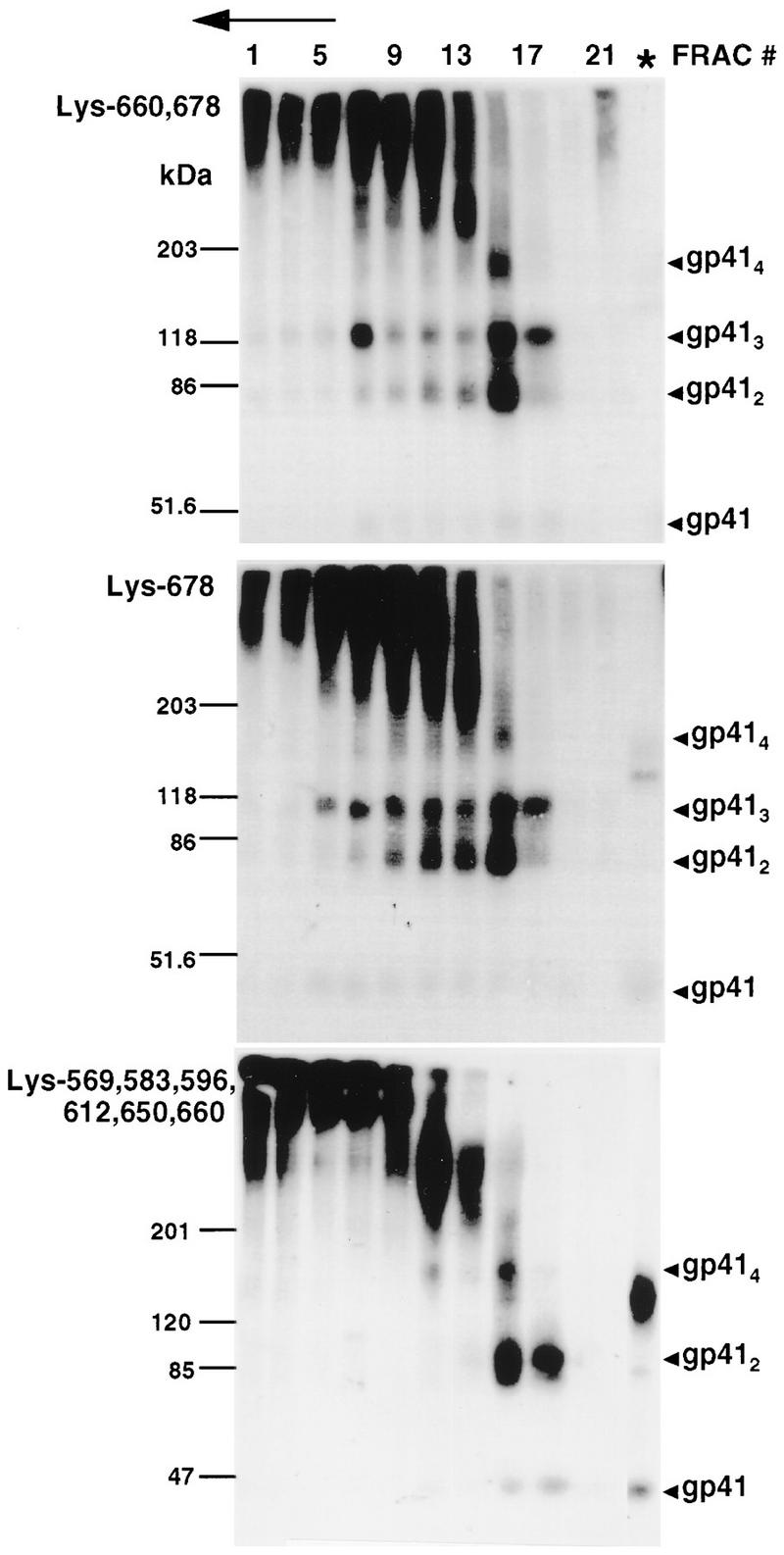
Role of Lys-678 in chemical cross-linkage of gp41 oligomers. Lysates from HeLa cells expressing the indicated Lys-to-Arg Env mutants were treated as described in the legend to Fig. 2. Env glycoproteins were visualized by immunoblotting with human α-588-599 antibody and 125I-protein A. The direction of sedimentation is right to left and is indicated by an arrow. ∗, unfractionated cell lysates; FRAC, fraction.
We therefore investigated if lysine pairs within one gp41 structural element were able to form intermolecular cross-links with lysine pairs within a heterologous element on separate monomers by maintaining combinations of four lysines. BS3 treatment of the mutant Lys-569,583,650,660 resulted in the covalent stabilization of gp41 oligomers in dimeric form only (Fig. 6). This result indicates that Lys-569 and Lys-583 in the N-terminal α-helix of one monomer are out of range of Lys-650 and Lys-660 in the C-terminal α-helix of another monomer. By contrast, BS3 treatment of the mutants Lys-569,583,596,612 and Lys-596,612,650,660 resulted in gp41 species corresponding to dimers and tetramers. These results indicate that Lys-596 within the disulfide-bonded loop and/or Lys-612 within the gp41 glycan cluster is within range of lysine residues present in both the N-terminal (Lys-569 and Lys-583) and C-terminal (Lys-650 and Lys-660) amphipathic α-helical sequences of adjacent monomers.
FIG. 6.
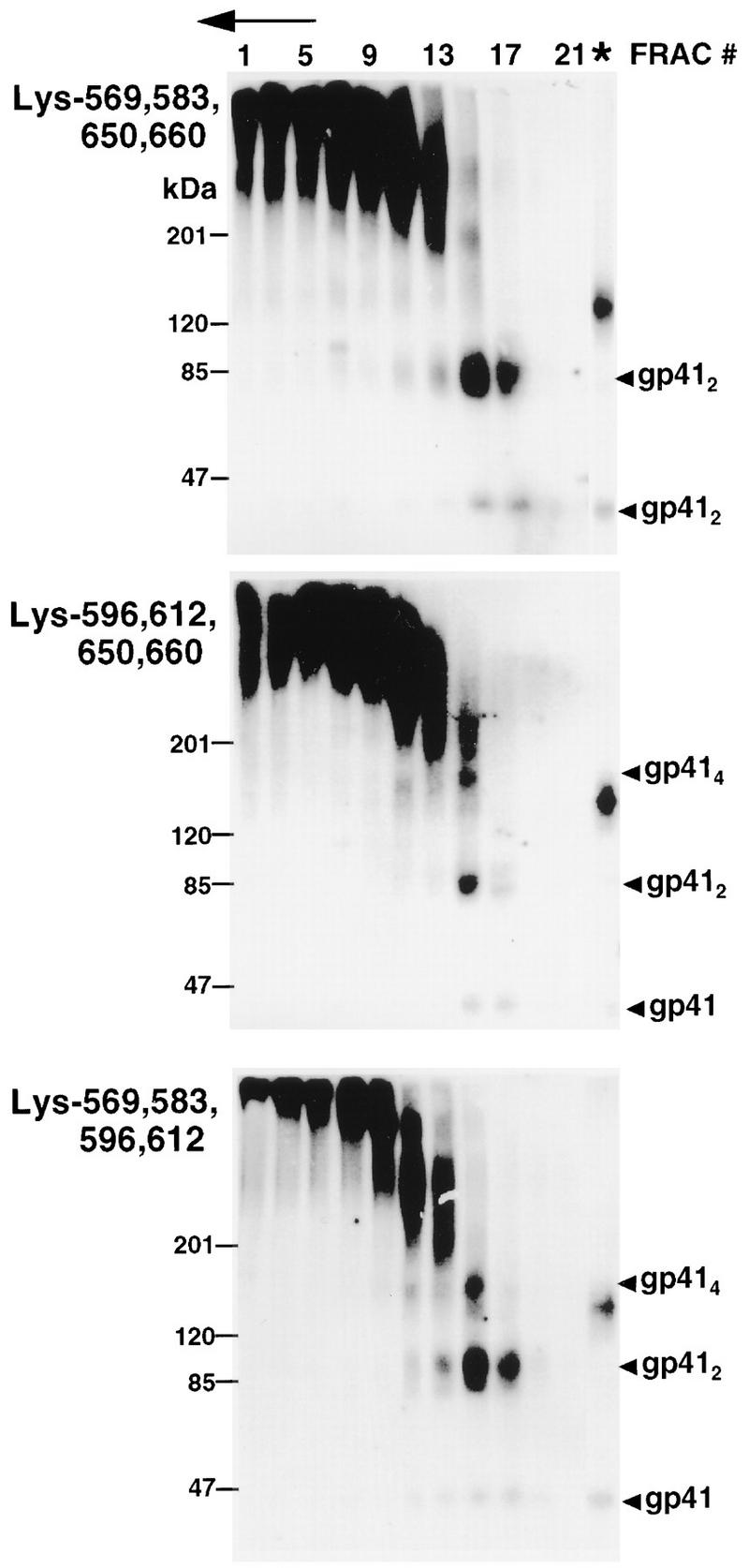
Sedimentation and BS3-cross-linking profiles of gp41 mutants containing four or more ectodomain lysines. Lysates from HeLa cells expressing the indicated Lys-to-Arg Env mutants were treated as described in the legend to Fig. 2. Env glycoproteins were visualized by immunoblotting with human α-588-599 antibody and 125I-protein A. The direction of sedimentation is right to left and is indicated by an arrow. ∗, unfractionated cell lysates; FRAC, fraction.
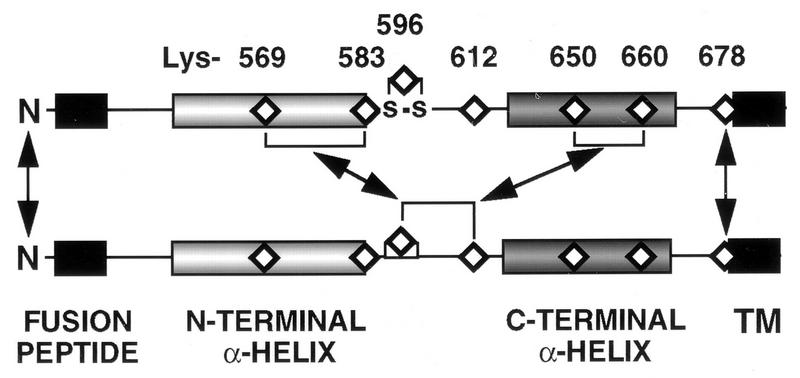
The cross-linking results are summarized in Fig. 1C and 7, with gp41 N termini closely juxtaposed in the oligomeric structure and Lys-678 in close proximity to its intermonomer homologous residue. Lys-596 and/or Lys 612 of one monomer is in close proximity to lysine residues within both the N- and C-terminal amphipathic α-helical sequences of an adjacent monomer(s) within the oligomer.
FIG. 7.
Diagramatic summary of intermonomer cross-links deduced from the BS3-cross-linking profiles of Lys-to-Arg mutants. Cross-links between two monomers are shown for clarity. N termini are closely juxtaposed, and Lys-678 is in close proximity to its intermonomer homologous residue. Lys-596 and/or Lys-612 of one monomer is in close proximity to lysine residues within both the N-terminal and C-terminal amphipathic α-helical sequences of an adjacent monomer(s). TM, transmembrane sequence.
The presence of the transmembrane sequence improves the efficiency of mutant–wild-type Env hetero-oligomer formation.
The formation of intermonomer cross-links through Lys-678 is consistent with transmembrane sequences being closely associated in Env oligomers. While truncation mutagenesis has shown that the transmembrane sequence is not required for Env oligomerization (19, 20), formation of hetero-oligomers between gp160 and a gp160 mutant truncated on the N-terminal side of the transmembrane sequence at Asn-672 occurs with a wild-type/mutant ratio of 1:0.4 (50), implying that the mutant lacks a C-terminal structural element(s) required for full oligomerization efficiency. To determine if the transmembrane sequence affects the efficiency of mutant–wild-type hetero-oligomer formation, vTF7.3-infected HeLa cells were transfected with dual T7-promoter-driven expression vectors (Fig. 8A) for coexpression of gp160 with a transmembrane sequence-lacking [gp160(1-672), pP672) or transmembrane sequence-containing [gp160(1-701), pP701) mutant. The efficiency of mutant–wild-type assembly was then determined following pulse-chase biosynthetic labeling and immunoprecipitation. Figure 8B shows that MAb 902 immunoprecipitated gp160 and gp120 from cells transfected with pPT7−, which directs expression of gp160 only. Transfection of cells with pP672 or pP701 gave rise to additional bands migrating to positions intermediate between those of gp160 and gp120 that correspond to the coexpressed truncation mutants. MAb C8, which is directed to the gp160 cytoplasmic tail and is absent from the truncation mutants, coimmunoprecipitated gp160 and the coexpressed truncation mutants, indicating hetero-oligomer assembly in both cases. Immunoprecipitation of the mutants by MAb C8 requires the presence of gp160, because this MAb did not recognize gp160(1-701) when expressed in the absence of gp160 from pS701 (Fig. 8B, pS701 lanes). Densitometric analysis of MAb C8-coimmunoprecipitated gp160-gp160(1-672) hetero-oligomers (Fig. 8B, MAb C8 panel, pP672 lane) indicates that they formed with a wild-type/mutant ratio of 1:0.29 ± 0.08 (mean ± standard deviation, n = 4). Inclusion of the transmembrane sequence in mutant gp160(1-701) resulted in a greater-than-twofold increase in the efficiency of hetero-oligomerization, with the gp160-gp160(1-701) hetero-oligomer forming at a wild-type/mutant ratio of 1:0.68 ± 0.15 (n = 4). To rule out the possibility that the presence of the transmembrane sequence in the gp160(1-701) mutant caused nonspecific aggregation with wild-type Env postsynthesis, immunoprecipitations were performed following an 18-min pulse-labeling, when gp160 is monomeric, and following a 4-h chase, when gp160 is oligomeric (21). Figure 8C indicates that both gp160 and gp160(1-701) were immunoprecipitated by MAb 902 following both labeling times, whereas MAb C8 immunoprecipitated only wild-type Env following the 18-min pulse but coimmunoprecipitated both glycoproteins following the 4-h chase. The negative control MAb IC3/86, directed to human glycophorin (7), did not recognize either glycoprotein (Fig. 8C, IC3 lanes). These results indicate that the transmembrane sequence enhances Env oligomerization.
FIG. 8.
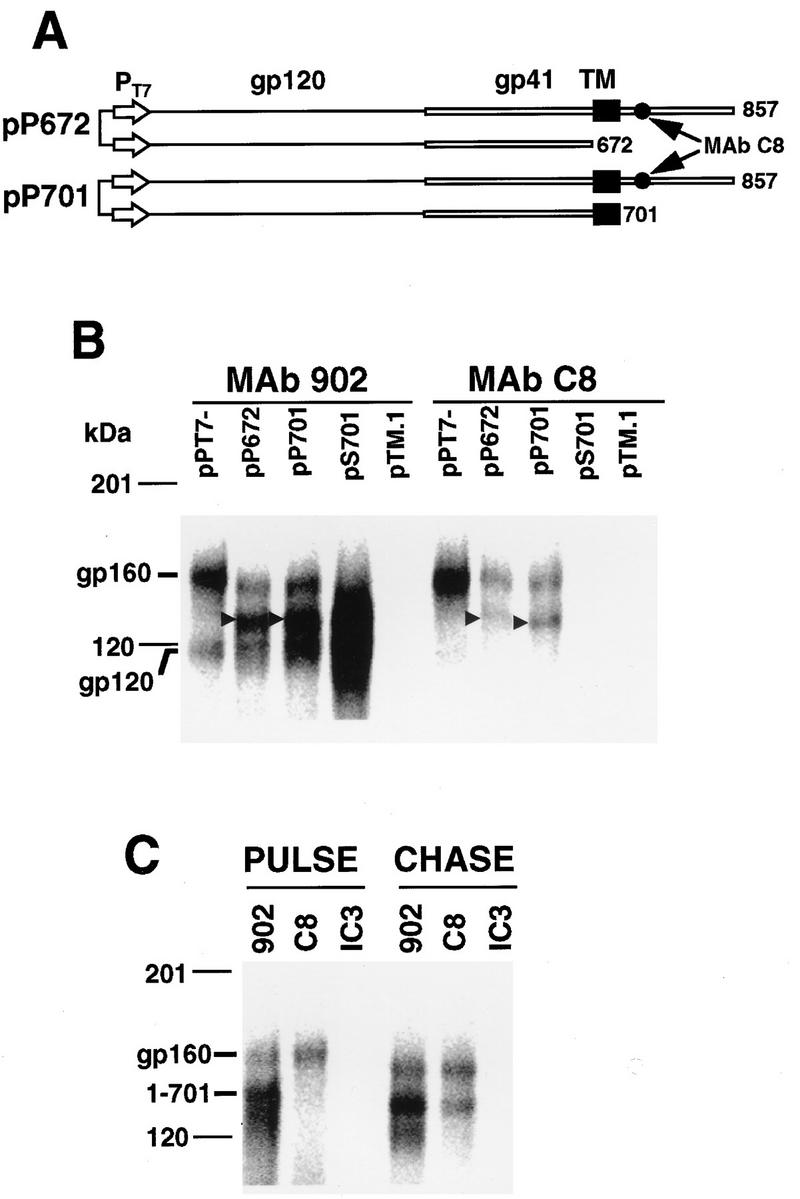
(A) Linear representation of the dual T7 promoter-driven expression vectors pP672 and pP701. PT7, T7 promoter; TM, transmembrane sequence. The location of the MAb C8 epitope is indicated. (B) Hetero-oligomerization between coexpressed wild-type and truncated Env proteins. Pulse-chase biosynthetically labeled wild-type and truncated Env proteins (indicated by arrowheads) were immunoprecipitated from HeLa cell lysates with MAb 902 or MAb C8. Samples were subjected to SDS-PAGE on 5 to 15% gradient gels and PhosphorImager SF analysis. Densitometry was performed with Imagequant software. (C) Formation of gp160-gp160(1-701) hetero-oligomers is delayed. Wild-type and truncated Env proteins were coexpressed in HeLa cells by transfection with pP701. At 21 h posttransfection, the expressed glycoproteins were biosynthetically labeled with 250 μCi of Tran35S-label for 18 min (PULSE) or labeled for 18 min and then chased for 4 h (CHASE) prior to cell lysis and immunoprecipitation with MAbs 902 and C8 or the control MAb IC3/86 (IC3). Samples were subjected to SDS-PAGE on 5 to 15% gradient gels and PhosphorImager SF analysis.
Effect of Lys-to-Arg mutations on Env structure and function.
It was of interest to evaluate whether the Lys-to-Arg substitutions altered Env protein functions, including sCD4-binding ability, gp41-gp120 association, precursor cleavage efficiency, shedding of cell surface gp120 in response to sCD4, and syncytium-forming function.
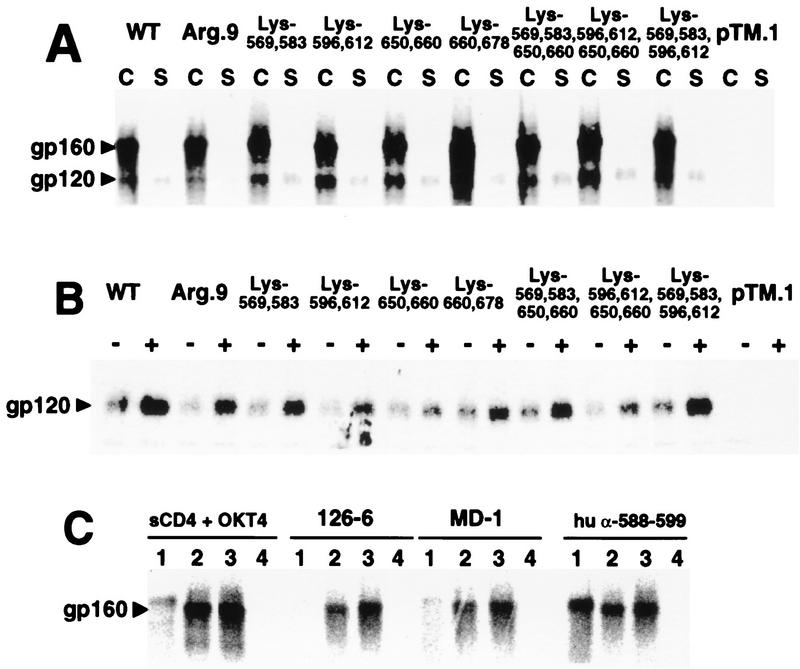
HeLa cells expressing wild-type and mutant Env proteins were pulsed with Tran35S-label for 30 min and then chased for 4 h to allow for intracellular transport and processing. Figure 9A indicates similar levels of cell-associated gp120 for all mutants tested. These results confirm that the mutations had not affected the ability of gp41 to anchor gp120 to cellular membranes and that precursor processing was normal. The ability of sCD4 to induce shedding of gp120 from cell surface gp41 was next assessed by comparing the amounts of gp120 immunoprecipitated from culture supernatants removed from mock-treated and sCD4-treated wild-type- or mutant Env-transfected cells. sCD4 treatment resulted in elevated levels of gp120 in culture supernatants for all mutants tested (Fig. 9B). These results indicate that the Lys-to-Arg mutations have not significantly affected the ability of cell surface, gp41-anchored gp120 to bind sCD4 and that this binding event induces conformational changes in gp120 and gp41 thought to result in shedding of gp120 (53).
FIG. 9.
(A) Effect of Lys-to-Arg substitutions on gp41-gp120 association. HeLa cells expressing the mutant or wild-type (WT) Env proteins were biosynthetically labeled with Tran35S-label and chased with complete medium for 4 h before lysis. Precleared cell lysates (lanes C) and culture supernatants (lanes S) were immunoprecipitated with human α-rgp120 antibody and protein A-Sepharose. gp120 and gp160 were visualized by autoradiography following SDS-PAGE on 5 to 15% gradient gels under reducing conditions. (B) sCD4-induced shedding of wild-type and mutant gp120. HeLa cells expressing the indicated mutant or wild-type Env proteins were biosynthetically labeled for 1 h with Tran35S-label. Following a 4-h chase, monolayers were washed and overlaid with medium alone (lanes −) or medium containing 15 μg of sCD4 (lanes +). Following a 2-h incubation, culture supernatants were immunoprecipitated with a mixture of human α-rgp120 antibody and IgG purified from pooled HIV-positive human plasma and protein A-Sepharose. Immune complexes were analyzed by autoradiography following SDS-PAGE on 5 to 15% gradient gels under reducing conditions. (C) Anti-gp41 antibodies recognize conformational epitopes on wild-type and Arg.9 mutant Env proteins. HeLa cells expressing wild-type or Arg.9 mutant Env proteins were biosynthetically labeled for 15 min and either lysed immediately or subjected to a 4-h chase with complete medium. Precleared cell lysates were then immunoprecipitated with protein A-Sepharose coated with the indicated antibodies or with sCD4 plus MAb OKT4. Immunoprecipitated proteins were electrophoresed on 5 to 15% gradient gels under reducing conditions and visualized by PhosphorImager SF analysis. Lanes 1, wild type, 15-min pulse; lanes 2, wild type; lanes 3, Arg.9; lanes 4, control plasmid pTM.1; lanes 2 to 4, 15-min pulse, 4-h chase. hu, human.
To assess if Lys-to-Arg mutations had altered gp41 tertiary structure, the ability of pulse-chase biosynthetically labeled Arg.9 to acquire conformational epitopes in the gp41 domain was tested. MAbs 126-6 and MD-1 recognize conformational epitopes in the gp41 ectodomain (43, 73). These MAbs immunoprecipitated minimal amounts of wild-type gp160 following a 15-min pulse-labeling (Fig. 9C, lanes 1). In contrast, high levels of both wild-type and Arg.9 mutant Env proteins were immunoprecipitated by both MAbs following a 4-h chase (Fig. 9C, lanes 2 and 3, respectively). Coimmunoprecipitation of Arg.9 gp160 with sCD4 and MAb OKT4 also confirms the acquisition of CD4-binding competence.
The effect of Lys-to-Arg mutations on Env fusion function was assessed in a HeLa-T4 cell syncytium assay. Table 1 shows that the WT′ mutant had syncytium-forming function that was indistinguishable from that of the wild type. This function was abolished following the replacement of all nine lysine residues with arginine (Arg.9) or maintenance of combinations of two (Lys-569,583, Lys-596,612, Lys-650,660, and Lys-660,678) or four (Lys-569,583,650,660 and Lys-596,612,650,660) ectodomain lysines. In contrast, maintenance of Lys-569,583,596,612 reduced the syncytium-forming function by 50%. We further investigated the role of lysines in the syncytium-forming function by preparing double- and single-substitution mutants on a wild-type (Lys-789 and Lys-803) gp41 background. The double substitutions K569/583R and K596/612R reduced syncytium-forming function by 81 and 50%, respectively. In contrast, the K650/660R mutant had a fusion function that was similar to that of wild-type Env. Normal syncytium-forming function was observed for the K583R, K596R, K612R, and K678R single-substitution mutants, while the K569R substitution resulted in an 80% decrease in relative syncytium-forming capacity. These results indicate that the Env fusion function can tolerate individual Lys-to-Arg substitutions at positions 583, 596, 612, 650, 660, and 678 and double substitutions at positions 650 plus 660 and 789 plus 803. Wild-type levels of Env fusion competence require lysine at position 569, and multiple substitutions involving Lys-596, -612, and -678 on a WT′ background are not tolerated. Although replacement of certain lysine residues with arginine adversely affected Env syncytium-forming capacity, there was no obvious correlation between loss of syncytium-forming function and disruption of the wild-type gp41 cross-linking profile. Since most properties of gp41 and gp120 are preserved in the Lys-to-Arg mutants, we infer that the cross-linking profiles observed reflect the native structure.
DISCUSSION
We have used mutation-directed chemical cross-linking to demonstrate that gp41 dimers are stabilized by cross-linking through N termini. Trimers and tetramers of gp41 are cross-linked provided that Lys-678 is present. In the absence of Lys-678, tetramers but not trimers can still be formed by cross-links between the intermonomer pairs: Lys-596,612 with Lys-569,583 or Lys-596,612 with Lys-650,660 (Fig. 7).
The N terminus of gp41 is generated as a result of the posttranslational cleavage of gp160 and precedes the fusion peptide (29, 30, 63, 70). The hydrophobic nature of the gp41 fusion peptide suggests that in the native prefusion state, it will be sequestered away from the surrounding solvent, buried within oligomeric Env. This arrangement has been suggested by antibody mapping studies of the oligomeric gp120-gp41 complex indicating that the sequence preceding the fusion peptide is occluded by gp120 (55). We find that gp41 N termini in the gp41 oligomer can be cross-linked by the soluble cross-linking agent following dissociation of gp41 from gp120 by detergent treatment. With reference to the structures of gp41 α-helical peptide complexes (9, 66), fusion peptides may be at the top of the coiled-coil oligomerization domain in close juxtaposition and in contact with solvent.
We observed that Lys-678, adjacent to the transmembrane sequence, is in close contact with its homologous residue in neighboring monomers. This arrangement may result from the close association between transmembrane sequences, since a mutant containing the transmembrane sequence was able to recruit coexpressed gp160 into hetero-oligomers more than twice as efficiently as a transmembrane sequence-negative mutant. Truncation of the transmembrane sequence and cytoplasmic tail does not adversely affect gp160 oligomerization or oligomer stability (19, 20), suggesting that the transmembrane sequence contributes to oligomerization but is not essential. Transmembrane sequences may serve as an initial docking site for gp160 monomers diffusing laterally in the endoplasmic reticulum membrane prior to oligomer stabilization through interactions in the gp41 ectodomain (20, 48, 50). Interactions between transmembrane sequences may also be important for Env fusion competence, since point mutations within the transmembrane sequence block the Env syncytium-forming function without affecting cell surface expression (45).
Recent work has shown that the N-terminal amphipathic α-helical sequence, Asn-548 to Gln-585, mediates gp160 and gp41 oligomerization (9, 50, 66). Formation of a coiled coil by this sequence would result in both Lys-569 and 583 occupying positions on the same face of an α-helix and projected in opposing directions to homologous residues on adjacent monomers, thereby precluding intermonomer cross-links through these amino acids. This is reflected in the results obtained with the Lys-569,583 mutant, which is not cross-linked at a higher order than dimer obtained by cross-linkage through N termini. Similar results were also obtained with mutants containing lysine pairs at positions 596 plus 612 or 650 plus 660, indicating that these residues are out of range of their homologous or heterologous residues on adjacent monomers.
However, Lys-596 within the disulfide-bonded loop and/or Lys-612 within the glycan cluster mediates intermonomer cross-links with lysines in both the N-terminal (Lys-569 and/or 583) and C-terminal (Lys-650 and/or 660) α-helical sequences of separate monomers. These data indicate that the sequence lying between the N- and C-terminal α-helical sequences contacts both helical domains in separate monomers such that Lys-596 and/or -612 comes into close contact with lysines within the helical domains. Crystallographic studies indicate that the N-terminal helix forms a coiled coil with the C-terminal helix buttressed against its exterior in an antiparallel orientation (9, 66). Comparison of these structures with that of a Moloney murine leukemia virus transmembrane fragment suggests that the disulfide loop may pack on the outside of the antiparallel α-helical core (22). These structures, which are likely to be in the fusion-active transmembrane conformation (5, 10, 66), indicate that cross-links between Lys-596 and/or -612 and those lysines within the α-helical domains are precluded. In the present study, gp41 was derived from an Env complex that had not been exposed to CD4 and chemokine receptors, which presumably trigger fusion activation (61, 62, 72). Cross-links between Lys-596 and/or -612 and lysines within the N- and C-terminal α-helical domains may therefore be possible only in the prefusogenic form of gp41. Furthermore, tetramers and dimers but not trimers were obtained with these mutants, suggesting that Lys-596 and -612 of one monomer may be within cross-linking range of Lys-569 and -583 or Lys-650 and -660 on separate monomers in the tetramer form only.
In agreement with other studies on the oligomerization of Env proteins expressed in mammalian cells (19, 20, 46–49, 56), we identified the highest-order quaternary structure for gp41 as a tetramer. This order of structure is also adopted by a synthetic peptide analog of the N-terminal amphipathic α-helical oligomerization domain (51) and by a chimera comprising the E. coli maltose-binding protein fused with the gp41 oligomerization domain (57). These findings, however, contrast with the trimeric structures observed for gp41 N- and C-terminal α-helical peptide complexes (9, 66) and for a gp41 ectodomain fragment expressed in insect cells lacking the hydrophobic fusion peptide and transmembrane sequence (67). These studies suggest that the gp41 amphipathic α-helical oligomerization domain has the propensity to form a four-stranded coiled coil but that this propensity is modified in the presence of the C-terminal α-helical sequence and in the absence of the fusion peptide and transmembrane sequence.
The introduced mutations did not adversely affect the overall structural integrity of gp41, since precursor processing efficiency, gp41-gp120 association, sCD4-induced shedding of gp120 from the cell surface, or recognition of mutant Env by conformation-dependent MAbs was unaffected. Variable levels of impairment of Env fusion function were observed for some mutants; however, this did not correlate with a cross-linking profile that was different from that obtained for wild-type gp41. For example, cross-linkage of the fusion-negative gp41 mutant, Lys-660,678, resulted in its migration in SDS-PAGE as tetramers, trimers, dimers, and monomers, as does the wild type. Fusion function impairment was observed for mutants containing the Lys-569-to-Arg mutation or the Lys-596,612-to-Arg mutations. Lysine at position 569 is conserved across clade B HIV-1 isolates (42). The aliphatic portion of Lys-569 forms part of a hydrophobic cavity on the surface of the coiled coil of gp41, while its ɛ-nitrogen forms a salt bridge with Asp-629 (9). Replacement of Lys-569 with Arg will alter the size and shape of this cavity and may modify the nature of the salt bridge with Asp-629. These changes may decrease the ability of gp41 to adopt the fusion-active conformation. In contrast, Arg and Gln occur at positions 596 and 612 in some clade B isolates. Replacement of either lysine with Arg does not affect fusion function, whereas simultaneous mutation of both partially impairs fusogenicity. Introduction of more Lys-to-Arg mutations caused increased impairment of fusion function and may result from an accumulation of altered hydrogen-bonding (4, 8) or ion-pairing interactions within the gp41-gp120 oligomeric complex. These results reflect the exquisite sensitivity of Env fusion function to conservative mutations that do not affect other aspects of Env structure (6, 18, 38, 50).
Our results provide information about the juxtaposition of structural elements in gp160-derived gp41 oligomers and may aid in the interpretation of structural information obtained for gp41 fragments expressed in heterologous systems.
ACKNOWLEDGMENTS
We thank Frosa Katsis for synthesis of oligonucleotides and peptides.
This work was supported by NH&MRC grant 960237. T.L.M. and P.P. were supported by CARG postdoctoral fellowships, and B.E.K. is an NH&MRC fellow.
REFERENCES
- 1.Abacioglu Y H, Fouts T R, Laman J D, Claasen E, Pincus S H, Moore J P, Roby C A, Kamin-Lewis R, Lewis G K. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a rantes, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Allan J S, Strauss J, Buck D W. Enhancement of SIV infection with soluble receptor molecules. Science. 1990;247:1084–1088. doi: 10.1126/science.2309120. [DOI] [PubMed] [Google Scholar]
- 4.Bode W, Walter J, Huber R, Wenzel H R, Tschesche H. The refined 2.2-Å (0.22-nm) X-ray crystal structure of the ternary complex formed by bovine trypsinogen, valine-valine and the Arg15 analogue of bovine pancreatic trypsin inhibitor. Eur J Biochem. 1984;144:185–190. doi: 10.1111/j.1432-1033.1984.tb08447.x. [DOI] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catimel B, Wilson K M, Kemp B E. Kinetics of the autologous red cell agglutination test. J Immunol Methods. 1993;165:183–192. doi: 10.1016/0022-1759(93)90344-7. [DOI] [PubMed] [Google Scholar]
- 8.Chacko S, Silverton E, Kam-Morgan L, Smith-Gill S, Cohen G, Davies D. Structure of an antibody-lysozyme complex: unexpected effect of conservative mutation. J Mol Biol. 1995;245:261–274. doi: 10.1006/jmbi.1994.0022. [DOI] [PubMed] [Google Scholar]
- 9.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Wharton S A, Weissenhorn W, Calder L J, Hughson F M, Skehel J J, Wiley D C. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc Natl Acad Sci USA. 1995;92:12205–12209. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 14.Dalgleish A G, Beverly P C L, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 15.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 16.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 17.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 18.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earl P L, Doms R W, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci USA. 1990;87:648–652. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earl P L, Moss B. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res Hum Retroviruses. 1993;9:589–594. doi: 10.1089/aid.1993.9.589. [DOI] [PubMed] [Google Scholar]
- 21.Earl P L, Moss B, Doms R W. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 Å resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 23.Fenouillet M, Gluckman J C, Jones I M. Functions of HIV envelope glycans. Trends Biochem Sci. 1994;19:65–70. doi: 10.1016/0968-0004(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 24.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 25.Fouts T R, Binley J M, Trkola A, Robinson J E, Moore J P. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freed E O, Delwart E L, Buchschacher G L, Jr, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freed E O, Martin M A. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J Biol Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 28.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freed E O, Myers D J, Risser R. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J Virol. 1989;63:4670–4675. doi: 10.1128/jvi.63.11.4670-4675.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed E O, Myers D J, Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc Natl Acad Sci USA. 1990;87:4650–4654. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 32.Gnann J W, Jr, Nelson J A, Oldstone M B A. Fine mapping of an immunodominant domain in the transmembrane glycoprotein of human immunodeficiency virus. J Virol. 1987;61:2639–2641. doi: 10.1128/jvi.61.8.2639-2641.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenwood F C, Hunter W M, Glover J S. The preparation of 131I-labelled human growth hormone of high specific radioactivity. Biochem J. 1963;89:114–123. doi: 10.1042/bj0890114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haffar O K, Dowbenko D J, Berman P W. The cytoplasmic tail of HIV-1 gp160 contains regions that associate with cellular membranes. Virology. 1991;180:439–441. doi: 10.1016/0042-6822(91)90054-f. [DOI] [PubMed] [Google Scholar]
- 35.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman R A, Kung P C, Hansen W P, Goldstein G. Simple and rapid measurement of human T lymphocytes and their subclasses in peripheral blood. Proc Natl Acad Sci USA. 1980;77:4914–4917. doi: 10.1073/pnas.77.8.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J-C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 38.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh W C, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 39.Layne S P, Merges M J, Dembo M, Spouge J L, Nara P L. HIV requires multiple gp120 molecules for CD4-mediated infection. Nature. 1990;346:277–279. doi: 10.1038/346277a0. [DOI] [PubMed] [Google Scholar]
- 40.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 41.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 42.Myers G, Korber B, Hahn B H, Jeang K-T, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N, editors. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 43.Myers R, Meiller T, Falkler W, Jr, Patel J, Joseph J. Abstracts of the 93rd General Meeting of the American Society for Microbiology 1993. Washington, D.C: American Society for Microbiology; 1993. A human monoclonal antibody to a cryptic gp41 epitope on HIV-1 infected cells, abstr. T-70; p. 444. [Google Scholar]
- 44.Otteken A, Earl P L, Moss B. Folding, assembly, and intracellular trafficking of the human immunodeficiency virus type 1 envelope glycoprotein analyzed with monoclonal antibodies recognizing maturational intermediates. J Virol. 1996;70:3407–3415. doi: 10.1128/jvi.70.6.3407-3415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Owens R J, Burke C, Rose J K. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owens R J, Compans R W. The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology. 1990;179:827–833. doi: 10.1016/0042-6822(90)90151-g. [DOI] [PubMed] [Google Scholar]
- 47.Pinter A, Honnen W J, Tilley S A, Bona C, Zaghouani H, Gorny M K, Zolla-Pazner S. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;63:2674–2679. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poumbourios P, El Ahmar W, McPhee D A, Kemp B E. Determinants of human immunodeficiency virus type 1 envelope glycoprotein oligomeric structure. J Virol. 1995;69:1209–1218. doi: 10.1128/jvi.69.2.1209-1218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poumbourios P, McPhee D A, Kemp B E. Antibody epitopes sensitive to the state of human immunodeficiency virus type 1 gp41 oligomerization map to a putative α-helical region. AIDS Res Hum Retroviruses. 1992;8:2055–2062. doi: 10.1089/aid.1992.8.2055. [DOI] [PubMed] [Google Scholar]
- 50.Poumbourios P, Wilson K A, Center R J, El Ahmar W, Kemp B E. Human immunodeficiency virus type 1 envelope glycoprotein oligomerization requires the gp41 amphipathic α-helical/leucine zipper-like sequence. J Virol. 1997;71:2041–2049. doi: 10.1128/jvi.71.3.2041-2049.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rabenstein M, Shin Y-K. A peptide from the heptad repeat of human immunodeficiency virus gp41 shows both membrane binding and coiled-coil formation. Biochemistry. 1995;34:13390–13397. doi: 10.1021/bi00041a016. [DOI] [PubMed] [Google Scholar]
- 52.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Jr, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 53.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sattentau Q J, Moore J P. human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sattentau Q J, Zolla-Pazner S, Poignard P. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology. 1995;206:713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 56.Schawaller M, Smith G E, Skehel J J, Wiley D C. Studies with crosslinking reagents on the oligomeric structure of the env glycoprotein of HIV. Virology. 1989;172:367–369. doi: 10.1016/0042-6822(89)90142-6. [DOI] [PubMed] [Google Scholar]
- 57.Shugars D C, Wild C T, Greenwell T K, Matthews T J. Biophysical characterization of recombinant proteins expressing the leucine zipper-like domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1996;70:2982–2991. doi: 10.1128/jvi.70.5.2982-2991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srinivas S K, Srinivas R V, Anantharamaiah G M, Segrest J P, Compans R W. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J Biol Chem. 1992;267:7121–7127. [PubMed] [Google Scholar]
- 59.Stamatatos L, Cheng-Mayer C. Structural modulations of the envelope gp120 glycoprotein of human immunodeficiency virus type 1 upon oligomerization and differential V3 loop epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor binding. J Virol. 1995;69:6191–6198. doi: 10.1128/jvi.69.10.6191-6198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staros J V. N-hydroxysulfosuccinimide active esters: bis(N-hydroxysulfosuccinimide) esters of two dicarboxylic acids are hydrophilic, membrane-impermeant protein cross-linkers. Biochemistry. 1982;21:3950–3955. doi: 10.1021/bi00260a008. [DOI] [PubMed] [Google Scholar]
- 61.Sullivan N, Sun Y, Li J, Hofman W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 63.Veronese F D, DeVico A L, Copeland T D, Oroszlan S, Gallo R C, Sarngadharan M G. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science. 1985;229:1402–1405. doi: 10.1126/science.2994223. [DOI] [PubMed] [Google Scholar]
- 64.Weiss C D, Levy J A, White J M. Oligomeric organization of gp120 on infectious human immunodeficiency virus type 1 particles. J Virol. 1990;64:5674–5677. doi: 10.1128/jvi.64.11.5674-5677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weissenhorn W, Calder L J, Dessen A, Laue T, Skehel J J, Wiley D C. Assembly of a rod-shaped chimera of a trimeric GCN4 zipper and the HIV-1 gp41 ectodomain expressed in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6065–6069. doi: 10.1073/pnas.94.12.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 67.Weissenhorn W, Wharton S A, Calder L J, Earl P L, Moss B, Aliprandis E, Skehel J J, Wiley D C. The ectodomain of HIV-1 env subunit gp41 forms a soluble, α-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 1996;15:1507–1514. [PMC free article] [PubMed] [Google Scholar]
- 68.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiley D C, Skehel J J, Waterfield M. Evidence from studies with a cross-linking reagent that the haemagglutinin of influenza virus is a trimer. Virology. 1977;79:446–448. doi: 10.1016/0042-6822(77)90371-3. [DOI] [PubMed] [Google Scholar]
- 70.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 72.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. [DOI] [PubMed]
- 73.Xu J-Y, Gorny M K, Palker T, Karwowska S, Zolla-Pazner S. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using 10 human monoclonal antibodies. J Virol. 1991;65:4832–4838. doi: 10.1128/jvi.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang H, Dornadula G, Alur P, Laughlin M A, Pomeratz R J. Amphipathic domains in the C terminus of the transmembrane protein (gp41) permeabilize HIV-1 virions: a molecular mechanism underlying natural endogenous reverse transcription. Proc Natl Acad Sci USA. 1996;93:12519–12524. doi: 10.1073/pnas.93.22.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]