Abstract
The matrix (MA) protein of human immunodeficiency virus type 1 (HIV-1) plays a critical role in virion morphogenesis and fulfills important functions during the early steps of infection. In an effort to identify cellular partners of MA, a Saccharomyces cerevisiae two-hybrid screen was utilized. A specific interaction between MA and HO3, a putative histidyl-tRNA synthetase, was demonstrated in this system. HO3-specific mRNA was detected in several tissues relevant for HIV infection, such as spleen, thymus, and peripheral blood lymphocytes, as well as in a number of T-lymphoid-cell lines. The binding of MA to HO3 was confirmed in transfected cells by coimmunoprecipitation. This interaction was abrogated by replacing two lysine residues at positions 26 and 27 of MA by threonine (MAKK27TT). HO3 localized both to the cytoplasm and to the nucleus of acutely transfected 293T cells. When overexpressed in HIV-1-producing cells, HO3 was incorporated into wild-type virions but not in ones containing the dilysine-mutated variant of MA. Correspondingly, overexpression of HO3 in virus producer cells enhanced the infectivity of wild-type but not MAKK27AA HIV-1 particles. The stimulating effect of HO3 was independent from the presence of Envelope, Vpr, or Vpu. Taken together, these results suggest that HO3, through its recognition of MA, plays a role in the life cycle of HIV-1.
The Gag protein of human immunodeficiency virus (HIV) is synthesized as a polyprotein precursor (p55Gag). Concomitant with or soon after virion budding, p55Gag is cleaved by the virally encoded protease (29) into its mature products p17 matrix (MA), p24 capsid (CA), p7 nucleocapsid (NC), the C-terminal p6, and several other small polypeptides, including p1 and p2 (15, 32). Several roles have been assigned to HIV type 1 (HIV-1) MA. These functions include targeting the Gag precursor to the plasma membrane (8, 16, 18, 37), promoting virus particle assembly and release (26, 31), governing envelope glycoprotein incorporation into virions (7, 9, 34), facilitating viral entry (35), and mediating the nuclear transport of the preintegration complex in nondividing cells (4, 10, 30).
MA is the N-terminal myristoylated cleavage product of the Gag precursor (26). MA myristoylation is required, although not sufficient, for the targeting of Gag to the plasma membrane and for particle release (17, 27, 33). Since MA does not direct Gag to intracellular membranes, it has been hypothesized that a membrane receptor capable of recognizing MA within the context of the Gag precursor might be involved in the plasma membrane targeting of Gag (14). In addition, the contribution of a specific intracellular trafficking pathway is suggested by mutations in MA that result in the accumulation of Gag at aberrant cytoplasmic sites (8, 16).
Several lines of evidence indicate that MA is also involved in recruiting the envelope (Env) glycoprotein into virions. Mutations in MA which prevent the particle incorporation of Env have been identified (7, 9, 34). Furthermore, in the absence of Env, the assembly and release of HIV-1 particles is detected at both the apical and basolateral sites of polarized MDCK cells, whereas in the presence of Env it occurs mostly in the basolateral region, where Env accumulates, suggesting that Env can redirect MA budding in infected cells (24). Finally, a direct interaction between HIV-1 MA and the cytoplasmic domain of gp41 was detected by using recombinant proteins (6).
MA also plays a role during the early steps of the infectious process by promoting the nuclear migration of the uncoated viral nucleoprotein complex, also known as the preintegration complex, and thereby facilitating infection of nondividing cells (3, 10, 12, 19, 25, 30). Together with Vpr, MA contributes to the karyophilic properties of the HIV-1 preintegration complex, via a conserved stretch of basic residues (amino acids [aa] 25 to 33) that resembles the prototypic nuclear localization signal (NLS) of simian virus 40 large antigen (4, 10). The MA NLS is recognized by members of the karyopherin-α family, which act as proximal mediators of nuclear import (11). In the absence of a functional Vpr gene, MA NLS mutant viruses have a decreased replicative capacity in terminally differentiated macrophages, owing to a block in nuclear import (4). Infection of such cells is likely crucial for HIV-1 transmission as well as for AIDS induction (21). Reflecting its multiple roles during infection, MA must be interacting with several viral and cellular proteins, including ones involved in its targeting or responsible for its posttranslational modifications.
Interaction of HIV-1 MA protein with the HO3 gene product in the Saccharomyces cerevisiae two-hybrid system.
The HIV-1 MA gene was fused to the Gal4 DNA binding domain in the pGBT9 plasmid (2) and was used as bait in the two-hybrid screen. The MA gene was PCR amplified from the plasmid R7 (containing the HIV-1HXB2 proviral DNA) by using the oligonucleotides Gag116 (5′GGG CCCGGATCCTCAGTAATTTGGCTGACCTGATT) and Gag115 (5′-CCCGGGGAATTCATGGGTGCGAGAGCGTCAGTA). The amplified DNA was cloned into the EcoRI and BamHI sites of pGBT9. The MAKK27TT mutant, containing two lysine-to-threonine changes at positions 26 and 27, was cloned following the same procedure but with the corresponding R7 mutant construct (11) used as a template in the PCR. The mutant MAY132F, in which the C-terminal tyrosine of MA is replaced by phenylalanine (10), was also cloned by PCR amplification of R7 DNA by using the oligonucleotides Gag115 and Gag117 (5′-GGGCCCGGATCCTCAGAAATTTTGGCTGACCTGATT). Expression in yeast of Gal4-MA fusion proteins was confirmed by Western analysis with MA-specific antibodies (Fig. 1A). Oligo(dT)-primed mRNA isolated from Jurkat cells was used to construct a cDNA library in the pACTII plasmid (Clontech Laboratories, Inc.). cDNAs (average size, 2.5 kb) were cloned into the EcoRI site of this plasmid and fused to the Gal4 activation domain. Library DNA (300 μg) was used together with the same amount of pGBT9-MA plasmid to cotransform Y190 yeast competent cells. Double transformants were plated on yeast drop out medium lacking Leu, Trp, and His and supplemented with 20 mM 3-amino-1,2,4-triazole (3-AT) to inhibit low levels of expression of His3p. After 2 weeks, fast-growing His+ colonies were picked up and tested for β-galactosidase (β-Gal) activity. Out of 107 double transformants, four clones were found to contain a 2.2-kb fragment identical in sequence to the putative histidyl-tRNA synthetase HO3 gene (23). These clones were designated MIP-C (for matrix interacting protein C), and each contained an open reading frame encoding aa 40 to 506 of the 506-aa HO3 protein (the HO3 cDNA clone sequence is available under GenBank accession no. U18937) in frame with the Gal4 activation domain. Other clones were found to contain an unknown gene which is currently under investigation.
FIG. 1.
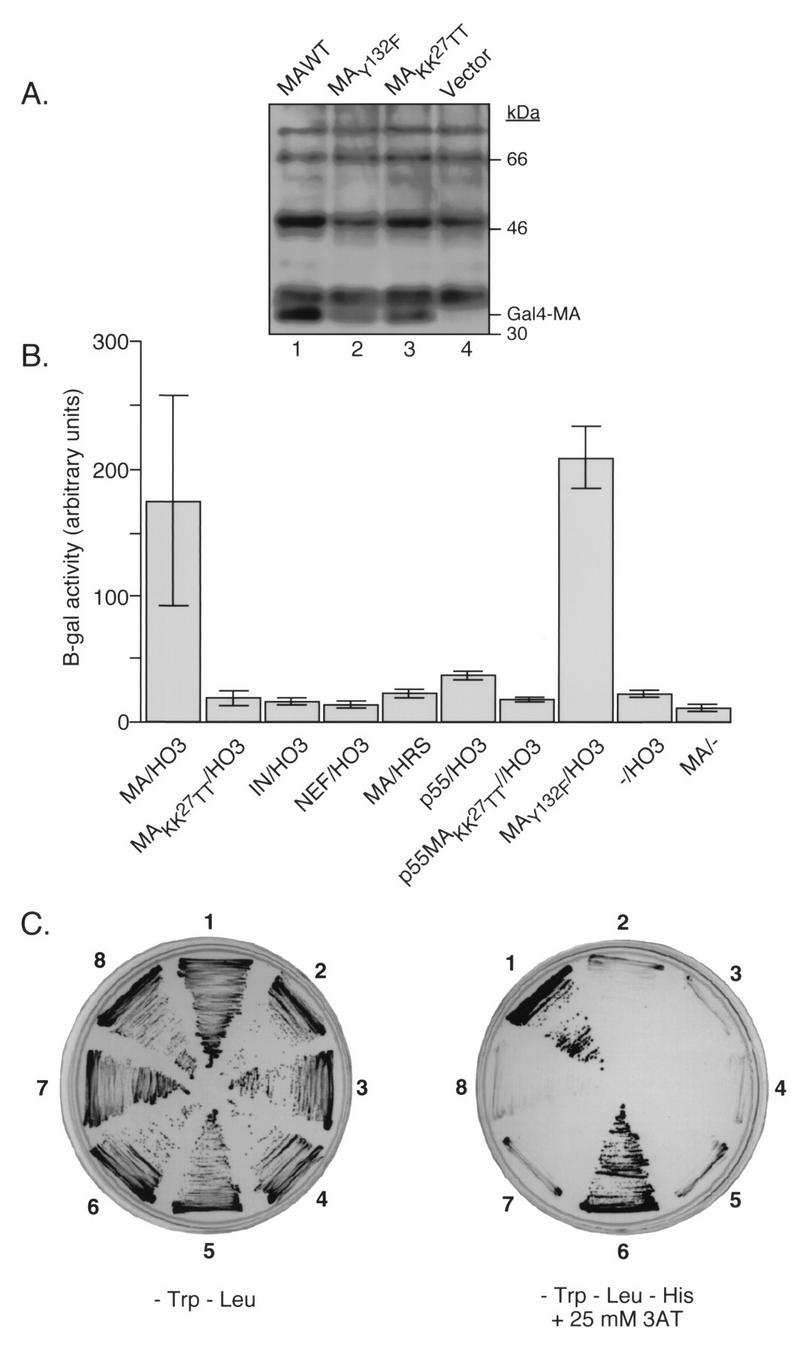
MA-HO3 interaction in the yeast two-hybrid system. (A) Lysates of yeast cells expressing the indicated pGBT9MA plasmids or the vector alone were analyzed by Western blotting with MA-specific antibodies. The position of the Gal4-MA fusion protein is indicated. (B) β-Gal activity in lysates of Y190 yeast cells expressing the indicated cDNAs. In each case, the first cDNA listed was fused to the sequence encoding the Gal4 DNA binding domain (in plasmid pGBT9) while the second was fused to the Gal4 activation domain (in plasmid pACT). −/HO3 denotes yeast clones containing the empty vector pGBT9 and the plasmid pACT-HO3. MA/− denotes yeast clones containing pGBT9-MA and the empty vector pACTII. Data represent the mean values ± standard errors of the means (error bars) from liquid cultures grown from three independent clones obtained after transformation with the corresponding DNAs. (C) growth of Y190 yeast clones on plates lacking either Trp and Leu (left) or Trp, Leu, and His (right) in the presence of 25 mM 3-AT. Sectors: 1, pGBT9-MAwt/pACT-HO3; 2, pGBT9-MAwt/pACT-HRS; 3, pGBT9-IN/pACT-HO3; 4, pGBT9-NEF/pACT-HO3; 5, pGBT9-MAKK27TT/pACT-MIPC; 6, pGBT9-MAwt/pACT-MIPC; 7, pGBT9-MAwt/pACT; 8, pGBT9-MAKK27TT/pACT-HO3.
To ascertain that HO3 was able to interact with HIV-1 MA, the full-length HO3 cDNA (encoding aa 1 to 506) was cloned into the SmaI-EcoRI sites of pACTII after PCR amplification using pCRII-HO3 as the template DNA and the oligonucleotides HO3-Sma (5′-TTCCCCCGGGAATGCCCCTGCTCGGACTTC TTCCCAGG) and HO3-Eco (5′-CCGGAATTCTCAAGACTCAGACAGTCG) as primers. The β-Gal activity of yeast double transformants containing different pGBT9-based plasmids was tested (Fig. 1B). Clones containing both pGBT9-MA and pACT-HO3 showed high levels of β-Gal activity. Yeast colonies containing only one of these constructs and the partner empty vector gave only background levels of β-Gal activity, revealing the specificity of the interaction between HO3 and MA. A MA mutant in which lysine residues 26 and 27 had been replaced by threonine (MAKK27TT) was unable to interact with HO3 in the two-hybrid system. The sequence mutated in this protein functions as an NLS for MA (4, 11, 30). To test whether HO3 was a nonspecific NLS-binding protein, we assayed its ability to interact with HIV-1 integrase (IN), which contains several NLS motifs (13). No interaction between these two proteins was found. HO3 was also unable to interact with the HIV-1 Nef protein. The HRS gene, encoding a bona fide human histidyl-tRNA synthetase (24), was cloned into the NcoI-BamHI sites of pACTII, after amplification from pCII-HRS with the oligonucleotides HRS-BglII (5′-GCCCGAGATCTTCAGCAGATGCAGAGGGGCTGG) and HRS-NcoI (5′-GCCCGCCATGGCAGAGCGTGCGCCGCTGG). HRS did not bind to HIV-1 MA despite a high degree of identity with HO3.
The p17 MA protein is synthesized in the infected cell as a precursor polypeptide (p55Gag) which is later cleaved by the viral protease (32). To ascertain whether the p55Gag precursor was able to bind HO3, pGBT9-p55 was constructed by cloning the AlwNI-EcoRI fragment (nt 1141 to 4647) of R7 in the AlwNI site of the MA gene of pGBT9-MA. pGBT9-p55MAKK27TT, containing the K26T and K27T mutations, was constructed in the same way but with pGBT9-MAKK27TT as the substrate for cloning. Compared with MA, p55Gag induced lower levels of β-Gal activity when coexpressed with HO3 in the yeast two-hybrid system. However, levels of p55Gag expression were significantly decreased compared to those of p17 MA (data not shown), making the comparison difficult. The MA dilysine mutation that abolished the interaction between MA and HO3 also abrogated the binding of p55Gag to HO3. This suggests that the induction of β-Gal activity observed with the Gag precursor was significant and confirms that this region of MA is important for HO3 recruitment. Finally, HO3 did interact with a mutant MA in which the C-terminal tyrosine has been replaced by phenylalanine (MAY132F). This protein served as a useful positive control, achieving levels of HO3 binding similar to those with wild-type MA. Results obtained through the β-Gal assay were confirmed by evaluating the ability of the different clones to grow in the absence of histidine (Fig. 1C). Taken together, these results argue for a specific interaction between HIV-1 MA and HO3.
HO3 is expressed in several human tissues and T-lymphoid-cell lines.
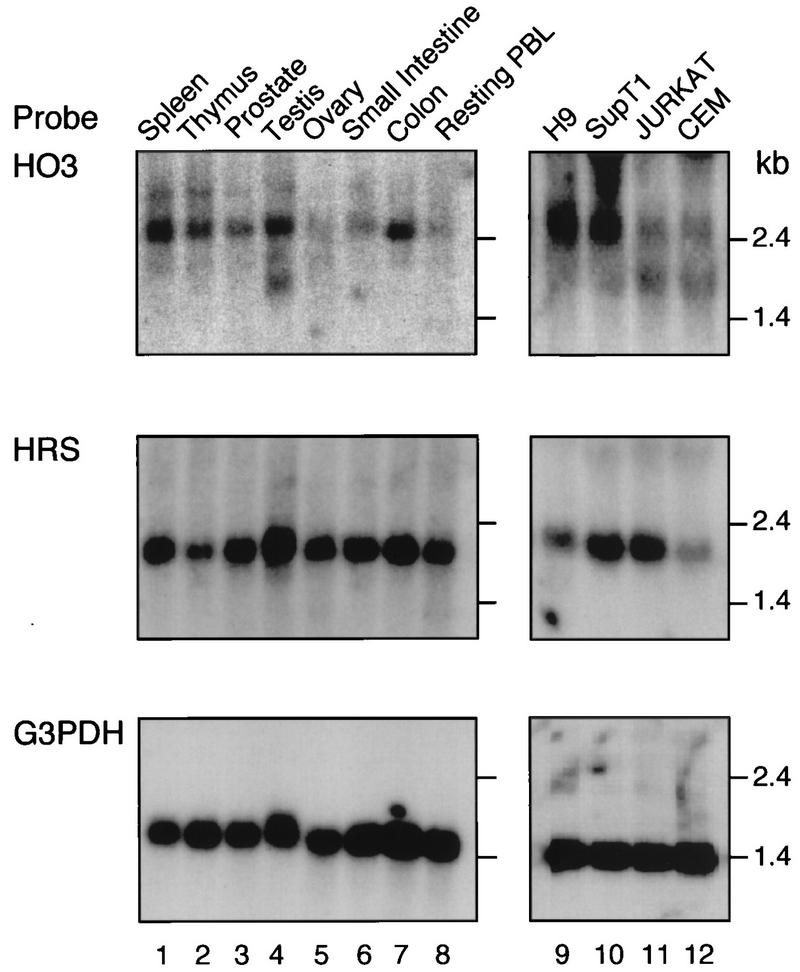
To analyze the distribution of HO3 in tissue, mRNAs representative of various human tissues were sequentially hybridized with cDNA probes specific for either HO3 or HRS (Fig. 2). RNA blot analysis was performed with a commercially prepared human multiple-tissue mRNA blot (Clontech Laboratories, Inc.). Probes with similar specific activities per nanogram of DNA were used, but longer exposure times were needed to detect HO3 (data not shown), suggesting that the levels of expression of this gene are lower than those of HRS. A 2.5-kb band was detected after hybridization with two different HO3 probes, in agreement with a previous report (23). HO3 was expressed in all tissues tested, with highest levels in spleen, testis, and colon and lower levels in thymus and resting peripheral blood lymphocytes (PBL). HO3 RNA was also detected in human T-lymphoid-cell lines commonly used to grow HIV-1, such as H9, SupT1, Jurkat, and CEM cells. Of note, in some of the tissues tested, such as testis (Fig. 2, lane 4), and in all T-cell lines (Fig. 2, lanes 9 to 12), a lower-molecular-weight RNA species (approximately 1.7 kb) was detected with the HO3 probes. This RNA might represent the result of alternative splicing.
FIG. 2.
HO3 and HRS mRNA expression in several human tissues and T-lymphoid-cell lines. Northern blot analysis of polyadenylated RNA (2 μg per lane) from human tissues (lanes 1 to 8) or a total RNA (10 μg per lane) from several T-lymphoid-cell lines (lanes 9 to 12) with cDNA probes specific for HO3 (top), HRS (middle), or glyceraldehyde-3-phosphate dehydrogenase (bottom). The sizes of the major HO3 and HRS transcripts are 2.5 and 2.0 kb, respectively.
Coimmunoprecipitation of HO3 and HIV-1 MA proteins in transfected 293T cells.
Because of low levels of endogenous HO3, all subsequent experiments were performed with transiently transfected 293T cells expressing high levels of exogenous HO3 protein. 293T cells were transfected by the calcium phosphate method as previously described (1). Expression of either HO3 or HRS in 293T cells was carried out by using the simian virus 40-based mammalian expression vector pCMX-PL2 (28). The EcoRI fragment of pCRII-HO3 was cloned into the EcoRI site of pCMX-PL2 to make pCMX-HO3. A SpeI fragment of pCII-HRS was cloned into the NheI site of pCMX-PL2 to make pCMX-HRS.
Transfection of 293T cells with a HO3 expression plasmid led to the accumulation of a protein with an apparent molecular mass of 52 kDa recognized by a polyclonal antiserum raised against the 57 N-terminal amino acids of HO3 (Fig. 3). Separation of cytoplasmic and nuclear fractions of these cells revealed that HO3 accumulated in both compartments. Western blotting with an α-tubulin-specific antibody (Boehringer Mannheim) verified that the nuclear fractions had not been contaminated with cytosolic proteins. Mock-transfected lysates showed a faint band comigrating with the exogenous HO3 protein expressed from pCMX-HO3. This band might represent the endogenous HO3 protein.
FIG. 3.
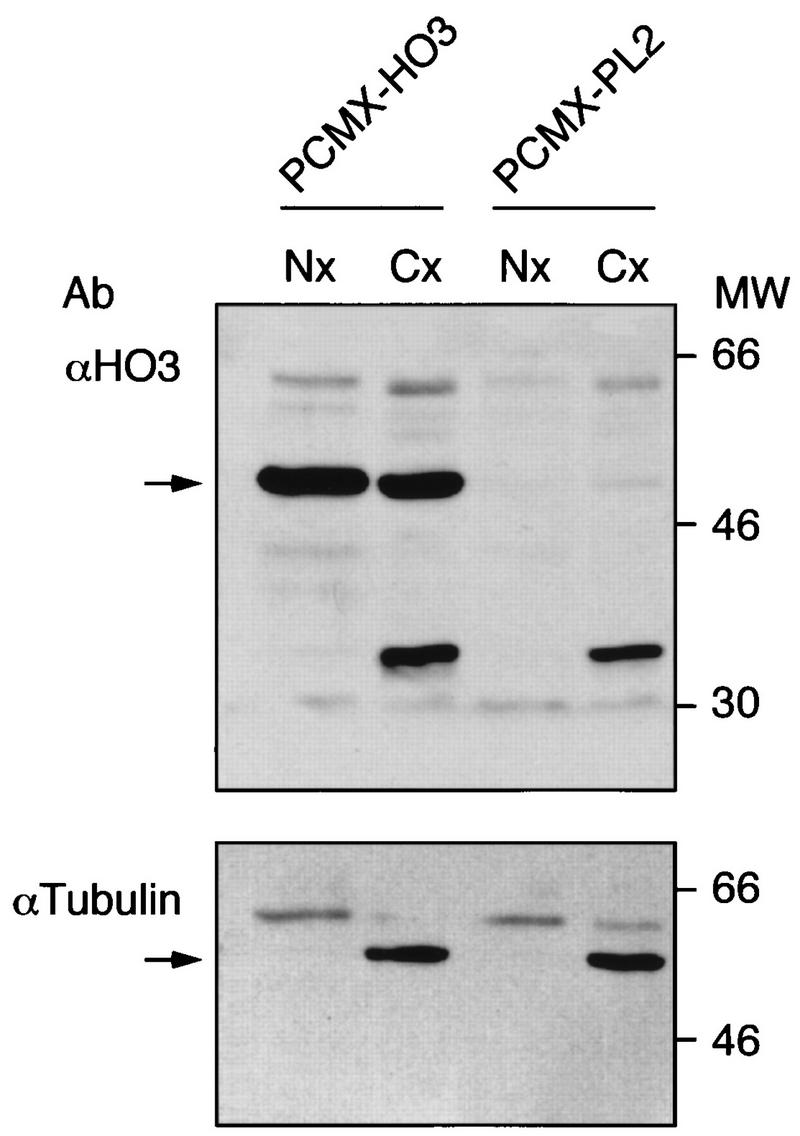
Subcellular localization of HO3 in 293T-transfected cells. 293T cells were transfected with 25 μg of either a HO3-expressing plasmid (pCMX-HO3) or a control vector (pCMX-PL2). After 48 h, nuclear (Nx) and cytoplasmic (Cx) extracts (each containing 30 μg of protein) were loaded on SDS–12% PAGE gels, transferred to polyvinylidene difluoride membrane, and probed with antibodies specific for HO3 (upper panel) or α-tubulin (lower panel). The positions of molecular size markers (in thousands) are indicated on the left.
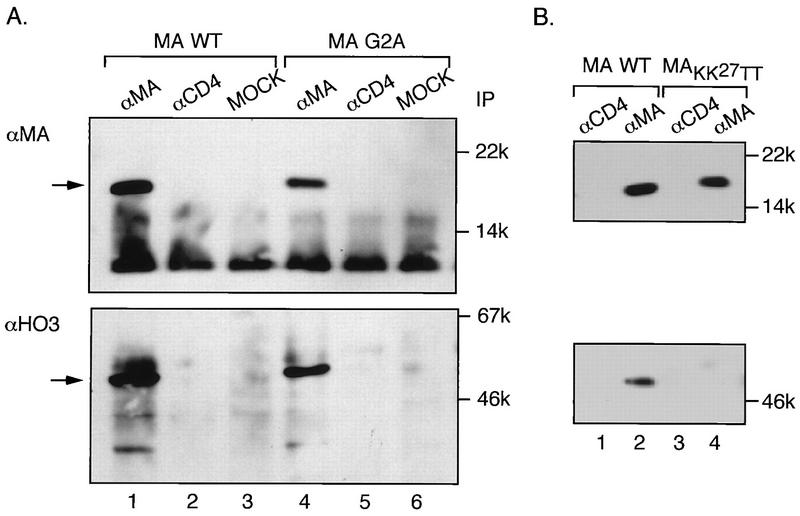
To test the interaction between HO3 and MA in mammalian cells, 293T cells were cotransfected with pCMX-HO3 and R7-MAstop, a modified R7 proviral construct in which a stop codon was placed immediately after the MA sequence (10), for high-level expression of HO3 and wild-type MA, respectively. After 48 h, the cells were washed with phosphate-buffered saline and resuspended in hypotonic buffer (20 mM potassium HEPES [pH 7.8], 5 mM potassium acetate, 0.5 mM MgCl2, 0.5 mM dithiothreitol, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and 1 μg each of leupeptin, aprotinin, and pepstatin per ml). Lysis was performed by several strokes with a Dounce homogenizer. Nuclear-cytoplasmic separation was carried out as previously described (10). Cytoplasmic lysates were diluted with 2 volumes of IP buffer, containing 20 mM sodium phosphate (pH 7.0), 50 mM NaCl, 1 mM MgCl2, 1 mM dithiothreitol, and 0.2% Tween 20, and coimmunoprecipitated with MA-specific antibodies (Advanced Biotechnologies, Inc.) bound to protein A and G agarose beads (Fig. 4). Anti-MA antibodies could coimmunoprecipitate HO3 with MA, as revealed by Western blot analysis of the immune complexes with a HO3-specific serum (dilution, 1:1,000) (Fig. 4A, lane 1). Neither CD4-specific antibodies (Pharmingen) nor protein A and G agarose beads alone (mock IPs) were able to capture HO3. A nonmyristoylated form of p17 MA (MAG2A), in which the first glycine of the p55Gag polyprotein has been changed to alanine, could also be immunoprecipitated with HO3 (Fig. 4A, lane 4). This result suggests that the binding of MA to membranes is not required for the interaction with HO3. Confirming the results obtained in the yeast two-hybrid system, a dilysine mutant of MA (MAKK27TT) did not interact with HO3 in the coimmunoprecipitation studies (Fig. 4B, lane 4).
FIG. 4.
Coimmunoprecipitation of HIV-1 MA and HO3 proteins. 293T cells were cotransfected with 10 μg of a construct expressing either wild-type (WT) (A and B), nonmyristoylated (MAG2A) (A), or dilysine-mutated (MAKK27TT) (B) forms of HIV-1 MA together with 25 μg of a HO3-expressing plasmid (pCMX-HO3). After 48 h, cytoplasmic lysates were prepared and immunoprecipitated with 2.5 μg of monoclonal antibodies against either MA or human CD4 or with a control antibody (mock). Immunoprecipitates were electrophoresed through SDS–12% PAGE gels, transferred to polyvinylidene difluoride membranes, and analyzed with polyclonal antibodies against either MA (upper panel) or HO3 (lower panel). Arrows on the left indicate positions of MA and HO3. The molecular weights of immunoprecipitates (IP) are given on the right.
Incorporation of HO3 into HIV-1 virions.
MA is one of the major structural proteins of HIV-1 virions. In mature viral particles, the majority of MA is localized to the inner face of the lipid bilayer (15, 16). Therefore, a protein interacting with MA is likely to be incorporated into virions. To test this hypothesis, 293T cells were cotransfected with HIV-1 proviral constructs expressing wild-type or dilysine-mutated MA together with either a HO3-expressing plasmid or a control vector. HO3 production was verified by Western blot analysis of cytoplasmic extracts (Fig. 5, top). No difference was found in the levels of p24 protein released in the culture medium (data not shown), indicating that the efficiency of viral particle formation was not affected by HO3 expression. Virions were harvested from the culture medium and purified by ultracentrifugation through a 20% (wt/vol) sucrose cushion at 26,000 rpm in an SW28 rotor (Beckman) for 1.5 h at 4°C, followed by centrifugation at 14,000 rpm for 90 min in a table-top microcentrifuge (Eppendorf). Normalized amounts of viral lysates were loaded on gels for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with MA-CA (Fig. 5, bottom)- or HO3 (Fig. 5, middle)-specific antibodies. HO3 was detected in wild-type virions but could not be found in particles containing the dilysine-mutated form of MA. No HO3 protein was detected out of mock-infected cells transfected with pCMX-HO3 alone. This corroborated the results of the binding studies and also ascertained that HO3 was incorporated in virions and not in vesicles that might have been nonspecifically released from transfected cells.
FIG. 5.
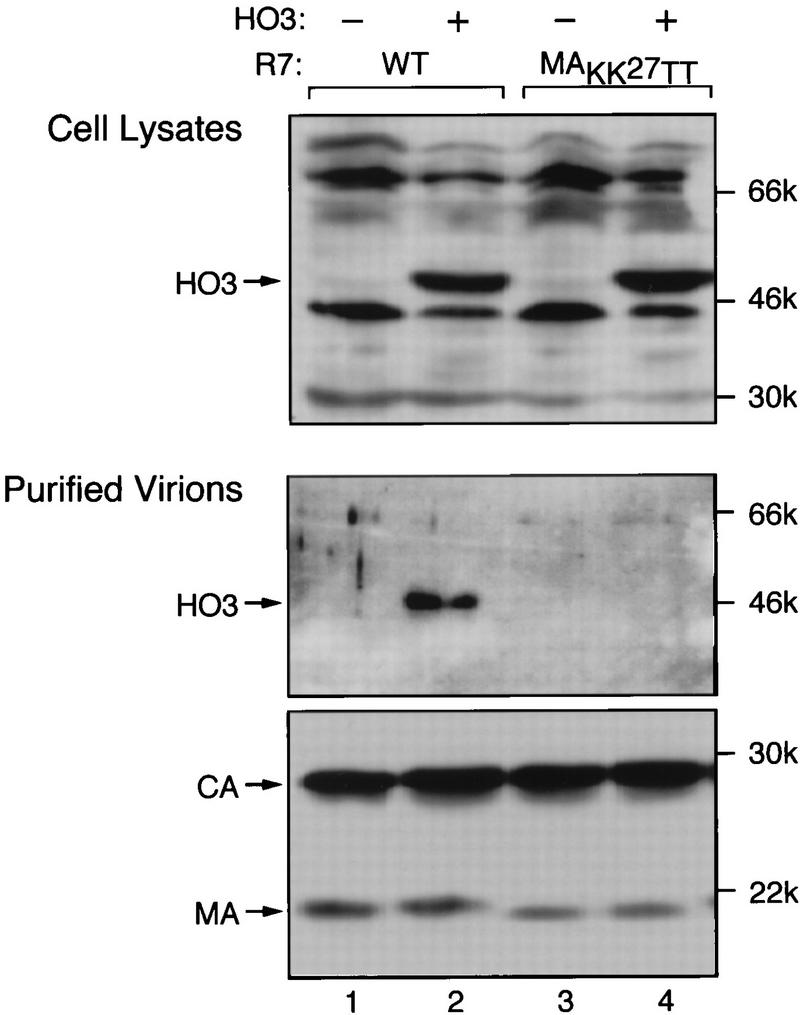
Specific association of HO3 with HIV-1 virions. 293T cells were transfected with 3 μg of proviral DNA constructs (R7) expressing wild-type (WT) or dilysine-mutated (MAKK27TT) versions of MA together with 30 μg of pCMX-HO3 (HO3 +) or pCMX-PL2 (HO3 −). Cellular extracts (top panel) and purified virions (lower two panels) were prepared at 48 h and analyzed by Western blotting with antibodies against HO3 (top two panels) or MA and CA (Bethesda Research Laboratories) (bottom panel). In lane 1, samples from cells transfected with pCMX-HO3 alone were analyzed. The molecular weights of immunoprecipitates are given on the right.
Enhancement of HIV-1 infectivity by HO3 overexpression in virus producer cells.
The particle incorporation of HO3 suggested that the MA-binding protein could affect the infectivity of HIV-1 virions. To probe this issue, the infectivity of the HIV-1 particles was tested on P4 cells, which contain a β-Gal gene under the control of the HIV-1 long terminal repeat (5) (Fig. 6A). HIV-1 particles made in HO3-expressing cells showed a two- to threefold increase in infectivity, as assessed by counting the number of blue foci induced per nanogram of p24 protein. In contrast, expression of HO3 in the producer cell did not significantly affect the infectivity of MAKK27TT HIV-1 particles, suggesting that the observed effect is dependent upon the interaction between HO3 and MA. The infectivity of HO3-containing particles was also tested on P4 cells that had been arrested by γ-irradiation at 8,000 rads from a 61Co source 1 day before infection. A similar increase in the infectivity of these virions was observed in arrested cells (Fig. 6A).
FIG. 6.
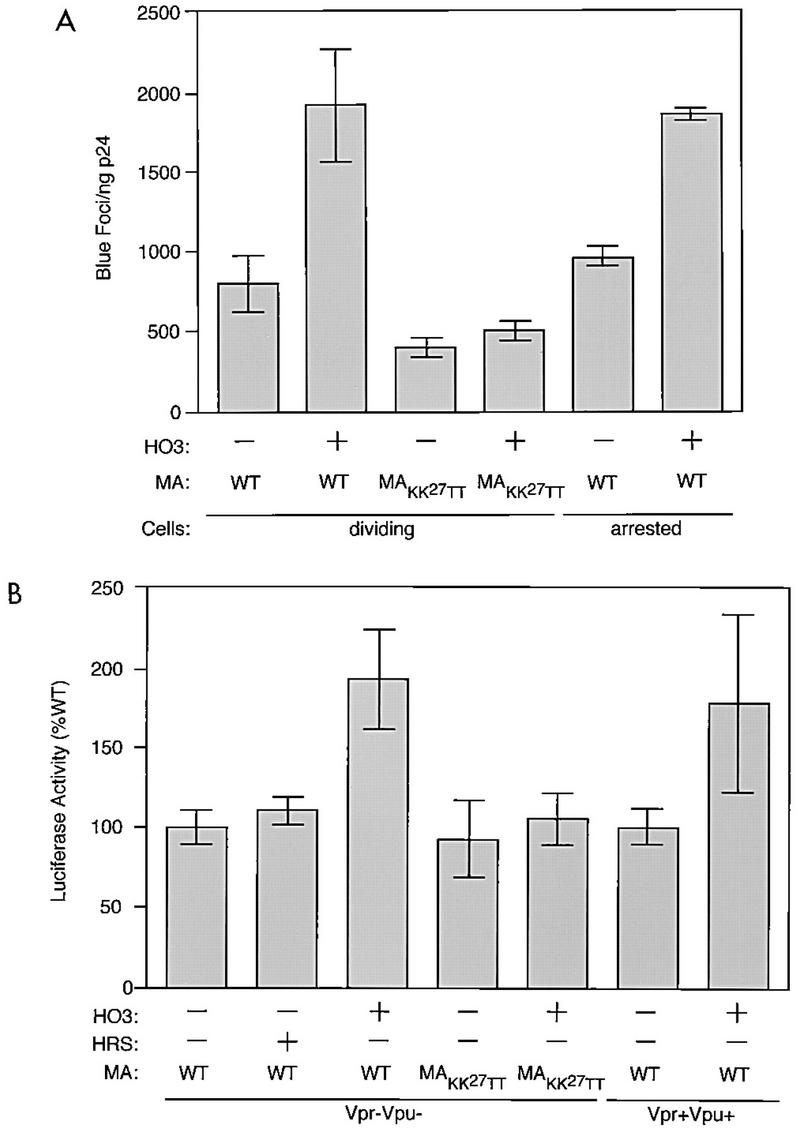
Infectivity of HIV-1 particles made in HO3-expressing cells. (A) 293T cells were cotransfected with 3 μg of proviral DNA constructs (R7) expressing wild-type (WT) or dilysine-mutated (MAKK27TT) versions of MA in the presence or the absence of 20 μg of pCMX-HO3 (the total amount of transfected DNA was kept constant). Infectivity of virions purified at 48 h by ultracentrifugation through a sucrose cushion was measured in a single-round assay using dividing or growth-arrested P4 cells as targets, as indicated. Data represent the mean values ± standard errors of the means (error bars) of four independent transfections or infections. Transduction efficiency of a murine leukemia virus-derived retroviral vector was only 5% in arrested targets, compared to that in dividing cells. (B) HIV-1-derived VSV G-pseudotyped retroviral particles containing a luciferase expression cassette were transiently produced in 293T cells cotransfected with plasmids expressing either HO3 or HRS or with a control vector. Different packaging constructs were utilized as follows: pCMV-ΔR8.2 (containing wild-type MA, Vpr, and Vpu genes), pCMV-ΔR9-ΔVpr (with a wild-type MA gene but defective for the production of Vpr and Vpu), or pCMV-ΔR9-ΔVpr-MAKK27TT (defective in the production of Vpr and Vpu and expressing a mutated MA gene). Equal amounts of transducing particles (as assessed by p24 content) were used to infect 293T cells. Luciferase activity in target cell lysates was measured 48 h later. Data represent the mean values ± standard errors of the means (error bars) from three transfections or infections and are given as the percentage of the luciferase activity obtained from cell lysates infected with wild-type MA viruses made in the absence of exogenous HO3 or HRS.
To confirm these results, HIV-1-based retroviral vector particles pseudotyped with the vesicular stomatitis virus (VSV) G protein and carrying a luciferase expression cassette were utilized (22). HIV-based retroviral particles were prepared by cotransfection of 293T cells with a mixture containing 2.5 μg of pMDG (encoding the G glycoprotein of VSV), 7.5 μg of packaging construct (encoding all HIV-1 proteins besides Env), 10 μg of the transfer vector pHR′CAluciferase (expressing luciferase under the control of a cytomegalovirus-actin hybrid promoter in a cassette flanked by HIV LTR sequences), and 15 μg of a pCMX-based expression vector. After 2 days, transducing particles were harvested from the supernatant and filtered through a 0.45-μm-pore-size filter. To determine the infectivity of the particles, these supernatants were used to infect 293T cells. After 2 days, cytoplasmic lysates were used to estimate the luciferase activity in a luminometer. Expression of HO3 enhanced the transduction efficiency (twofold) of HIV-1 vector particles containing wild-type MA but not MAKK27TT (Fig. 6B). This effect was independent from the presence of functional vpr and vpu genes in the packaging construct. Therefore, the increased infectivity of HIV-1-derived particles observed when HO3 is overexpressed in virus producer cells requires neither the HIV envelope nor the Vpr and Vpu proteins.
Using the two-hybrid system, we have identified the HO3 gene product as a putative cellular partner of HIV-1 MA. The HO3 gene was originally identified as a novel gene oriented in a head-to-head configuration with the human histidyl-tRNA synthetase gene (HRS) (24), and it encodes a polyprotein which has a high degree of homology with HRS (72% amino acid sequence identity). However, no evidence indicating that HO3 is endowed with enzymatic activity is yet available. HO3 expression appears to be ubiquitous, and HO3 RNA was detected in tissues relevant for HIV infection, such as spleen, thymus, and PBL, as well as in human T-lymphoid-cell lines.
In the yeast system, HO3 interacted specifically with the MA protein of HIV-1. A double substitution of lysine residues 26 and 27 of MA abrogated the interaction between the two proteins. The viral proteins IN and Nef failed to interact with HO3, and HRS did not bind to MA. Taken together, these results suggest a highly specific interaction between HO3 and MA.
The MA-HO3 interaction was also demonstrated by coimmunoprecipitation studies on cytoplasmic extracts of transfected 293T cells and through the detection of HO3 in HIV-1 virions produced from these cells. Replacing lysine 26 and 27 of MA by threonine abolished the MA-HO3 interaction and prevented the particle association of HO3. Our attempts to demonstrate the same phenomena in the absence of HO3 overexpression failed, likely due to the low levels of endogenous HO3 and to the limitations of our detection technique.
The virion incorporation of HO3 demonstrates that it must recognize the Gag precursor, since the latter is cleaved into its final products, including MA, only at the time of or shortly after budding. Yeast data showed a weak but significant interaction between HO3 and p55Gag.
HO3 was found in both cytoplasmic and nuclear fractions of transfected 293T cells, and its subcellular localization was unaffected in the presence of MA. Reciprocally, HO3 expression did not trigger a redistribution of MA (data not shown). However, the high levels of protein expression achieved in this system may have masked subtle differences. Residues important for HO3 binding have been proposed to participate in increasing the affinity of the Gag precursor for the plasma membrane (37). This model is supported by structural studies, which indicate that these amino acids are juxtaposed to the virus lipid bilayer while the C-terminal end of the molecule protrudes towards the inner regions of the virion (20). However, neither mutating lysine 26 and 27 of MA nor overexpressing HO3 significantly influenced the efficiency of viral particle formation and budding. In contrast, HO3 overexpression stimulated the infectivity of HIV-1 virions, albeit to a low degree (two- to threefold). HO3 did not act through an enhancement of CD4-mediated HIV-1 entry, since it could augment the infectivity of VSV G-pseudotyped HIV-1-derived retroviral vector particles as well. Nevertheless, this effect appeared to be specific, because the infectivity of virions containing MA molecules mutated at positions 26 and 27 was not stimulated by HO3.
The low degree of HO3-induced stimulation of HIV-1 infectivity might be interpreted as indicating that the low levels of endogenous HO3 are sufficient to carry out the functions of this protein, with overexpression having only a minor additional influence. However, it also correlates with the normal replication of the MAKK27TT HIV-1 mutant in the cell systems used here. In that regard, it is noteworthy that lysines 26 and 27 of MA are also critical for MA recognition by members of the karyopherin-α family (11), an interaction which facilitates HIV-1 replication in terminally differentiated macrophages and the establishment of a stable infection intermediate in quiescent T lymphocytes (4, 10, 30). This strongly suggests that the MA-HO3 interaction might be more relevant in the context of these cells. Future studies, in particular those aimed at suppressing endogenous HO3 production and at studying its effect in primary cells, may help solve this problem.
Acknowledgments
We thank Yen Chen, Diana Camaur, Isabel Pérez-Otaño, and our colleagues at the Infectious Disease Laboratory for many helpful suggestions; Jurg Ruf for providing the Jurkat cDNA library; Nina Raben and Ralph C. Nichols for the gift of HO3 and HRS cDNAs and HO3-specific antiserum; and L. Barden for the artwork.
J.L. is the holder of a Human Frontier Science Program Organization Fellowship. This work was supported by NIH award RO1 AI37510.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 2.Bartel P, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1993;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 3.Bukrinsky M I, Sharova N, Dempsey M P, Stanwick T L, Bukrinskaya A G, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubei A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charneau P, Mirambeau G, Roux P, Paulus S, Buc H, Clavel F. HIV-1 reverse transcription. A termination step at the center of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 6.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 7.Dorfman T, Mammano F, Haseltine W A, Gottlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Facke M, Janetzko A, Shoeman R L, Krausslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 11.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 13.Gallay, P., and D. Trono. Unpublished data.
- 14.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature. 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 15.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 16.Gelderblom H R, Hausmann E H S, Ozel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 17.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis P F, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massiah M A, Starich M R, Paschall C, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 21.Meltzer M S, Gendelman H E. Mononuclear phagocytes as targets, tissue reservoirs, and immunoregulatory cells in human immunodeficiency virus disease. Curr Top Microbiol Immunol. 1992;181:239–263. doi: 10.1007/978-3-642-77377-8_9. [DOI] [PubMed] [Google Scholar]
- 22.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 23.O’Hanlon T P, Raben N, Miller F. A novel gene oriented in a head-to-head configuration with the human histidyl-tRNA synthetase (HRS) gene encodes an mRNA that predicts a polypeptide homologous to HRS. Gene. 1995;210:556–566. doi: 10.1006/bbrc.1995.1696. [DOI] [PubMed] [Google Scholar]
- 24.Owens R J, Dubay J, Hunter E, Compans R W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz A M, Henderson L E, Oroszlan S. Fatty acylation of proteins. Annu Rev Cell Biol. 1988;4:611–647. doi: 10.1146/annurev.cb.04.110188.003143. [DOI] [PubMed] [Google Scholar]
- 27.Spearman P, Wang J-J, Heyden N V, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umesono K, Murakami K, Thompson C, Evans R. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varmus H E, Swanstrom R. Replication of retroviruses. In: Weiss R, et al., editors. RNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. pp. 369–512. [Google Scholar]
- 30.von Schwedler U, Kornbluth R S, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wills J, Craven R. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 33.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie R C. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X, Yuan X, Matsuda Z, Lee T H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Yu Q C, Lee T H, Essex M. The C terminus of human immunodeficiency virus type 1 matrix protein is involved in early steps of the virus life cycle. J Virol. 1992;66:5667–5670. doi: 10.1128/jvi.66.9.5667-5670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan X, Yu X, Lee T H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]