Abstract
RNA 2 of soil-borne wheat mosaic virus (SBWMV), the type species of the genus Furovirus, encodes a protein previously hypothesized to be initiated at an in-frame non-AUG codon upstream of the AUG initiation codon (nucleotide positions 334 to 336) for the 19-kDa capsid protein. Site-directed mutagenesis and in vitro transcription and translation analysis indicated that CUG (nucleotides 214 to 216) is the initiation codon for a protein with a calculated molecular mass of 25 kDa composed of a 40-amino-acid extension to the N terminus of the 19-kDa capsid protein. A stable deletion mutant, which was isolated after extensive passages of a wild-type SBWMV, contained a mixture of two deleted RNA 2’s, only one of which coded for the 25-kDa protein. The amino acid sequence of the N-terminal extension was moderately conserved and the CUG initiation codon was preserved among three SBWMV isolates from Japan and the United States. This amino acid sequence conservation, as well as the retention of expression of the 25-kDa protein in the stable deletion mutant, suggests that the 25-kDa protein is functional in the life cycle of SBWMV. This is the first report of a non-AUG translation initiation in a plant RNA virus genome.
Soil-borne wheat mosaic virus (SBWMV) is the type species of the genus Furovirus (8). The group is characterized by transmission through plasmodiophoraceous fungus in soil and a divided plus-sense RNA genome, with each RNA individually encapsidated into rigid rod-shaped particles (7, 35). The SBWMV genome consists of two RNA species, RNA 1 (7,099 nucleotides) and RNA 2 (3,593 nucleotides), both of which have been sequenced from a U.S. (Nebraska) isolate (34). From the 5′ terminus, RNA 1 codes for N-terminal overlapping 150-kDa and 209-kDa proteins thought to be the viral RNA polymerase and a 37-kDa putative cell-to-cell-movement protein. Similarly, RNA 2 codes for the 19-kDa capsid protein, the UGA termination codon of which is periodically read through, at 10 to 20% efficiency, to produce an 84-kDa protein thought to be required for virus transmission by the plasmodiophoraceous fungus Polymyxa graminis (32). In the case of beet necrotic yellow vein virus, it has been experimentally shown that the capsid readthrough region is required for transmission by fungus (36, 37). A 19-kDa cysteine-rich protein that may function in regulation of RNA replication based on the putative function of similar proteins from related viruses (14, 16, 27) is encoded by the 3′-proximal open reading frame (see Fig. 1A).
FIG. 1.
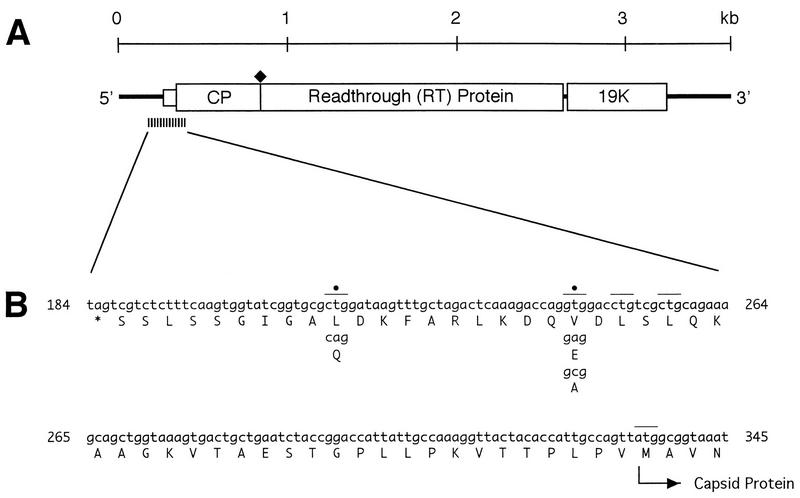
(A) Genome organization of SBWMV RNA 2. Horizontal boxes indicate open reading frames. A filled diamond above the horizontal box identifies the location of a UGA codon at the end of the capsid protein gene, which is periodically read through to produce an 84-kDa protein. (B) Nucleotide (lowercase) and amino acid (uppercase, below each codon) sequences from nucleotides 184 to 345 of the wild-type RNA 2 of the U.S.-Nebraska isolate. Three CTG triplets, one GTG triplet, and the ATG capsid initiation codon are indicated by upper lines. A T at position 215 was replaced with A, and a T at position 245 (identified with black dots) was replaced with A or C by site-directed, in vitro mutagenesis. An asterisk under positions 184 to 186 represents a termination codon. The initiation codon for the capsid protein is at positions 334 to 336.
In addition to the proteins defined from the nucleotide sequences, a protein of 28 kDa (28K protein) (as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) was produced by the SBWMV RNA 2 in vitro as well as in virus-infected wheat tissues (17, 33). The protein reacted with antisera against SBWMV virions, suggesting that the protein is a component of the virion or shares epitopes with the capsid protein. In vitro transcription and translation analysis showed that transcripts starting at RNA 2 nucleotide position 159 expressed the 28K protein in addition to the 19-kDa capsid and 84-kDa readthrough proteins, whereas those starting at nucleotide 256 did not produce the 28K protein but did express the capsid and readthrough proteins (34). This result implied that the 28K protein arose not from posttranslational modification of the 19-kDa capsid protein but from translation initiation at an in-frame non-AUG upstream from the capsid protein initiation codon at nucleotides 334 to 336. Since there is an upstream in-frame UAG termination codon at nucleotides 184 to 186, it was hypothesized that a non-AUG initiation codon for the 28K protein was located between nucleotides 187 and 256 (34).
Non-AUG translation initiation has been reported for both procaryotic and eucaryotic cellular gene expression (21, 22, 26), as well as from adeno-associated parvovirus (2), murine leukemia retrovirus (28), and Sendai paramyxovirus (13) genes. GUG, ACG, and CUG codons are generally used as non-AUG initiation codons in animal cells (3). It was reported that efficiency of translation initiation at non-AUG codons depends on the context just as for the AUG codon, i.e., a purine at −3 and a G at +4 (22, 23). Within this context, a GUG codon was reported to be the most efficient initiator in mammalian cells. The importance of positions +5 (A) and +6 (U) has also been reported (3). On the other hand, in plant cells transient-expression analysis showed that CUG was the next most efficient initiator after AUG (15). Furthermore, it has been shown that AUU also functions as a translation initiator in plant and insect cells (1, 30).
This study was performed to identify the non-AUG initiation codon for the 28K polypeptide observed in vivo and in vitro initiating upstream from the SBWMV capsid protein gene.
In vitro mutagenesis of CUG and GUG codons upstream from the capsid protein gene.
A full-length SBWMV RNA 2 cDNA of the Nebraska isolate (1988) was cloned downstream from the bacteriophage T7 RNA polymerase promoter in pGEM3Z (Promega) as follows. RNA 2 cDNA was synthesized with avian myeloblastosis virus reverse transcriptase (Life Sciences) from the 3′ end with the primer 5′-TGCTCTAGATGGGCCGGATAACCCTCCGG-3′, in which the 6 underlined bases identify an XbaI site for linearization of the template DNA and the 20 bases in boldface type are complementary to the 3′-terminal sequence of SBWMV RNA 2. The 3.6-kb cDNA was made double stranded with Escherichia coli DNA polymerase I (Pharmacia) and RNase H (Pharmacia), flush-ended with T4 DNA polymerase (Pharmacia), digested with XbaI, and ligated into SmaI-XbaI-restricted pGEM3Z. For cloning the 5′-terminal region, including an upstream T7 RNA polymerase promoter sequence, cDNA was synthesized with a minus-sense primer annealing to nucleotides 113 to 131 and amplified by PCR with Taq DNA polymerase (Promega) by use of an additional plus-sense, upstream primer, 5′-GGCATATG TAATACGAC TCAC TATAG TAT T TC T TC T TCACATACGACA-3′, in which the 6 underlined bases are a unique NdeI site, the 17 italicized bases are the T7 RNA polymerase promoter sequence, and the 23 boldface bases are the 5′-terminal 23 nucleotides of SBWMV RNA 2. The PCR products were digested with NdeI and HpaI at nucleotide 26 and ligated into an NdeI-HpaI-cut vector DNA which contains the cDNA insert ranging from the HpaI site to the 3′ terminus with an extra XbaI site. The nucleotide sequence of the resulting full-length cDNA clone, pSW2.4, was determined by dideoxy sequencing and confirmed to be identical to the sequence of the 1981 Nebraska isolate (34).
Figure 1A shows the genome organization of SBWMV RNA 2. Figure 1B shows the nucleotide and amino acid sequences between nucleotides 184 and 345, including the 5′-terminal region of the capsid protein gene. Between nucleotides 187 and 256, there are three CUG triplets at positions 214 to 216, 250 to 252, and 256 to 258, one GUG triplet at position 244 to 246, and no ACG or AUU codon (Fig. 1B). Of the three CUG codons, only the one at positions 214 to 216 is in the preferred context, i.e., G at both −3 and +4 positions as well as A at +5 and U at +6. Given this, the U at position 215 was replaced with an A, changing the potential CUG start codon to CAG, by in vitro mutagenesis (24). Similarly, U at position 245 was replaced with A or C to replace the GUG codon at position 244 to 246 with GAG or GCG, respectively. Mutant plasmid DNA was linearized with XbaI and transcribed in vitro with T7 RNA polymerase (Pharmacia) in the presence of a cap analog (New England Biolabs). In vitro transcripts were translated in vitro in rabbit reticulocyte lysates (Promega) in the presence of [35S]methionine (1,200 Ci/mmol; Amersham). Translation products were separated in an SDS–12.5% polyacrylamide gel and visualized by fluorography using sodium salicylate (9).
Figure 2 shows in vitro translation products directed by in vitro transcripts from wild-type and different mutant constructs. From the wild-type transcripts, the 19-kDa capsid protein, 28K protein, and 84-kDa capsid readthrough protein (84-kDa CP/RT) were expressed (lane 1). When nucleotide 215 was changed from a U to an A, resulting in replacement of the CUG with CAG, the capsid and 84-kDa readthrough proteins were produced but 28K protein was no longer expressed (lane 2). By contrast, when U at position 245 was substituted with A or C, resulting in replacement of the GUG with GAG or GCG, respectively, the 28K protein was expressed in addition to the capsid and 84-kDa readthrough proteins (lanes 3 and 4). Two or three extra, faint products detected between the 28K protein and the 84-kDa readthrough protein probably initiated internally at AUG codons downstream from the opal (UGA) termination codon for the capsid protein gene as previously described (34). Accumulation of these additional translation products varied among different translations and depended on batches of reticulocyte lysates as well as in vitro transcripts. This result indicates that the CUG codon at position 214 to 216 is the initiation codon for the 28K polypeptide, with the calculated molecular mass of 25 kDa. Hereafter the 28K protein is referred to as the 25-kDa protein.
FIG. 2.
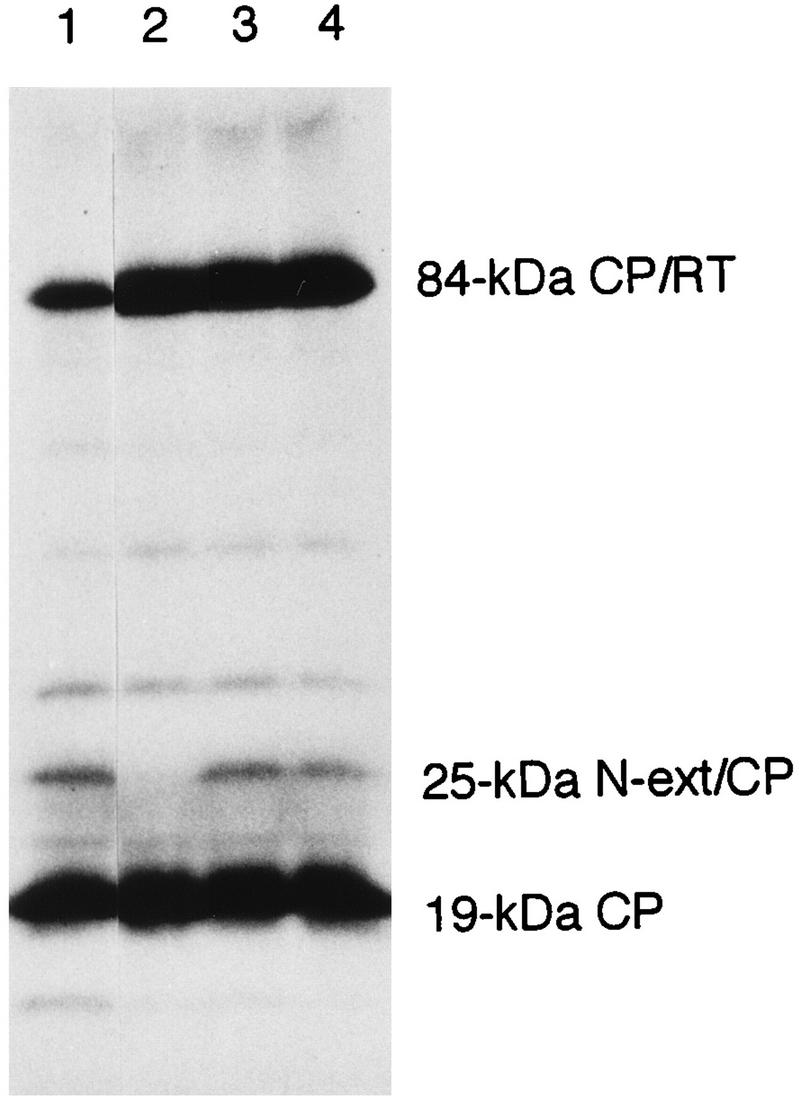
[35S]methionine-labelled in vitro translation products in rabbit reticulocyte lysates analyzed by SDS–12.5% PAGE and fluorography. Translation was directed with in vitro transcripts from the wild-type full-length cDNA construct pSW2.4 (lane 1) and from mutant constructs in the pSW2.4 background with T215→A (lane 2), T245→A (lane 3), and T245→C (lane 4) substitutions. The 25-kDa N-terminal extension and capsid protein marker (N-ext/CP) in the right margin indicates the position of the 28K protein. RT, readthrough protein.
Synthesis of the 25-kDa protein from the Lab 1 deletion mutant RNA 2.
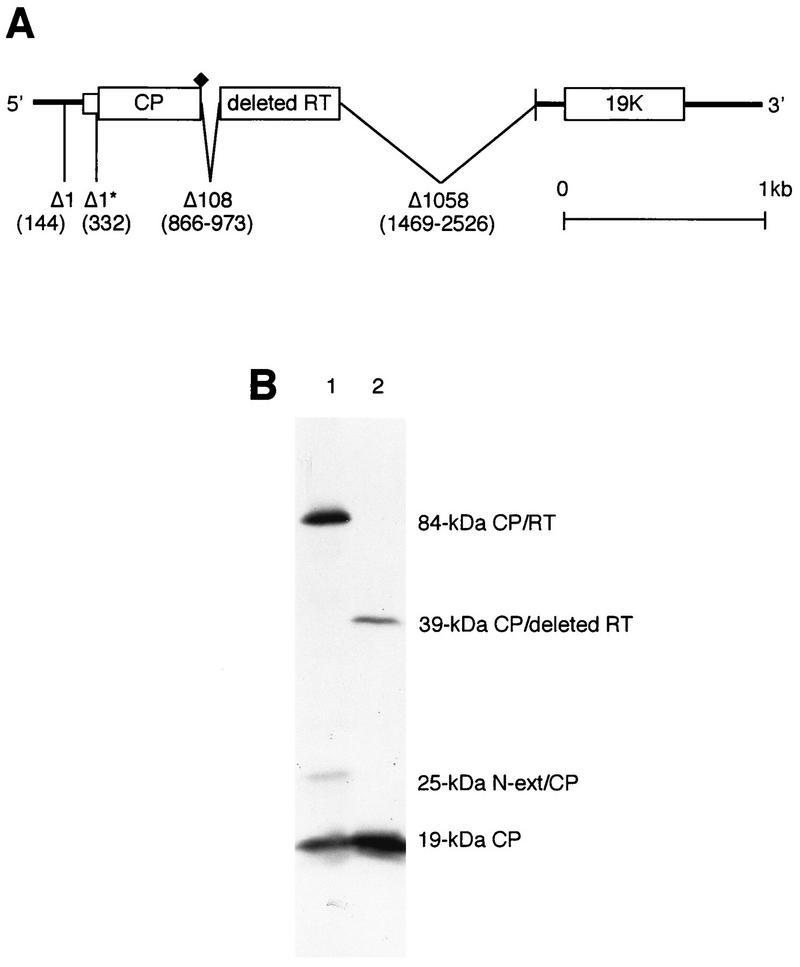
SBWMV generates spontaneous deletion mutants when wild-type virus is maintained or passaged in wheat plants for prolonged periods at around 17°C (10–12, 17, 31–33). A stable deletion mutant, the Lab 1 isolate, was isolated from a wild-type infection originating in an infested field in Nebraska in 1975 after successive mechanical transfers in wheat plants (Triticum aestivum L., cultivar Michigan Amber) for 2 years in a growth cabinet (5, 6). Previously, it was shown that the 25-kDa protein is produced by the Lab 1 isolate in amounts similar to those of the wild-type virus both in vivo and in vitro (17). To verify the 25-kDa protein-encoding sequence and to identify deletion sites in the Lab 1 mutant genome, the complete nucleotide sequence of the Lab 1 RNA 2 was determined from two cDNA clones, p706 and p715, and from viral RNA for the 5′-terminal region as previously described (34) (Fig. 3A). In the 5′-terminal region upstream from the capsid protein gene, there were two single nucleotide deletions, a U from a U trimer (wild-type nucleotides 144 to 146) and another U from a U dimer (nucleotides 332 to 333), the latter of which was located immediately upstream of the capsid protein gene initiation codon. When another 10 independent cDNA clones were further examined for these two deletions, a U was deleted from nucleotides 144 to 146 in all 10 clones. Three clones retained the U dimer at nucleotides 332 to 333, whereas 7 clones had the U deletion, as was shown in clones p706 and p715. On the other hand, in the capsid-readthrough region, there were a 108-nucleotide (wild-type nucleotide positions 866 to 973), in-frame deletion, beginning one C residue after the capsid protein UGA termination codon, and a 1,058-nucleotide deletion (wild-type nucleotide positions 1469 to 2526) in the C-terminal region, resulting in truncation of the wild-type 84-kDa readthrough protein to 39 kDa (Fig. 3A). Thus, in the deleted capsid-readthrough polypeptide, only the wild-type amino acid at positions 214 to 378 (the position 1 is the capsid N terminus) was fused to the C terminus of the capsid protein. There were no further nucleotide deletions in the 3′-terminal region.
FIG. 3.
Comparison of the Lab 1 deletion mutant RNA 2 and the wild-type RNA 2. (A) Genome organization of the Lab 1 deletion mutant RNA 2 compared with that of the wild-type RNA 2. The nucleotide sequence was determined from two independent cDNA clones, and both had single-nucleotide deletions at positions 144 and 332 and 108-nucleotide and 1,058-nucleotide deletions from the wild-type sequence. The single-nucleotide deletion at position 332 (asterisk) was found in 7 of 10 independent cDNA clones, whereas that at position 144 occurred in all 10 clones. (B) [35S]methionine-labelled in vitro translation products of the wild-type RNA 2 transcripts from pSW2.4 (lane 1) and the Lab 1 deletion mutant RNA transcripts from p706 (lane 2) in rabbit reticulocyte lysates analyzed by SDS–12.5% PAGE and fluorography. N-ext/CP, N-terminal extension and capsid protein. RT, readthrough protein.
The Lab 1 cDNA clone p706, which had deletions of the two single U’s, was transcribed in vitro with the upstream T7 promoter, and the resulting transcripts were translated in vitro in rabbit reticulocyte lysates. Compared with the wild-type translation products (Fig. 3B, lane 1), the p706 transcripts directed synthesis of only the 19-kDa capsid protein and the capsid readthrough protein with a calculated molecular mass of 39 kDa but not the 25-kDa polypeptide (lane 2). Deletion of the U immediately upstream of the AUG capsid initiation codon causes a frameshift of the translation initiated at the CUG codon, resulting in termination of translation at the third amino acid after the deletion. From these results, it appears that the Lab 1 isolate is a mixture of two mutants, only one of which codes for the 25-kDa protein.
Conservation of the amino acid sequences in the N-terminal extension.
To examine conservation of the amino acid sequences in the N-terminal extension among isolates of SBWMV from different geographic regions, nucleotide sequences of the 5′-terminal 0.9 kb of RNA 2 were determined from the wild-type, nondeleted forms of an Illinois isolate from 1990 (a kind gift from Adriana Hewings, University of Illinois, Urbana) and a Japanese isolate, JT, from 1982 (33). Figure 4 shows nucleotide and amino acid sequence alignments of the three isolates in the 5′-terminal untranslated region (5′ UTR, panel A), the 120-nucleotide N-terminal extension region (panel B), and the capsid protein gene (panel C). Table 1 shows percent identities among the three isolates in each region at the nucleotide and amino acid levels. In the three regions combined, the Illinois isolate was 89% identical to the Nebraska isolate at the nucleotide level while there was 68% identity between the Japanese isolates and either of the two U.S. isolates. In both the Illinois and Japanese isolates, there was a CUG codon 120 nucleotides upstream from the capsid protein AUG initiation codon, similar to the Nebraska isolate. Significant amino acid sequence identities were found in the N-terminal extension region among the three isolates (Table 1). The N-terminal extension was rich in charged amino acids, particularly in the N-terminal half, whereas the C-terminal region preceding the capsid protein possessed clusters of hydrophobic amino acids in all three isolates (Fig. 4B). On the other hand, the capsid protein was more conserved than the N-terminal extension at both nucleotide and amino acid levels among the three isolates. The capsid proteins of the two U.S. isolates were identical at the amino acid level, while that of the Japanese isolate was 82% identical to those of the two U.S. isolates (Fig. 4C and Table 1).
FIG. 4.

Alignments of nucleotide (lowercase) and translated amino acid (uppercase) sequences in the 5′-terminal 0.9-kb region among three SBWMV geographical isolates from the United States and Japan. (A) The 5′-terminal untranslated (UTR) region. (B) The N-terminal extension region to the capsid protein gene. (C) The capsid protein gene. NE, the U.S. Nebraska isolate, isolated in 1988 in Havelock, Nebr.; IL, the U.S. Illinois isolate, isolated in 1990 in Urbana, Ill.; JT, the Japanese isolate, isolated in 1982 in Tochigi, Japan. Nucleotides and amino acids in IL and JT sequences identical to those in NE are represented as dots.
TABLE 1.
Percent identities at the nucleotide and amino acid levels in the 5′-untranslated region, the N-terminal extension region, and the capsid protein-coding region among Nebraska (NE), Illinois (IL), and Japanese (JT) isolates of SBWMV
| Region | % Identity at the nucleotide level/amino acid level
|
||
|---|---|---|---|
| NE vs IL | NE vs JT | IL vs JT | |
| 5′-Untranslated regiona | 96 | 59 | 61 |
| N-terminal extensionb | 87/90 | 60/65 | 58/58 |
| Capsid protein | 87/100 | 74/82 | 74/82 |
Nucleotide positions 1 to 213 of NE and IL and positions 1 to 209 of JT were compared. Gaps were included to maximize the alignment as shown in Fig. 4A.
Nucleotide positions 214 to 333 of NE and IL and positions 210 to 329 of JT were compared.
The 25-kDa protein is possibly functional in the SBWMV life cycle.
Previously, it was shown that the 25-kDa protein and equivalent proteins were produced not only by wild-type SBWMV but also by extensively passaged stable deletion mutants (17, 33). Although the Nebraska Lab 1 isolate used in this study was a mixture of two mutants based on the deletion of a U at positions 331 to 332, it appeared that there was a selection pressure preventing the deletion of the U in the RNA 2 population. It is not likely that the U is required for efficient translation initiation of the capsid protein gene because the deletion mutant RNA 2 (clone p706) produced a similar amount of capsid protein compared with the wild type (Fig. 3B, lanes 1 and 2), and the context surrounding the capsid initiation codon still retained the preferred consensus sequence with a purine at the −3 position even after the deletion. Rather, the translated 25-kDa protein itself is probably required for replication, movement, and/or spreading of the virus. The significant amino acid sequence conservation in the N-terminal extension among the three isolates from different geographic regions also suggests that the 25-kDa protein is functional in the SBWMV life cycle. Similarities in the N-terminal extension at the amino acid level among the three isolates were fewer than those in the capsid protein, and the N-terminal half of the extension is highly charged, suggesting that the N-terminal extension is not as highly structured as the capsid protein.
It is noteworthy that contexts surrounding the CUG codon as well as the capsid initiation codon in the Japanese isolate do not have the preferred sequence (3), with a U at −3 in both codons (Fig. 4A and B). However, it was previously shown that both the 25-kDa and capsid proteins are translated efficiently in wheat germ extracts (33), also supporting the argument that production of the 25-kDa protein may be substantial in the virus life cycle.
The N-terminal extension of the capsid protein has been found only in cereal plant-infecting furoviruses such as SBWMV (17, 33) and sorghum chlorotic spot virus (19). Use of an in-frame non-AUG translation initiation upstream of the AUG codon allows for the production of two kinds of C-terminally identical proteins in different amounts due to different efficiencies in translation initiation at the two sites. On the other hand, the capsid proteins of the fungus-transmitted rod-shaped viruses can also be extended C-terminally by partial readthrough of the termination codon (4, 18, 20, 34) or by frameshift (25). The C-terminal extension to the capsid protein of beet necrotic yellow vein virus was shown to be required for virion assembly (29) and transmission by the fungus Polymyxa betae (36, 37). In the case of the SBWMV Lab 1 isolate, which was isolated after successive mechanical transfers without a fungal vector, only an N-terminal portion (165 amino acids long) in the readthrough region was retained and fused to the C terminus of the capsid protein. The deleted 39-kDa capsid-readthrough polypeptide is probably required during virus replication or spread.
Further investigation of function(s) of the N-terminal extension as well as the C-terminal extension in the SBWMV life cycle should be made when infectious transcripts produced in vitro from full-length cDNAs for both RNA 1 and RNA 2 became available.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the following accession numbers: D86320 (US-NE Lab1 RNA 2 complete sequence), AB002812 (US-IL wild-type RNA 2, 5′-terminal 0.9 kb), and AB002813 (Japan-JT wild-type RNA 2, 5′-terminal 0.9 kb).
Acknowledgments
I am grateful to James Strauss of the California Institute of Technology and Peter Day of Rutgers University for their support in this study and to Myron Brakke for critical reading of the manuscript.
REFERENCES
- 1.Beames B, Braunagel S, Summers M D, Lanford R E. Polyhedrin initiator codon altered to AUU yields unexpected fusion protein from a baculovirus vector. BioTechniques. 1991;11:378–383. [PubMed] [Google Scholar]
- 2.Becerra S P, Rose J A, Hardy M, Baroudy B M, Anderson C W. Direct mapping of adeno-associated virus capsid proteins B and C: a possible ACG initiation codon. Proc Natl Acad Sci USA. 1985;82:7919–7923. doi: 10.1073/pnas.82.23.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeck R, Kolakofsky D. Positions +5 and +6 can be major determinants of the efficiency of non-AUG initiation codons for protein synthesis. EMBO J. 1994;13:3608–3617. doi: 10.1002/j.1460-2075.1994.tb06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouzoubaa S, Ziegler V, Beck D, Guilley H, Richards K, Jonard G. Nucleotide sequence of beet necrotic yellow vein virus RNA-2. J Gen Virol. 1986;67:1689–1700. [Google Scholar]
- 5.Brakke M K. Sedimentation coefficients of the virions of soil-borne wheat mosaic virus. Phytopathology. 1977;67:1433–1438. [Google Scholar]
- 6.Brakke, M. K. Personal communication.
- 7.Brakke M K, Estes A P, Schuster M L. Transmission of soil-borne wheat mosaic virus. Phytopathology. 1965;55:79–86. [Google Scholar]
- 8.Brunt A A. Furovirus. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 445–448. [Google Scholar]
- 9.Chamberlain J P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, MacFarlane S A, Wilson T M A. Detection and sequence analysis of a spontaneous deletion mutant of soil-borne wheat mosaic virus RNA 2 associated with increased symptom severity. Virology. 1994;202:921–929. doi: 10.1006/viro.1994.1414. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, MacFarlane S A, Wilson T M A. An analysis of spontaneous deletion sites in soil-borne wheat mosaic virus RNA 2. Virology. 1995;209:213–217. doi: 10.1006/viro.1995.1245. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, MacFarlane S A, Wilson T M A. Effect of cultivation temperature on the spontaneous development of deletions in soilborne wheat mosaic furovirus RNA 2. Phytopathology. 1995;85:299–306. [Google Scholar]
- 13.Curran J, Kolakofsky D. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 1988;7:245–251. doi: 10.1002/j.1460-2075.1988.tb02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donald R G K, Jackson A O. RNA-binding activities of barley stripe mosaic virus γb fusion proteins. J Gen Virol. 1996;77:879–888. doi: 10.1099/0022-1317-77-5-879. [DOI] [PubMed] [Google Scholar]
- 15.Gordon K, Fütterer J, Hohn T. Efficient initiation of translation at non-AUG triplets in plant cells. Plant J. 1992;2:809–813. [PubMed] [Google Scholar]
- 16.Hehn A, Bouzoubaa S, Bate N, Twell D, Marbach J, Richards K, Guilley H, Jonard G. The small cysteine-rich protein P14 of beet necrotic yellow vein virus regulates accumulation of RNA 2 in cis and coat protein in trans. Virology. 1994;210:73–81. doi: 10.1006/viro.1995.1318. [DOI] [PubMed] [Google Scholar]
- 17.Hsu Y H, Brakke M K. Cell-free translation of soil-borne wheat mosaic virus RNAs. Virology. 1985;143:272–279. doi: 10.1016/0042-6822(85)90114-x. [DOI] [PubMed] [Google Scholar]
- 18.Kashiwazaki S, Scott K P, Reavy B, Harrison B D. Sequence analysis and gene content of potato mop-top virus RNA 3: further evidence of heterogeneity in the genome organization of furoviruses. Virology. 1995;206:701–706. doi: 10.1016/s0042-6822(95)80092-1. [DOI] [PubMed] [Google Scholar]
- 19.Kendall T L, Langenberg W G, Lommel S A. Molecular characterization of sorghum chlorotic spot virus, a proposed furovirus. J Gen Virol. 1988;69:2335–2345. [Google Scholar]
- 20.Koenig R, Commandeur U, Loss S, Beier C, Kaufmann A, Lesemann D-E. Beet soil-borne virus RNA 2: similarities and dissimilarities to the coat protein gene-carrying RNAs of other furoviruses. J Gen Virol. 1997;78:469–477. doi: 10.1099/0022-1317-78-2-469. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 24.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manohar S K, Guilley H, Dollet M, Richards K, Jonard G. Nucleotide sequence and genetic organization of peanut clump virus RNA 2 and partial characterization of deleted forms. Virology. 1993;195:33–41. doi: 10.1006/viro.1993.1343. [DOI] [PubMed] [Google Scholar]
- 26.Peabody D S. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 27.Petty I T D, French R, Jones R W, Jackson A O. Identification of barley stripe mosaic virus genes involved in viral RNA replication and systemic movement. EMBO J. 1990;9:3453–3457. doi: 10.1002/j.1460-2075.1990.tb07553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prats A-C, De Billy G, Wang P, Darlix J-L. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 1989;205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt C, Balmori E, Jonard G, Richards K E, Guilley H. In vitro mutagenesis of biologically active transcripts of beet necrotic yellow vein virus RNA 2: evidence that a domain of the 75-kDa readthrough protein is important for efficient virus assembly. Proc Natl Acad Sci USA. 1992;89:5715–5719. doi: 10.1073/pnas.89.13.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz J, Prüfer D, Rohde W, Tacke E. Non-canonical translation mechanisms in plants: efficient in vitro and in planta initiation at AUU codons of the tobacco mosaic virus enhancer sequence. Nucleic Acids Res. 1996;24:257–263. doi: 10.1093/nar/24.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirako Y, Brakke M K. Two purified RNAs of soil-borne wheat mosaic virus are needed for infection. J Gen Virol. 1984;65:119–127. [Google Scholar]
- 32.Shirako Y, Brakke M K. Spontaneous deletion mutation of soil-borne wheat mosaic virus RNA II. J Gen Virol. 1984;65:855–858. [Google Scholar]
- 33.Shirako Y, Ehara Y. Comparison of the in vitro translation products of wild-type and a deletion mutant of soil-borne wheat mosaic virus. J Gen Virol. 1986;67:1237–1245. [Google Scholar]
- 34.Shirako Y, Wilson T M A. Complete nucleotide sequence and organization of the bipartite RNA genome of soil-borne wheat mosaic virus. Virology. 1993;195:16–32. doi: 10.1006/viro.1993.1342. [DOI] [PubMed] [Google Scholar]
- 35.Shirako Y, Wilson T M A. Furoviruses. In: Webster R G, Granoff A, editors. Encyclopedia of virology. London, United Kingdom: Academic Press Ltd.; 1994. pp. 508–516. [Google Scholar]
- 36.Tamada T, Kusume T. Evidence that the 75K readthrough protein of beet necrotic yellow vein virus RNA-2 is essential for transmission by the fungus Polymyxa betae. J Gen Virol. 1991;72:1497–1504. doi: 10.1099/0022-1317-72-7-1497. [DOI] [PubMed] [Google Scholar]
- 37.Tamada T, Schmitt C, Saito M, Guilley H, Richards K, Jonard G. High resolution analysis of the readthrough domain of beet necrotic yellow vein virus readthrough protein: a KTER motif is important for efficient transmission of the virus by Polymyxa betae. J Gen Virol. 1996;77:1359–1367. doi: 10.1099/0022-1317-77-7-1359. [DOI] [PubMed] [Google Scholar]