Abstract
Minimum requirements have been determined for synthesis and secretion of the Pediococcus antimicrobial peptide, pediocin AcH, in Escherichia coli. The functional mature domain of pediocin AcH (Lys+1 to Cys+44) is targeted into the E. coli sec machinery and secreted to the periplasm in active form when fused in frame to the COOH terminus of the secretory protein maltose-binding protein (MBP). The PapC-PapD specialized secretion machinery is not required for secretion of the MBP-pediocin AcH chimeric protein, indicating that in Pediococcus, PapC and PapD probably are required for recognition and processing of the leader peptide rather than for translocation of the mature pediocin AcH domain across the cytoplasmic membrane. The chimeric protein displays bactericidal activity, suggesting that the NH2 terminus of pediocin AcH does not span the phospholipid bilayer in the membrane-interactive form of the molecule. However, the conserved Lys+1-Tyr-Tyr-Gly-Asn-Gly-Val+7-sequence at the NH2 terminus is important because deletion of this sequence abolishes activity. The secreted chimeric protein is released into the culture medium when expressed in a periplasmic leaky E. coli host. The MBP fusion-periplasmic leaky expression system should be generally advantageous for production and screening of the activity of bioactive peptides.
Pediocin AcH is a 44-amino-acid antimicrobial peptide that belongs to the class IIa family of nonlanthionine bacteriocins synthesized by several species of lactic acid bacteria (15, 17). The peptide is produced by Pediococcus acidilactici H (2, 25) and has an amino acid sequence identical to that of pediocin PA-1, which is produced by P. acidilactici PAC1.0 (21). Several other bacteriocins in this family have been characterized, including leucocin A (12) and sakacin P (38). Pediocin AcH displays broad-spectrum bactericidal activity against gram-positive and stressed gram-negative bacteria associated with food spoilage and human pathogenesis (16, 28–30). Bacteriocins also have potential applications in controlling topical infections caused by bacterial pathogens (19, 39). For these reasons, it is important to overproduce bacteriocins such as pediocin AcH in a suitable bacterial host and determine amino acids required for activity.
As is the case for nisin, a class I lantibiotic-type bacteriocin (15, 17), pediocins PA-1 and AcH kill bacteria by forming pore complexes in the cytoplasmic membrane, resulting in dissipation of the membrane electrochemical potential (6, 23, 24). Although binding of class IIa bacteriocins to membranes and nucleation of pore complex assembly may be promoted in vivo by membrane-associated receptor proteins (1), recent biophysical studies show that pediocin PA-1 can form pore complexes in pure Listeria phospholipid vesicles in the absence of membrane proteins (6). On the basis of in vitro studies, it is proposed that binding of pediocin PA-1 to membranes is mediated by interactions between positively charged amino acids in the peptide and negatively charged phospholipid head groups (6).
The pre-pediocin AcH structural gene (papA, encoding 62 amino acids) is the first gene in the pap operon carried on the P. acidilactici plasmid pSMB74 (25). Also present in the operon are genes required in the producer strain for immunity (papB, encoding 118 amino acids) and membrane translocation (papC, encoding 217 amino acids, and papD, encoding 718 amino acids) (5, 40). The PapD gene product also is required for removal of the 18-amino-acid leader peptide from the inactive pre-pediocin AcH precursor and generation of the active mature form of the peptide during membrane translocation (5, 40). The sequence of the leader peptide differs markedly from those of signal peptides of gram-positive (35) and gram-negative (41) bacterial standard secretory proteins and is presumed to target the precursor into a specialized secretion machinery composed of PapC and PapD (15). PapC and PapD are homologous to the respective membrane fusion protein (HlyD) and ATPase (HlyB) components of the Escherichia coli hemolysin secretion machinery and other ABC export systems (9). PapD shares a double-glycine protease domain with other ABC export proteins active in transport of bacteriocins such as pediocin AcH (12a). Pre-pediocin AcH can be secreted and processed in E. coli if the PapC and/or PapD protein is coexpressed in the host (5, 40). The immunity function of PapB is not required for E. coli expression (5).
In this article, we report that the mature sequence region of pediocin AcH can be produced and secreted in an active state in E. coli without coexpression of PapC and PapD if it is fused to the secretory protein maltose-binding protein (MBP). The MBP-pediocin AcH chimeric protein is released into the culture medium when expressed in a periplasmic leaky host in which the gene encoding the outer membrane protein Braun’s lipoprotein (4) has been disrupted. The implications of the results are discussed with respect to the function of the leader peptide during pre-pediocin AcH secretion in Pediococcus and the structure of the membrane-interactive form of the peptide.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. The malE plasmids pPR682 and pIH821 were used to construct MBP-pediocin AcH expression plasmids. In both plasmids, transcription of fusion genes is controlled by the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter. DNA encoding pediocin AcH was obtained by PCR amplification of plasmid pMBR1.0 DNA (5). E. coli E609 (46) and its isogenic derivative E609L, which contains a Tn10 insertion in the lpp gene, were used to study expression of MBP-pediocin AcH chimeric proteins. E. coli strains were grown at 37°C in Luria-Bertani broth or agar, and 12.5 μg of tetracycline per ml was added to the media for strain E609L. Ampicillin (100 μg/ml) was added to the media when E609 and E609L were transformed with expression plasmids. MBP-pediocin AcH activity was tested against the indicator strain Listeria innocua Lin 11, grown at 30°C in tryptone-glucose-yeast extract (TGE) broth or agar (5).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Reference(s) or source |
|---|---|---|
| L. innocua Lin 11 | Nonpathogenic; pediocin AcH sensitive | Jean Richard, INRA, Paris, France |
| E. coli | ||
| E609 | HfrC pps | Henry C. Wu 46 |
| E609L | E609 lpp::Tn10; periplasmic leaky; Tcr | Henry C. Wu |
| pMBR1.0 | pap gene cluster in pHPS9; Cmr Emr | 5, 11 |
| pPR682 | malE plasmid; Apr | New England Biolabs |
| pIH821 | malE(Δ2-26) plasmid; Apr | New England Biolabs |
| pPR6821 | malE-papA plasmid; Apr | This study |
| pIH8211 | malE(Δ2-26)-papA plasmid; Apr | This study |
| pPR6822 | malE-papA(Δ1-7) plasmid; Apr | This study |
Construction of MBP-pediocin AcH expression plasmids.
A DNA fragment encoding the mature domain of pediocin AcH was obtained by PCR amplification of pMBR1.0 DNA. The sequence of the 5′ primer used for amplification (5′-AAATACTACGGTAATGGGGTTACTTGTG-3′) is identical to codons Lys+1 to Cys+9 of the papA gene (25). The 3′ primer (5′-GGGTCGACCTAGCATTTATGATTACCT-3′) begins with a GG dinucleotide clamp followed by a SalI restriction enzyme site (GTCGAC) and ends with a 19-nucleotide sequence complementary to the stop codon and final six codons of papA. PCR amplification was performed with 25 U of Taq DNA polymerase (Gibco-BRL) per ml, 0.1 μg of pMBR1.0 template per ml, 6 mM MgCl2, 10 mM each nucleoside triphosphate, and 400 μg of each primer per ml. A 30-cycle repeated protocol consisting of 90 s of strand denaturation (94°C), 60 s of primer annealing (55°C), and 60 s of primer extension (72°C) was used to amplify DNA.
The PCR product was purified by agarose gel electrophoresis (48), subjected to a Klenow polymerase reaction to fill in potentially ragged ends, phosphorylated with T4 polynucleotide kinase, and digested with SalI restriction endonuclease (31). The resulting fragment was gel purified again and ligated between the StuI (blunt-end) and SalI restriction sites within the multiple cloning sites of pPR682 and pIH821. Translational fusion genes, in which the malE and papA coding sequences are joined in frame, were created by cloning procedures. Ligation mixtures were transformed into competent E609L cells prepared by treatment with CaCl2 (31). The MBP-pediocin AcH expression plasmids, designated pPR6821 and pIH8211, were identified by restriction enzyme digestion, and their papA coding regions were confirmed by double-stranded DNA sequencing with Sequenase DNA polymerase (U.S. Biochemicals).
The pPR6822 plasmid, which lacks the coding sequence for amino acids Lys+1 to Val+7 of pediocin AcH, was constructed by similar procedures. The 5′ PCR primer (5′-ACTTGTGGCAAACATTCCTGCTCTGT-3′) used for amplification was identical to codons Thr+8 to Val+16 of papA. The 3′ PCR primer was the same as that used to construct full-length fusion genes. In pPR6822, the malE sequence is fused in frame to codons 8 to 44 of papA. The MBP and pediocin AcH domains of all three chimeric proteins are connected by a 14-amino-acid linker peptide (see below).
Detection of MBP-pediocin AcH release from E. coli by colony overlay screening.
Release of chimeric proteins from E. coli was monitored by overlaying producer colonies with agar containing strain L. innocua Lin 11 and examining overlay lawns for zones of growth inhibition (5). Cultures of E. coli expression strains were grown to saturation overnight in liquid media plus required antibiotics. On the following day, cells were serially diluted into fresh media lacking antibiotics, 10 μl of selected dilutions containing ∼50 to 100 cells was added to 5 ml of molten 0.8% TGE soft agar, and the mixtures were poured into petri plates. After the agar solidified, 3 ml of melted TGE soft agar without cells was poured over its surface and allowed to solidify, and the plates were incubated at 37°C for 24 h until colonies formed. On the next day, 5 ml of melted TGE soft agar containing 5 μl of an overnight culture of L. innocua Lin 11 was overlaid on each plate. In some cases, 1 mM IPTG was added to the agar overlay to induce synthesis of MBP-pediocin AcH proteins. The plates were incubated overnight at 37°C and examined for zones of growth inhibition around E. coli colonies. Representative plates were photographed.
SDS-polyacrylamide gel electrophoresis analysis of MBP-pediocin AcH production and activity.
Expression strain cultures were grown to mid-log phase at 37°C in liquid media plus antibiotics. Preinduction samples were taken, separated by centrifugation into cell pellet and supernatant fractions, and processed for sodium dodecyl sulfate (SDS)-polyacrylamide gel analysis as described below. After collection of preinduction samples, IPTG (1 mM final concentration) was added to the cultures, which were grown for an additional 3 h until postinduction sampling.
Pelleted cells were prepared for SDS-polyacrylamide gel electrophoresis by being solubilized directly in sample loading buffer containing SDS. Proteins in culture supernatants were precipitated with ice-cold 10% trichloroacetic acid. The precipitates were washed once with 5% trichloroacetic acid and once with 80% acetone and then were dried under a vacuum and solubilized in sample loading buffer. Samples were run on 10% acrylamide-bisacrylamide–SDS gels (18), and MBP-pediocin AcH production levels were quantitated by laser scanning densitometry after Coomassie blue dye staining of gels (1a). The same methods and gel system were used to analyze production by Western immunoblotting. MBP-pediocin AcH bands were visualized on immunoblots by staining with rabbit anti-MBP primary antiserum and goat anti-rabbit immunoglobulin G–alkaline phosphatase secondary-antibody complex (22).
The bactericidal activity of chimeric proteins was analyzed by polyacrylamide gel electrophoresis and gel overlay screening. Cell pellet and culture supernatant samples first were run on 16% acrylamide-bisacrylamide–SDS gels (32). Subsequently, the gels were washed in sterile water for 3 h to remove SDS, placed on prepoured TGE agar plates, and covered with a 20-ml TGE soft-agar overlay containing L. innocua Lin 11 cells by previously reported methods (2, 45). The plates were incubated at 37°C overnight and examined for zones of growth inhibition associated with proteins in the samples.
RESULTS
Colony overlay screening analysis of MBP-pediocin AcH production in E. coli.
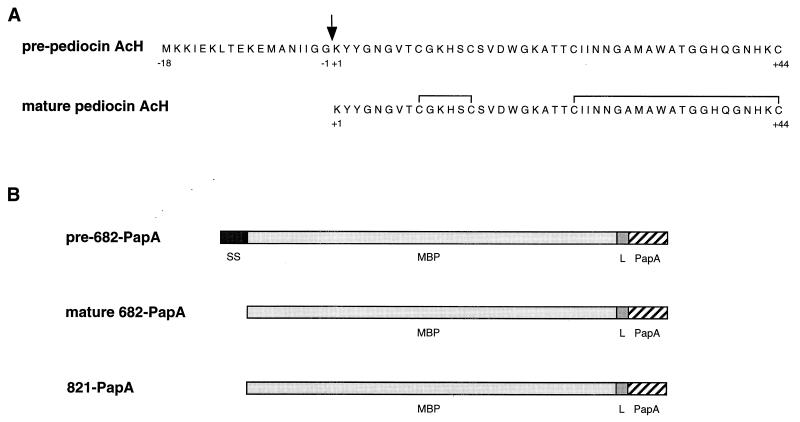
The secretion of translational fusion proteins in which the mature region of pediocin AcH (Lys+1 to Cys+44) is fused to the COOH terminus of MBP was investigated to determine if the PapC-PapD specialized secretion machinery is required for pediocin AcH production in E. coli. Two types of MBP-pediocin AcH chimeric proteins were constructed and analyzed (Fig. 1). In one (designated pre-682-PapA), a wild-type MBP domain (43 kDa) containing a functional signal sequence is joined to pediocin AcH. The mature 682-PapA protein should be secreted to the periplasm after processing of the signal sequence during secretion (7) if the E. coli sec machinery can accommodate the pediocin AcH chain. In the second chimeric protein (designated 821-PapA), an MBP domain lacking a signal sequence [MBP(Δ2-26) (42)] is joined to pediocin AcH. The MBP(Δ2-26) domain is unable to functionally interact with components of the E. coli sec machinery (42), and the 821-PapA protein should be trapped in the cytoplasm (7, 13).
FIG. 1.
(A) Amino acid sequences of pre-pediocin AcH and of mature pediocin AcH formed after processing of the 18-amino-acid leader peptide and oxidation of the four cysteines (15). (B) Properties of MBP-pediocin AcH chimeric proteins. The pre-682-PapA primary translation product contains the wild-type MBP domain with its signal sequence (SS), a 14-amino-acid linker (L) peptide (-Asn-Ser-Ser-Ser-Val-Pro-Gly-Arg-Gly-Ser-Ile-Asp-Gly-Arg-), and the 44-amino-acid mature domain of pediocin AcH (PapA). The signal sequence is removed and mature 682-PapA is formed during secretion. The 821-PapA protein is identical to mature 682-PapA (see text). Chimeric proteins were named after the pPR682 and pIH821 plasmids (Table 1).
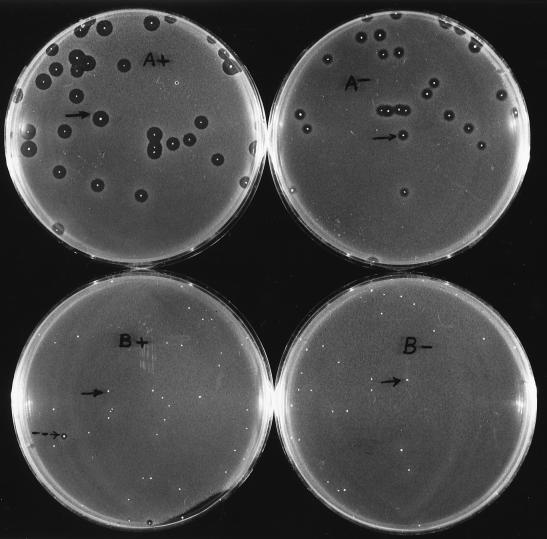
A periplasmic leaky E. coli host, strain E609L (Table 1), was used to investigate secretion of the two chimeric proteins. E609L colonies synthesizing the proteins were overlaid with agar containing L. innocua Lin 11 and were examined for release of antimicrobial activity. Colonies of strain E609L/pPR6821 formed large zones of growth inhibition (average diameter = 7 mm) when 682-PapA synthesis was induced with IPTG and smaller zones when synthesis was not induced (Fig. 2A). Smaller zones were formed in the absence of IPTG due to the lower level of expression taking place under noninducing conditions than under inducing conditions (data not shown).
FIG. 2.
Colony overlay screening of MBP-pediocin AcH release from strains E609L/pPR6821 (A) and E609L/pIH8211 (B). The agar overlays either contained (+) or lacked (−) 1 mM IPTG. Colonies that formed representative zones of growth inhibition under the two growth conditions (arrows) (A) and a rare colony that produced a small zone of growth inhibition (see text) (dashed arrow) and representative colonies that did not produce zones of growth inhibition (solid arrows) (B) are indicated.
In contrast, only a few colonies of strain E609L/pIH8211 formed small zones of growth inhibition under inducing growth conditions (Fig. 2B). Release of the cytoplasmic 821-PapA protein probably was due to lysis of dead cells in these colonies. It has been observed that induction of synthesis of the 821-PapA protein is more toxic to the host than 682-PapA. For example, strain E609L/pIH8211 undergoes a >90% reduction in viable-cell number after 24-h IPTG induction, whereas only a 10% reduction in viability occurs after 24-h IPTG induction of strain E609L/pPR6821 (data not shown).
The 682-PapA protein is secreted via the E. coli sec machinery.
The mechanism by which 682-PapA is released from the periplasmic leaky host was investigated by examining the status of processing of the MBP signal sequence. In this regard, processing of the MBP signal sequence is indicative that the protein has been secreted via the sec machinery, because the catalytic domain of the processing enzyme, signal peptidase I, is located in the periplasm (3, 27). A 3-h induction period was selected for these experiments because the maximal level of synthesis of chimeric proteins in both E609L/pPR6821 and E609L/pIH8211 strains is achieved within 3 h, as is release of the 682-PapA protein from the former strain. The toxic effects (loss of viability and cell lysis) of the chimeric proteins on the strains are minimal during this time (data not shown).
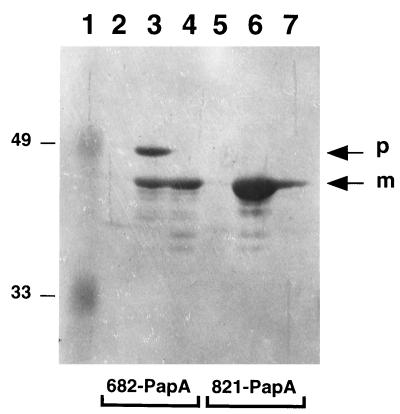
Two forms of the 682-PapA protein were present in the cell fraction of the 3-h-induced E609L/pPR6821 strain (Fig. 3, lane 3). The larger of these proteins is the unprocessed precursor, and the smaller is the processed mature form in which the MBP signal sequence has been removed. Band assignments are based on observations that only the mature form of 682-PapA was released into the culture supernatant (Fig. 3, lane 4), and released 682-PapA molecules were the same size as the cytoplasmic 821-PapA protein, which lacks a signal sequence (Fig. 3, lane 6). Because the 682-PapA protein present in the culture supernatant is processed, it has been secreted from the strain via the sec machinery. Minimal amounts of the protein appear to be released by cell lysis, because very little 682-PapA precursor ever is detected in the culture supernatant. In contrast, only a small fraction of 821-PapA molecules was released from cells (Fig. 3, lane 7). As noted above, release is correlated with toxicity and cell death caused by overexpression of the intracellular protein.
FIG. 3.
Western immunoblot analysis of MBP-pediocin AcH synthesis and secretion in strains E609L/pPR6821 (lanes 2 to 4) and E609L/pIH8211 (lanes 5 to 7). Samples were prepared from uninduced cells (lanes 2 and 5), 3-h-induced cells (lanes 3 and 6), and culture supernatants from 3-h-induced cells (lanes 4 and 7). Equivalent amounts of cell pellets and supernatants were loaded in the lanes. Precursor (p) and processed mature (m) forms of 682-PapA are indicated. The 821-PapA protein comigrates with the processed form of 682-PapA. Prestained molecular weight standards were loaded in lane 1, and their molecular weights (in thousands) are indicated on the left.
It should be noted that not all 682-PapA molecules were secreted, as indicated by the fact that the unprocessed precursor form of the protein also was detected in cells (Fig. 3, lane 3). Some precursors may be trapped in the cytoplasm because the sec machinery is unable to keep pace with fusion protein synthesis under inducing conditions (22). Lastly, secreted mature-form 682-PapA molecules that remain associated with cells (Fig. 3, lane 3) have not leaked out of the periplasm. In this regard, periplasmic leaky strains typically release only 10 to 50% of their periplasmic proteins (20).
MBP-pediocin AcH chimeric proteins possess bactericidal activity.
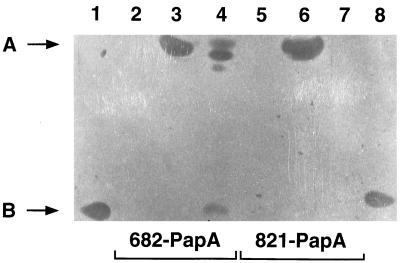
Chimeric proteins were assayed for activity by SDS-polyacrylamide gel electrophoresis and gel overlay screening against L. innocua Lin 11 (Fig. 4). The 821-PapA chimeric protein present in the cytoplasmic fraction of strain E609L/pIH8211 exhibited activity, as indicated by the zone of growth inhibition formed in the high-molecular-weight region of the gel (Fig. 4, lane 6). Zones of growth inhibition attributable to the 682-PapA chimeric protein also appeared in the high-molecular-weight region of the gel in both cell and culture supernatant fractions (Fig. 4, lanes 3 and 4). It should be noted that the unprocessed and processed forms of 682-PapA are not resolved well on the 16% acrylamide gels used to separate chimeric proteins from pediocin AcH-size molecules. These results indicate that pediocin AcH is active when its NH2 terminus is blocked.
FIG. 4.
Analysis of MBP-pediocin AcH bactericidal activity by SDS-polyacrylamide gel electrophoresis and gel overlay screening. Results are shown for strain E609L/pPR6821 (lanes 2 to 4) and E609L/pIH8211 (lanes 5 to 7) uninduced cells (lanes 2 and 5), 3-h-induced cells (lanes 3 and 6), and culture supernatants from 3-h-induced cells (lanes 4 and 7). Purified pediocin AcH from strain P. acidilactici LB42-923 was also analyzed (lanes 1 and 8). The migration positions of the active 682-PapA (lane 3) and 821-PapA (lane 6) chimeric proteins (molecular mass = 47 kDa) (A) and the migration positions of wild-type pediocin AcH (molecular mass = 4.6 kDa) (lanes 1 and 8) and the active breakdown product derived from the 682-PapA protein (lane 4) (B) are indicated. See text for explanation of other zones of growth inhibition near A in lane 4.
Some 682-PapA molecules in the culture supernatant were degraded, as indicated by the appearance of a growth inhibition zone in the low-molecular-weight region and several zones in the high-molecular-weight region of the gel (Fig. 4, lane 4). The low-molecular-weight active species migrated comparably to wild-type pediocin AcH obtained from P. acidilactici (Fig. 4, lanes 1 and 8). Proteolysis of 682-PapA occurred only if it had passed through the periplasm and outer membrane of the host and into the medium. Possibly, a protease(s) residing in the membrane or periplasmic compartments of the cells (14, 36) is responsible for cleaving the secreted chimeric protein.
The NH2 terminus of pediocin AcH is required for bactericidal activity.
A truncated derivative of 682-PapA that lacks amino acids Lys+1 to Val+7 [designated 682-PapA(Δ1-7)] was constructed to determine if the presence of the NH2-terminal region of pediocin AcH is required for bactericidal activity. The region deleted contains a sequence (Lys+1-Tyr-Tyr-Gly-Asn-Gly-Val+7-) of unknown function that is conserved in class IIa bacteriocins (15, 17). The 682-PapA(Δ1-7) protein was synthesized in an amount comparable to that of 682-PapA, and 682-PapA(Δ1-7) was efficiently processed and released from the leaky host (data not shown). However, the truncated protein did not display activity against L. innocua Lin 11 by colony overlay or gel overlay screening. The results show that the deleted pediocin AcH sequence is required for bactericidal activity and suggest that the 682-PapA low-molecular-weight active degradation product discussed above is produced by cleavage of the chimeric protein upstream of the pediocin AcH domain.
The 682-PapA protein is efficiently released from the periplasmic leaky E. coli host.
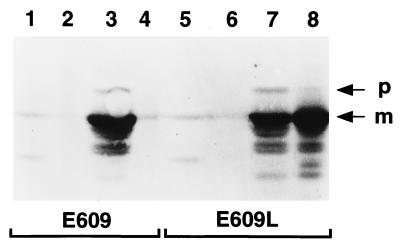
To confirm that the periplasmic leaky host is advantageous for pediocin AcH production, we compared the levels of 682-PapA released from strain E609L and the nonleaky wild-type E609 strain (Table 1). As shown in Fig. 5, the two strains synthesized comparable amounts of the protein. However, E609 released only a small fraction of secreted 682-PapA molecules into the culture medium, whereas E609L released about half (Fig. 5, compare lanes 3 and 4 and lanes 7 and 8). The results show that the periplasmic leaky host is better than the wild-type strain for production and release of the 682-PapA protein.
FIG. 5.
Western immunoblot analysis of 682-PapA synthesis and release in strains E609/pPR6821 (lanes 1 to 4) and E609L/pPR6821 (lanes 5 to 8). Results are shown for uninduced cells (lanes 1 and 5), culture supernatants from uninduced cells (lanes 2 and 6), 3-h-induced cells (lanes 3 and 7), and culture supernatants from 3-h-induced cells (lanes 4 and 8). Equivalent amounts of cell pellets and supernatants were loaded in the lanes. Precursor (p) and processed mature (m) forms of 682-PapA are indicated.
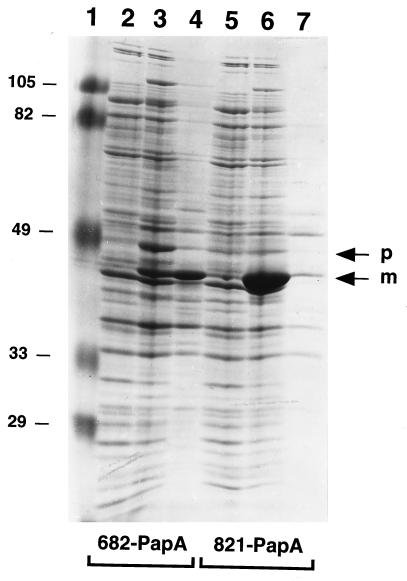
The amounts of 682-PapA synthesized and released from strain E609L were estimated by laser scanning densitometry of Coomassie blue-stained polyacrylamide gel samples (Fig. 6). On the basis of densitometry, the combined level of precursor and processed forms of 682-PapA associated with cells was ∼12% of the total cell protein (Fig. 6, lane 3). Because about half of the processed molecules were released into the medium (Fig. 6, compare lanes 3 and 4), the total 682-PapA expression level was on the order of 18%. The 821-PapA protein also was expressed at a high level (∼15% of the total cell protein) (Fig. 6, lane 6), but as discussed above, little of this protein was released from the host (Fig. 6, lane 7).
FIG. 6.
SDS-polyacrylamide gel analysis of MBP-pediocin AcH synthesis and release in strains E609L/pPR6821 (lanes 2 to 4) and E609L/pIH8211 (lanes 5 to 7). Samples were prepared from uninduced cells (lanes 2 and 5), 3-h-induced cells (lanes 3 and 6), and culture supernatants from 3-h-induced cells (lanes 4 and 7). Equivalent amounts of cell pellets and supernatants were loaded in the lanes. Precursor (p) and processed mature (m) forms of 682-PapA are indicated. Prestained molecular weight standards were loaded in lane 1, and their molecular weights (in thousands) are indicated on the left.
DISCUSSION
A potential problem associated with achieving high-level production of native bacteriocins in E. coli is the requirement for balanced coexpression of the specialized secretion machinery needed for recognition and processing of their prepeptides. While powerful systems such as T7 RNA polymerase vectors (37) could be used for expression, it is likely that cellular toxicity will result from high-level synthesis of these integral membrane proteins. For example, the NisT ATPase component of the nisin specialized secretion machinery (9) and other bacterial integral membrane proteins (8, 22) have been shown to interfere with E. coli growth and viability when overexpressed. For this reason, we elected to overexpress and secrete pediocin AcH as a chimeric protein. In this form, pediocin AcH is targeted into the standard E. coli sec machinery, and potential problems associated with overexpression of PapC and PapD can be avoided.
The results demonstrate that this approach eliminates the requirement for coexpression of the PapC-PapD specialized secretion machinery. High-level synthesis of MBP-pediocin AcH proteins was achieved because of the powerful transcription and efficient translation initiation signals present in malE vectors (7). Although the rate of synthesis of the 682-PapA protein exceeded the capacity of the E. coli sec machinery for secretion, about two-thirds of the molecules nonetheless were processed and secreted, and about half of the secreted molecules were released into the culture medium from the periplasmic leaky host. On the basis of the staining intensity of the processed 682-PapA protein in the culture medium, we estimate the level of the protein in the medium to be ∼57 mg/g of total cell protein in the cultures. It should be possible to increase the yield by releasing molecules trapped in the periplasm by osmotic shock treatment of cells (7, 26).
Our results demonstrate for the first time that a class IIa bacteriocin can be secreted via the standard sec machinery of a gram-negative bacterium. Apparently, the peptide remains sufficiently unfolded prior to translocation to be accommodated by the core translocation machinery composed of the E. coli SecA, SecE, and SecY proteins (33, 43), for which there are homologs in gram-positive bacteria (35). Only two other, unrelated bacteriocins have been shown to be secreted by the cellular sec machinery. One member of the colicin family of bacteriocins, colicin V, can be secreted in E. coli when fused to the OmpA signal peptide (47). Another bacteriocin, divergicin A, produced by Carnobacterium divergens LV13, contains a standard NH2-terminal signal peptide and is secreted via the sec machinery of this gram-positive bacterium (44).
The results raise the question of why an ABC export system is used for secretion of pediocin AcH in Pediococcus. The peptide chain can be accommodated by the sec machinery, there is no need for a membrane fusion protein (i.e., PapC) to bridge inner and outer membranes, and the peptide need not be targeted into a modification machinery associated with the ABC export system, as occurs with nisin (34). The data also eliminate the formal possibilities that the ABC export machinery participates in catalysis of disulfide bond formation or folding of pediocin AcH after translocation across the membrane. Instead, the data suggest that the Pediococcus specialized secretion machinery is required primarily for recognition and processing of the pre-pediocin AcH leader peptide. Why the standard sec pathway is not used for secretion of this bacteriocin remains unknown.
The finding that MBP-pediocin AcH chimeric proteins retain activity provides insight into the general structural features of the membrane-interactive form of the peptide. First, the results suggest that the NH2 terminus of the peptide normally does not occupy the interior of a pore complex, because MBP domains do not sterically interfere with the activity of chimeric proteins. Second, the α-amino group of Lys+1 is not required in the native peptide for a salt bridge within the pore complex. However, it remains possible that the Lys+1 α-amino group normally does participate in interactions with phospholipid head groups and that the arginine in the linker region immediately upstream of Lys+1 substitutes in this capacity in chimeric proteins (Fig. 1). Third, the results support the prediction that the COOH-terminal half of pediocin AcH forms the transmembrane portion of the peptide (10). It is unlikely that the NH2-terminal region of MBP-pediocin AcH proteins could insert deeply into bilayers, because insertion would require transfer of polar amino acids in the linker and/or the MBP domain into the membrane. Fourth, the NH2-terminal sequence (Lys+1 to Val+7) is important for the bactericidal activity of the peptide. Deletion of these residues may remove a putative β turn formed by amino acids 4 to 7 (6).
In conclusion, the MBP fusion-periplasmic leaky expression system should be generally useful for production and screening of the activity of bioactive peptides such as bacteriocins. The system also should be useful to facilitate purification (7), because relatively few proteins are released into the culture medium along with MBP chimeras. In the future, the system will be used to isolate mutants with alterations in the activity of pediocin AcH and other antimicrobial peptides. Because large amounts of chimeric proteins are released from colonies and zones of growth inhibition in overlays are large, mutants with low specific activities should be detectable.
ACKNOWLEDGMENTS
We thank investigators for providing bacterial strains.
We acknowledge financial support from the National Science Foundation, the State of Wyoming, and the Michigan Biotechnology Institute.
Footnotes
This paper is dedicated to the memory of Henry C. Wu.
REFERENCES
- 1.Abee T. Pore-forming bacteriocins of Gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol Lett. 1995;129:1–10. doi: 10.1016/0378-1097(95)00137-T. [DOI] [PubMed] [Google Scholar]
- 1a.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 2.Bhunia A, Johnson M C, Ray B. Direct detection of an antimicrobial peptide of Pediococcus acidilactici in sodium dodecyl sulphate-polyacrylamide gel electrophoresis. J Ind Microbiol. 1987;2:319–322. [Google Scholar]
- 3.Bilgin N, Lee J I, Zhu H-Y, Dalbey R, von Heijne G. Mapping of catalytically important domains in Escherichia coli leader peptidase. EMBO J. 1990;9:2717–2722. doi: 10.1002/j.1460-2075.1990.tb07458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun V, Rehn K. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem. 1969;10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 5.Bukhtiyarova M, Yang R, Ray B. Analysis of the pediocin AcH gene cluster from plasmid pSMB74 and its expression in a pediocin-negative Pediococcus acidilactici strain. Appl Environ Microbiol. 1994;60:3405–3408. doi: 10.1128/aem.60.9.3405-3408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Shapira R, Eisenstein M, Montville T J. Functional characterization of pediocin PA-1 binding to liposomes in the absence of a protein receptor and its relationship to a predicted tertiary structure. Appl Environ Microbiol. 1997;63:524–531. doi: 10.1128/aem.63.2.524-531.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.di Guan C, Li P, Riggs P D, Inouye H. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene. 1988;67:21–30. doi: 10.1016/0378-1119(88)90004-2. [DOI] [PubMed] [Google Scholar]
- 8.Eckert B, Beck C F. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J Bacteriol. 1989;171:3557–3559. doi: 10.1128/jb.171.6.3557-3559.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fath M J, Kolter R. ABC-transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fimland G, Blingsmo O R, Sletten K, Jung G, Nes I F, Nissen-Meyer J. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol. 1996;62:3313–3318. doi: 10.1128/aem.62.9.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haima P, van Sinderen D, Schotting H, Bron S, Venema G. Development of a β-galactosidase α-complementation system for molecular cloning in Bacillus subtilis. Gene. 1990;86:63–69. doi: 10.1016/0378-1119(90)90114-7. [DOI] [PubMed] [Google Scholar]
- 12.Hastings J W, Sailer M, Johnson K, Roy K L, Vederas J C, Stiles M E. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J Bacteriol. 1991;173:7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Havarstein L S, Holo H, Nes I F. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by Gram-positive bacteria. Microbiology. 1994;140:2383–2389. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- 13.Hook, V. Y. H., K. Moran, R. Kannan, A. Kohn, M. O. Lively, A. Azaryan, M. Schiller, and K. Miller. High level expression of the prohormones proenkephalin, pro-neuropeptide Y, proopiomelanocortin, and β-protachykinin for in vitro prohormone processing. Protein Express. Purification, in press. [DOI] [PubMed]
- 14.Ichihara S, Matsubara Y, Kato C, Akasaka K, Mizushima S. Molecular cloning, sequencing, and mapping of the gene encoding protease I and characterization of proteinase and proteinase-defective Escherichia coli mutants. J Bacteriol. 1993;175:1032–1037. doi: 10.1128/jb.175.4.1032-1037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack R W, Tagg J R, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalchayanand N, Hanlin M B, Ray B. Sublethal injury makes Gram-negative and resistant Gram-positive bacteria sensitive to the bacteriocins, pediocin AcH and nisin. Lett Appl Microbiol. 1992;15:239–243. [Google Scholar]
- 17.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Liu W, Hansen J N. Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J Biol Chem. 1992;267:25078–25085. [PubMed] [Google Scholar]
- 20.Lopes J, Gottfried S, Rothfield L. Leakage of periplasmic enzymes by mutants of Escherichia coli and Salmonella typhimurium: isolation of “periplasmic leaky” mutants. J Bacteriol. 1972;109:520–525. doi: 10.1128/jb.109.2.520-525.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marugg J D, Gonzalez C F, Kunka B S, Ledeboer A M, Pucci M J, Toonen M Y, Walker S A, Zoetmulder L C M, Vandenbergh P A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller K W, Konen P L, Olson J, Ratanavanich K M. Membrane protein topology determination by proteolysis of maltose binding protein fusions. Anal Biochem. 1993;215:118–128. doi: 10.1006/abio.1993.1563. [DOI] [PubMed] [Google Scholar]
- 23.Moll G N, Roberts G C K, Konings W N, Driessen A J M. Mechanism of lantibiotic-induced pore-formation. Antonie Leeuwenhoek. 1996;69:185–191. doi: 10.1007/BF00399423. [DOI] [PubMed] [Google Scholar]
- 24.Moll G N, Clark J, Chan W C, Bycroft B W, Roberts G C K, Konings W N, Driessen A J M. Role of transmembrane pH gradient and membrane binding in nisin pore formation. J Bacteriol. 1997;179:135–140. doi: 10.1128/jb.179.1.135-140.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motlagh A M, Bukhtiyarova M, Ray B. Complete nucleotide sequence of pSMB74, a plasmid encoding the production of pediocin AcH in Pediococcus acidilactici. Lett Appl Microbiol. 1994;18:305–312. doi: 10.1111/j.1472-765x.1994.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 26.Neu H C, Heppel L A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 27.Randall L L, Hardy S J S. Export of protein in bacteria. Microbiol Rev. 1984;48:290–298. doi: 10.1128/mr.48.4.290-298.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray B. Pediocin(s) of Pediococcus acidilactici as food biopreservatives. In: Ray B, Daeschul M A, editors. Food biopreservatives of microbial origin. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 265–322. [Google Scholar]
- 29.Ray B. Sublethal injury, bacteriocins, and food microbiology. ASM News. 1993;59:285–291. [Google Scholar]
- 30.Ray B. Characteristics and application of pediocin(s) of Pediococcus acidilactici: pediocin PA-1/AcH. In: Bozuglu T F, Ray B, editors. Lactic acid bacteria: current advances in metabolism, genetics, and applications. New York, N.Y: Springer; 1996. pp. 155–203. [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 33.Schatz P J, Beckwith J. Genetic analysis of protein export in Escherichia coli. Annu Rev Genet. 1990;24:215–248. doi: 10.1146/annurev.ge.24.120190.001243. [DOI] [PubMed] [Google Scholar]
- 34.Siegers K, Heinzmann A, Entian K-D. Biosynthesis of lantibiotic nisin. J Biol Chem. 1996;271:12294–12301. doi: 10.1074/jbc.271.21.12294. [DOI] [PubMed] [Google Scholar]
- 35.Simonen M, Palva I. Protein secretion in Bacillus species. Microbiol Rev. 1993;57:109–137. doi: 10.1128/mr.57.1.109-137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strauch K L, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Studier F S, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct the expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 38.Tichaczek P S, Vogel R F, Hammes W. Cloning and sequencing sakP encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH673. Microbiology. 1994;140:361–367. doi: 10.1099/13500872-140-2-361. [DOI] [PubMed] [Google Scholar]
- 39.Valenta C, Bernkop-Schnurch A, Rigler H P. The antistaphylococcal effect of nisin in a suitable vehicle: a potential therapy for atopic dermatitis in man. J Pharm Pharmacol. 1996;48:988–991. doi: 10.1111/j.2042-7158.1996.tb06019.x. [DOI] [PubMed] [Google Scholar]
- 40.Venema K, Kok J, Marugg J D, Toonen M Y, Ledeboer A M, Venema G. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC 1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol. 1995;17:515–522. doi: 10.1111/j.1365-2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]
- 41.von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 42.Weiss J B, Bassford P J., Jr The folding properties of the Escherichia coli maltose-binding protein influence its interaction with SecB in vitro. J Bacteriol. 1990;172:3023–3029. doi: 10.1128/jb.172.6.3023-3029.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wickner W, Leonard M R. Escherichia coli preprotein translocase. J Biol Chem. 1996;271:29514–29516. doi: 10.1074/jbc.271.47.29514. [DOI] [PubMed] [Google Scholar]
- 44.Worobo R W, van Belkum M J, Sailer M, Roy K L, Vederas J C, Stiles M E. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol. 1995;177:3143–3149. doi: 10.1128/jb.177.11.3143-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang R, Johnson M C, Ray B. Novel method to extract large amounts of bacteriocins from lactic acid bacteria. Appl Environ Microbiol. 1992;58:3355–3359. doi: 10.1128/aem.58.10.3355-3359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yem D W, Wu H C. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J Bacteriol. 1978;133:1419–1426. doi: 10.1128/jb.133.3.1419-1426.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L H, Fath M J, Mahanty H K, Tai P C, Kolter R. Genetic analysis of the colicin V secretion pathway. Genetics. 1995;141:25–32. doi: 10.1093/genetics/141.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhen L, Swank R T. A simple and high yield method for recovering DNA from agarose gels. BioTechniques. 1993;14:894–898. [PubMed] [Google Scholar]